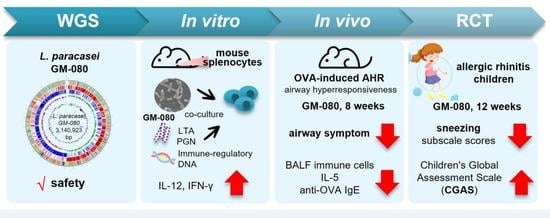
Lacticaseibacillus paracasei GM-080 Ameliorates Allergic Airway Inflammation in Children with Allergic Rhinitis: From an Animal Model to a Double-Blind, Randomized, Placebo-Controlled Trial
Abstract
:1. Introduction
2. Materials and Methods
2.1. L. paracasei Strain and Cell-Wall Component Preparation
2.2. Mouse Splenocyte Stimulation and Enzyme-Linked Immunosorbent Assay–Based Detection of Cytokines
2.3. Antimicrobial Susceptibility Profiling
2.4. DNA Extraction, Whole-Genome Sequencing, and Hybrid Genome Assembly
2.5. Annotation of Protein-Coding Genes, Virulence Factors, and Antibiotic Resistance
2.6. OVA-Induced AHR Mouse Model
2.7. ELISA Determination of OVA-Specific Immunoglobulins
2.8. AHR Determination
2.9. BALF Collection and White Blood Cell Count
2.10. Patient Recruitment
2.11. Randomized, Double-Blind, Placebo-Controlled Trial Design
2.12. Skin Prick Tests and Serum Biomarkers
2.13. Statistical Analysis
3. Results
3.1. GM-080 Induces Th1 Cytokine Production in Mouse Splenocytes
3.2. WGS Analysis Revealed That GM-080 Is a Safe L. paracasei Strain
3.3. GM-080 Genome Contains Immunosuppressive Motifs and CpG-Containing Oligonucleotides That Induce Th1 Cytokines in Mouse Splenocytes
3.4. Orally Gavage GM-080 Alleviates OVA-Induced Allergic Airway Inflammation in Mice
3.5. GM-080 Alleviates PAR in Children
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lipworth, B.J.; White, P.S. Allergic inflammation in the unified airway: Start with the nose. Thorax 2000, 55, 878–881. [Google Scholar] [CrossRef] [Green Version]
- Cohn, L.; Homer, R.J.; MacLeod, H.; Mohrs, M.; Brombacher, F.; Bottomly, K. Th2-induced airway mucus production is dependent on IL-4Ralpha, but not on eosinophils. J. Immunol. 1999, 162, 6178–6183. [Google Scholar] [CrossRef]
- Cohn, L.; Homer, R.J.; Marinov, A.; Rankin, J.; Bottomly, K. Induction of airway mucus production By T helper 2 (Th2) cells: A critical role for interleukin 4 in cell recruitment but not mucus production. J. Exp. Med. 1997, 186, 1737–1747. [Google Scholar] [CrossRef]
- Brusselle, G.G.; Kips, J.C.; Tavernier, J.H.; van der Heyden, J.G.; Cuvelier, C.A.; Pauwels, R.A.; Bluethmann, H. Attenuation of allergic airway inflammation in IL-4 deficient mice. Clin. Exp. Allergy 1994, 24, 73–80. [Google Scholar] [CrossRef]
- Hogan, S.P.; Koskinen, A.; Foster, P.S. Interleukin-5 and eosinophils induce airway damage and bronchial hyperreactivity during allergic airway inflammation in BALB/c mice. Immunol. Cell Biol. 1997, 75, 284–288. [Google Scholar] [CrossRef]
- Taube, C.; Duez, C.; Cui, Z.H.; Takeda, K.; Rha, Y.H.; Park, J.W.; Balhorn, A.; Donaldson, D.D.; Dakhama, A.; Gelfand, E.W. The role of IL-13 in established allergic airway disease. J. Immunol. 2002, 169, 6482–6489. [Google Scholar] [CrossRef] [Green Version]
- Strober, B.E. Why Biologic Therapies Sometimes Lose Efficacy. Semin. Cutan. Med. Surg. 2016, 35, S78–S80. [Google Scholar] [CrossRef]
- McFarland, L.V. From yaks to yogurt: The history, development, and current use of probiotics. Clin. Infect. Dis. 2015, 60, S85–S90. [Google Scholar] [CrossRef] [Green Version]
- Das, R.R.; Singh, M.; Shafiq, N. Probiotics in treatment of allergic rhinitis. World Allergy Organ. J. 2010, 3, 239–244. [Google Scholar] [CrossRef] [Green Version]
- Fiocchi, A.; Pawankar, R.; Cuello-Garcia, C.; Ahn, K.; Al-Hammadi, S.; Agarwal, A.; Beyer, K.; Burks, W.; Canonica, G.W.; Ebisawa, M.; et al. World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Probiotics. World Allergy Organ. J. 2015, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Sharma, G.; Im, S.H. Probiotics as a Potential Immunomodulating Pharmabiotics in Allergic Diseases: Current Status and Future Prospects. Allergy Asthma Immunol. Res. 2018, 10, 575–590. [Google Scholar] [CrossRef]
- Voo, P.Y.; Wu, C.T.; Sun, H.L.; Ko, J.L.; Lue, K.H. Effect of combination treatment with Lactobacillus rhamnosus and corticosteroid in reducing airway inflammation in a mouse asthma model. J. Microbiol. Immunol. Infect. 2022, 55, 766–776. [Google Scholar] [CrossRef]
- Lan, H.; Gui, Z.; Zeng, Z.; Li, D.; Qian, B.; Qin, L.Y.; Dai, L.; Song, J.L. Oral administration of Lactobacillus plantarum CQPC11 attenuated the airway inflammation in an ovalbumin (OVA)-induced Balb/c mouse model of asthma. J. Food Biochem. 2022, 46, e14036. [Google Scholar] [CrossRef]
- Yan, S.; Ai, S.; Huang, L.; Qiu, C.; Zhang, F.; He, N.; Zhuang, X.; Zheng, J. Systematic review and meta-analysis of probiotics in the treatment of allergic rhinitis. Allergol. Immunopathol. (Madr) 2022, 50, 24–37. [Google Scholar] [CrossRef]
- Forsythe, P.; Inman, M.D.; Bienenstock, J. Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am. J. Respir. Crit. Care Med. 2007, 175, 561–569. [Google Scholar] [CrossRef]
- Nawaz, M.; Ma, C.; Basra, M.A.; Wang, J.; Xu, J. Amelioration of ovalbumin induced allergic symptoms in Balb/c mice by potentially probiotic strains of lactobacilli. Benef. Microbes 2015, 6, 669–678. [Google Scholar] [CrossRef]
- Yan, D.C.; Hung, C.H.; Sy, L.B.; Lue, K.H.; Shih, I.H.; Yang, C.Y.; Chen, L.C.; Sun, H.L.; Lee, M.S.; Chambard, J.; et al. A Randomized, Double-Blind, Placebo-Controlled Trial Assessing the Oral Administration of a Heat-Treated Lactobacillus paracasei Supplement in Infants with Atopic Dermatitis Receiving Topical Corticosteroid Therapy. Skin Pharmacol. Physiol. 2019, 32, 201–211. [Google Scholar] [CrossRef]
- Costa, D.J.; Marteau, P.; Amouyal, M.; Poulsen, L.K.; Hamelmann, E.; Cazaubiel, M.; Housez, B.; Leuillet, S.; Stavnsbjerg, M.; Molimard, P.; et al. Efficacy and safety of the probiotic Lactobacillus paracasei LP-33 in allergic rhinitis: A double-blind, randomized, placebo-controlled trial (GA2LEN Study). Eur. J. Clin. Nutr. 2014, 68, 602–607. [Google Scholar] [CrossRef]
- Ahmed, M.; Billoo, A.G.; Iqbal, K. Efficacy of probiotic in perennial allergic rhinitis under five year children: A randomized controlled trial. Pak. J. Med. Sci. 2019, 35, 1538–1543. [Google Scholar] [CrossRef] [Green Version]
- Tsai, W.H.; Chou, C.H.; Chiang, Y.J.; Lin, C.G.; Lee, C.H. Regulatory effects of Lactobacillus plantarum-GMNL6 on human skin health by improving skin microbiome. Int. J. Med. Sci. 2021, 18, 1114–1120. [Google Scholar] [CrossRef]
- Additives, E.P.o.; Feed, P.o.S.u.i.A. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar]
- Jhong, J.H.; Tsai, W.H.; Yang, L.C.; Chou, C.H.; Lee, T.Y.; Yeh, Y.T.; Huang, C.H.; Luo, Y.H. Heat-Killed Lacticaseibacillus paracasei GMNL-653 Exerts Antiosteoporotic Effects by Restoring the Gut Microbiota Dysbiosis in Ovariectomized Mice. Front. Nutr. 2022, 9, 804210. [Google Scholar] [CrossRef]
- Zimin, A.V.; Puiu, D.; Luo, M.C.; Zhu, T.; Koren, S.; Marçais, G.; Yorke, J.A.; Dvořák, J.; Salzberg, S.L. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res. 2017, 27, 787–792. [Google Scholar] [CrossRef] [Green Version]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol. Biol. (Clifton N.J.) 2019, 1962, 227–245. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics (Oxford Engl.) 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety, A. EFSA statement on the requirements for whole genome sequence analysis of microorganisms intentionally used in the food chain. EFSA J. 2021, 19, e06506. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saleh, H.A.; Durham, S.R. Perennial rhinitis. BMJ 2007, 335, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Zeger, S.L.; Liang, K.Y.; Albert, P.S. Models for longitudinal data: A generalized estimating equation approach. Biometrics 1988, 44, 1049–1060. [Google Scholar] [CrossRef] [Green Version]
- Ring, J.; Behrendt, H.; de Weck, A. History and classification of anaphylaxis. Chem. Immunol. Allergy 2010, 95, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lack, G.; Bradley, K.L.; Hamelmann, E.; Renz, H.; Loader, J.; Leung, D.Y.; Larsen, G.; Gelfand, E.W. Nebulized IFN-gamma inhibits the development of secondary allergic responses in mice. J. Immunol. 1996, 157, 1432–1439. [Google Scholar] [CrossRef]
- Kips, J.C.; Brusselle, G.J.; Joos, G.F.; Peleman, R.A.; Tavernier, J.H.; Devos, R.R.; Pauwels, R.A. Interleukin-12 inhibits antigen-induced airway hyperresponsiveness in mice. Am. J. Respir. Crit. Care Med. 1996, 153, 535–539. [Google Scholar] [CrossRef]
- Bouladoux, N.; Hall, J.A.; Grainger, J.R.; dos Santos, L.M.; Kann, M.G.; Nagarajan, V.; Verthelyi, D.; Belkaid, Y. Regulatory role of suppressive motifs from commensal DNA. Mucosal Immunol. 2012, 5, 623–634. [Google Scholar] [CrossRef] [Green Version]
- Montamat, G.; Leonard, C.; Poli, A.; Klimek, L.; Ollert, M. CpG Adjuvant in Allergen-Specific Immunotherapy: Finding the Sweet Spot for the Induction of Immune Tolerance. Front. Immunol. 2021, 12, 590054. [Google Scholar] [CrossRef]
- Karimi, K.; Inman, M.D.; Bienenstock, J.; Forsythe, P. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am. J. Respir. Crit. Care Med. 2009, 179, 186–193. [Google Scholar] [CrossRef]
- Zhong, Y.; Huang, J.; Tang, W.; Chen, B.; Cai, W. Effects of probiotics, probiotic DNA and the CpG oligodeoxynucleotides on ovalbumin-sensitized Brown-Norway rats via TLR9/NF-kappaB pathway. FEMS Immunol. Med. Microbiol. 2012, 66, 71–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iliev, I.D.; Tohno, M.; Kurosaki, D.; Shimosato, T.; He, F.; Hosoda, M.; Saito, T.; Kitazawa, H. Immunostimulatory oligodeoxynucleotide containing TTTCGTTT motif from Lactobacillus rhamnosus GG DNA potentially suppresses OVA-specific IgE production in mice. Scand J. Immunol. 2008, 67, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Mazhary, Z.; Fard, N.A.; Minuchehr, Z.; Javanshir, N. Package of anti-allergic probiotic Lactobacillus by focusing on the regulatory role of immunosuppressive motifs in allergy. Inform. Med. Unlocked 2020, 18, 100280. [Google Scholar] [CrossRef]
- Li, A.L.; Sun, Y.Q.; Du, P.; Meng, X.C.; Guo, L.; Li, S.; Zhang, C. The Effect of Lactobacillus actobacillus Peptidoglycan on Bovine beta-Lactoglobulin-Sensitized Mice via TLR2/NF-kappaB Pathway. Iran. J. Allergy Asthma Immunol. 2017, 16, 147–158. [Google Scholar] [PubMed]
- Mat, Z.; Grensemann, B.; Yakin, Y.; Knobloch, J.; Koch, A. Effect of lipoteichoic acid on IL-2 and IL-5 release from T lymphocytes in asthma and COPD. Int. Immunopharmacol. 2012, 13, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.H.; Pan, T.M.; Wu, Y.J.; Chang, S.J.; Chang, M.S.; Hu, C.Y. Exopolysaccharide activities from probiotic bifidobacterium: Immunomodulatory effects (on J774A.1 macrophages) and antimicrobial properties. Int. J. Food Microbiol. 2010, 144, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Bacerio, J.; Izquierdo, M.; Aguado, M.E.; Varela, A.C.; Gonzalez-Matos, M.; Del Rivero, M.A. Using microbial metalo-aminopeptidases as targets in human infectious diseases. Microb. Cell 2021, 8, 239–246. [Google Scholar] [CrossRef]
- Sahiner, U.M.; Birben, E.; Erzurum, S.; Sackesen, C.; Kalayci, O. Oxidative stress in asthma. World Allergy Organ. J. 2011, 4, 151–158. [Google Scholar] [CrossRef] [Green Version]
- Aponte, M.; Murru, N.; Shoukat, M. Therapeutic, Prophylactic, and Functional Use of Probiotics: A Current Perspective. Front. Microbiol. 2020, 11, 562048. [Google Scholar] [CrossRef]
- Stevens, T.L.; Bossie, A.; Sanders, V.M.; Fernandez-Botran, R.; Coffman, R.L.; Mosmann, T.R.; Vitetta, E.S. Regulation of antibody isotype secretion by subsets of antigen-specific helper T cells. Nature 1988, 334, 255–258. [Google Scholar] [CrossRef]
- Yamaki, K.; Uchida, H.; Harada, Y.; Li, X.; Yanagisawa, R.; Takano, H.; Hayashi, H.; Taneda, S.; Mori, Y.; Yoshino, S. Effect of methotrexate on Th1 and Th2 immune responses in mice. J. Pharm. Pharmacol. 2003, 55, 1661–1666. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.H.; Hong, S.G.; Mun, S.K.; Kim, S.J.; Lee, S.J.; Kim, J.J.; Kang, K.Y.; Yee, S.T. The Protective Effects of Astaxanthin on the OVA-Induced Asthma Mice Model. Molecules 2017, 22, 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, W.Y.; Fu, L.S.; Lin, H.K.; Shen, C.Y.; Chen, Y.J. Evaluation of the effect of Lactobacillus paracasei (HF.A00232) in children (6–13 years old) with perennial allergic rhinitis: A 12-week, double-blind, randomized, placebo-controlled study. Pediatr. Neonatol. 2014, 55, 181–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| COG Functional Categories a | Same Gene No. | Unique Genes in GM-080 (Total No.) | Unique Genes in BCRC 16100 (Total No.) |
|---|---|---|---|
| Cell wall/membrane/envelope biogenesis (M) | 72 | rfbP, rgpAc, mprF (3) | kdsD, tuaG, tagF, ywqC, tagE, gtf1 (6) |
| Replication, recombination, and repair (L) | 89 | recT, pi112, tnpA1, tnp1216, cas2, cas1, cas9, int3, pi346, tnpR, is18, yqaJ, rusA (13) | - |
| Posttranslational modification, protein turnover, and chaperones (O) | 44 | gst (1) | - |
| Carbohydrate transport and metabolism (G) | 139 | agaD, kduI, kdgK, ahaA, xylP, lacE, lacG, lacF (8) | pts32BC, gatY, mnaA (3) |
| Amino acid transport and metabolism (E) | 132 | dppA, pepA, yxeO (3) | - |
| Coenzyme transport and metabolism (H) | 53 | - | pdxA (1) |
| Inorganic ion transport and metabolism (P) | 92 | feoA, ytmL (2) | kdgT, sfuB, fbpC (3) |
| Secondary metabolites biosynthesis, transport, and catabolism (Q) | 15 | kduD (1) | - |
| Intracellular trafficking, secretion, and vesicular transport (U) | 36 | chaT1, clpP (2) | secY2, secA2 (2) |
| Code | Core Sequence | Strain | Sequence | IFN-r (pg/mL) b | IL-12 (pg/mL) c |
|---|---|---|---|---|---|
| IM3 | TCAAGCTTGA | TCAAGCTTGA | ND d | ND | |
| IM4 | TCAAGCTTGA | GM-080 | CAAGCGTCAAGCTTGAATGA | ND | ND |
| IM5 | GM-080 | AAAAATTCAAGCTTGATAGT | ND | 609.8 ± 227.69 | |
| IM6 | GM-080 | CCATCGTCAAGCTTGACTTG | ND | ND | |
| IM7 | GM-080 | CCCTAATCAAGCTTGATTAA | ND | ND | |
| IM8 | BCRC 16100 | GCAGCTTCAAGCTTGAAAAA | ND | ND | |
| IM9 | BCRC 16100 | CCGGCCTCAAGCTTGAATTG | ND | ND | |
| IM10 | BCRC 16100 | TTTCATTCAAGCTTGACGCT | ND | ND | |
| IM11 | BCRC 16100 | CCTTAATCAAGCTTGATTAG | ND | ND | |
| ODN1 | GACGATCGTC | GM-080 | GCTTGACGATCGTCTCTGGA | ND | 38.8 ± 65.2 |
| ODN2 | ACGACGTCGT | GM-080 | GGTCACGACGTCGTTTACAAA | 490 ± 272.74 | 46.1 ± 32.88 |
| ODN3 | GACGATCGTC | BCRC 16100 | AATTGACGATCGTCTAATTC | ND | ND |
| ODN4 | BCRC 16100 | TGTCGACGATCGTCGTCTGT | ND | ND | |
| ODN5 | BCRC 16100 | CAGAGACGATCGTCAAGCGA | 77.381 ± 5.4 | ND | |
| ODN6 | ACGACGTCGT | BCRC 16100 | CGTCACGACGTCGTGACCGGC | ND | ND |
| Characteristics | A Group (n = 28) | B Group (n = 31) | C Group (n = 31) | PLC Group (n = 32) | p |
|---|---|---|---|---|---|
| Male b | 20 (71.4%) | 18 (58.1%) | 18 (58.1%) | 21 (65.6%) | 0.660 |
| Age (year) c | 8.25 (2.78) | 8.74 (2.77) | 8.42 (2.55) | 8.56 (2.95) | 0.917 |
| Sneezing c | 1.69 (0.77) | 1.61 (0.67) | 1.49 (0.79) | 1.75 (0.64) | 0.527 |
| Rhinorrhea c | 1.83 (0.55) | 1.81 (0.68) | 1.69 (0.70) | 1.95 (0.60) | 0.435 |
| Nasal pruritus c | 1.73 (0.70) | 1.72 (0.53) | 1.69 (0.67) | 1.70 (0.79) | 0.997 |
| Nasal congestion c | 2.10 (0.63) | 1.97 (0.66) | 2.04 (0.71) | 2.04 (0.61) | 0.880 |
| TNSS c | 7.34 (1.44) | 7.11 (1.74) | 6.92 (1.57) | 7.44 (1.56) | 0.559 |
| Total serum IgE (kU/L) c | 652.58 (1366.19) | 547.66 (530.99) | 614.81 (661.08) | 405.68 (384.19) | 0.642 |
| Combine with asthma b | 11 (39.3%) | 14 (45.2%) | 11 (35.3%) | 12 (37.5%) | 0.878 |
| Combine with atopic dermatitis b | 12 (42.9%) | 11 (35.5%) | 8 (25.8%) | 11 (34.4%) | 0.590 |
| Combine with conjunctivitis b | 5 (17.9%) | 6 (19.4%) | 5 (16.1%) | 9 (28.1%) | 0.648 |
| Allergic sensitization b | |||||
| Mite (Df) | 23 (82.1%) | 25 (80.6%) | 20 (64.5%) | 25 (78.1%) | 0.350 |
| Mite (Dp) | 26 (92.9%) | 30 (96.8%) | 28 (90.3%) | 31 (96.9%) | 0.624 |
| Cockroach | 6 (21.4%) | 7 (22.6%) | 10 (32.3%) | 5 (15.6%) | 0.470 |
| Animal dander (Cats) | 6 (21.4%) | 4 (12.9%) | 7 (22.6%) | 7 (21.9%) | 0.749 |
| Animal dander (Dogs) | 7 (25.0%) | 6 (19.4%) | 7 (22.6%) | 9 (28.1%) | 0.869 |
| Mold | 1 (3.6%) | 2 (6.5%) | 2 (6.5%) | 1 (3.1%) | 0.887 |
| Subscale | Examination | A Group (n = 28) | B Group (n = 31) | C Group (n = 31) | PLC Group (n = 32) | p |
|---|---|---|---|---|---|---|
| Sneezing | Visit 2 | 1.69 (0.77) | 1.61 (0.67) | 1.49 (0.79) | 1.75 (0.64) | 0.527 |
| Visit 3 | 1.61 (0.71) | 1.20 (0.57) a | 1.43 (0.83) | 1.67 (0.67) | 0.045 e | |
| Visit 4 | 1.68 (0.75) | 1.19 (0.70) a | 1.41 (0.83) | 1.40 (0.65) | 0.090 | |
| Visit 5 | 1.51 (0.74) | 1.02 (0.66) a | 1.35 (0.96) | 1.50 (0.66) | 0.049 e | |
| Visit 6 | 1.33 (0.76) | 1.08 (0.68) a | 1.06 (0.82) ab | 1.56 (0.82) | 0.033 e | |
| Rhinorrhea | Visit 2 | 1.83 (0.55) | 1.81 (0.68) | 1.69 (0.70) | 1.95 (0.60) | 0.435 |
| Visit 3 | 1.71 (0.67) | 1.73 (0.83) | 1.51 (0.86) | 1.95 (0.75) | 0.170 | |
| Visit 4 | 1.69 (0.71) | 1.42 (0.80) a | 1.39 (0.76) | 1.65 (0.66) | 0.252 | |
| Visit 5 | 1.37 (0.73) | 1.20 (0.72) ab | 1.24 (1.00) | 1.59 (0.69) | 0.203 | |
| Visit 6 | 1.42 (0.81) | 1.11 (0.67) ab | 1.24 (0.99) | 1.44 (0.75) | 0.320 | |
| Nasal pruritus | Visit 2 | 1.73 (0.70) | 1.72 (0.53) | 1.69 (0.67) | 1.70 (0.79) | 0.997 |
| Visit 3 | 1.54 (0.83) | 1.42 (0.73) | 1.35 (0.67) | 1.65 (0.80) | 0.415 | |
| Visit 4 | 1.43 (0.81) | 1.34 (0.78) a | 1.24 (0.59) a | 1.51 (0.72) | 0.466 | |
| Visit 5 | 1.24 (0.64) a | 1.09 (0.64) a | 1.11 (0.78) a | 1.47 (0.75) | 0.123 | |
| Visit 6 | 1.08 (0.77) a | 1.08 (0.80) a | 1.09 (0.75) a | 1.33 (0.89) | 0.520 | |
| Nasal congestion | Visit 2 | 2.10 (0.63) | 1.97 (0.66) | 2.04 (0.71) | 2.04 (0.61) | 0.880 |
| Visit 3 | 1.87 (0.74) | 1.87 (0.82) | 1.86 (0.88) | 2.07 (0.76) | 0.663 | |
| Visit 4 | 1.87 (0.81) | 1.60 (0.78) | 1.72 (0.64) | 1.84 (0.72) | 0.476 | |
| Visit 5 | 1.33 (0.71) ab | 1.35 (0.88) a | 1.47 (0.89) a | 1.77 (0.84) | 0.154 | |
| Visit 6 | 1.29 (0.73) abc | 1.15 (0.88) abc | 1.19 (0.90) abc | 1.48 (0.84) a | 0.414 | |
| TNSS | Visit 2 | 7.34 (1.44) | 7.11 (1.74) | 6.92 (1.57) | 7.44 (1.56) | 0.559 |
| Visit 3 | 6.74 (2.01) | 6.23 (2.21) | 6.15 (2.51) | 7.35 (2.24) | 0.135 | |
| Visit 4 | 6.68 (2.45) | 5.54 (2.50) a | 5.77 (1.95) a | 6.40 (2.05) | 0.174 | |
| Visit 5 | 5.44 (2.13) a | 4.67 (2.41) ab | 5.18 (2.97) a | 6.33 (2.36) | 0.066 | |
| Visit 6 | 5.12 (2.44) abc | 4.42 (2.39) ab | 4.57 (3.00) abc | 5.56 (2.49) | 0.286 |
| A Group (n = 28) | B Group (n = 31) | C Group (n = 31) | PLC Group (n = 32) | p | |||||
|---|---|---|---|---|---|---|---|---|---|
| Subscale | n | Value (95% CI a) | n | Value (95% CI) | n | Value (95% CI) | n | Value (95% CI) | |
| Sneezing | 28 | 0.028 (−0.321, 0.377) | 31 | −0.229 (−0.536, 0.077) | 31 | −0.250 (−0.604, 0.104) | 32 | Referent | 0.041 b |
| Rhinorrhea | 28 | −0.014 (−0.404, 0.376) | 31 | −0.332 (−0.677, 0.012) | 31 | −0.202 (−0.627, 0.223) | 32 | Referent | 0.104 |
| Nasal pruritus | 28 | −0.258 (−0.671, 0.155) | 31 | −0.250 (−0.662, 0.161) | 31 | −0.242 (−0.643, 0.159) | 32 | Referent | 0.414 |
| Nasal congestion | 28 | −0.192 (−0.583, 0.199) | 31 | −0.330 (−0.750, 0.089) | 31 | −0.289 (−0.713, 0.136) | 32 | Referent | 0.441 |
| TNSS | 28 | −0.436 (−1.663, 0.791) | 31 | −1.142 (−2.328, 0.044) | 31 | −0.983 (−2.324, 0.357) | 32 | Referent | 0.070 |
| Subscale | Examination | A Group (n = 28) | B Group (n = 31) | C Group (n = 31) | PLC Group (n = 32) | p-Value 4 Groups |
|---|---|---|---|---|---|---|
| Global Assessment | Visit 3 | 2.43 (0.88) | 2.42 (0.62) | 2.58 (0.56) | 2.47 (0.72) | 0.795 |
| Visit 4 | 2.61 (0.63) | 2.61 (0.67) | 2.71 (0.69) | 2.44 (0.76) | 0.471 | |
| Visit 5 | 3.00 (0.61) bc | 2.94 (0.81) b | 2.77 (0.76) | 2.53 (0.84) | 0.083 | |
| Visit 6 | 2.96 (0.74) c | 2.97 (0.71) b | 3.16 (0.86) d | 2.59 (0.91) | 0.049 e |
| Biomarker | Examination | A group (n = 28) | B group (n = 31) | C group (n = 31) | PLC group (n = 32) | p |
|---|---|---|---|---|---|---|
| Total IgE (kU/L) a | Baseline | 652.58 (1366.19) | 547.66 (530.99) | 614.81 (661.08) | 405.68 (384.19) | 0.642 |
| Visit 6 | 648.73 (1466.52) | 449.69 (469.01) | 599.39 (677.36) | 419.52 (529.22) | 0.833 | |
| IFN-γ (ng/mL) a | Baseline | 436.96 (366.48) | 494.73 (760.74) | 302.30 (442.22) | 383.25 (498.97) | 0.331 |
| Visit 6 | 467.10 (915.50) | 1042.03 (3006.39) b | 740.28 (2020.84) | 515.89 (1830.42) | 0.480 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, E.-K.; Chang, W.-W.; Jhong, J.-H.; Tsai, W.-H.; Chou, C.-H.; Wang, I.-J. Lacticaseibacillus paracasei GM-080 Ameliorates Allergic Airway Inflammation in Children with Allergic Rhinitis: From an Animal Model to a Double-Blind, Randomized, Placebo-Controlled Trial. Cells 2023, 12, 768. https://doi.org/10.3390/cells12050768
Lin E-K, Chang W-W, Jhong J-H, Tsai W-H, Chou C-H, Wang I-J. Lacticaseibacillus paracasei GM-080 Ameliorates Allergic Airway Inflammation in Children with Allergic Rhinitis: From an Animal Model to a Double-Blind, Randomized, Placebo-Controlled Trial. Cells. 2023; 12(5):768. https://doi.org/10.3390/cells12050768
Chicago/Turabian StyleLin, En-Kwang, Wen-Wei Chang, Jhih-Hua Jhong, Wan-Hua Tsai, Chia-Hsuan Chou, and I-Jen Wang. 2023. "Lacticaseibacillus paracasei GM-080 Ameliorates Allergic Airway Inflammation in Children with Allergic Rhinitis: From an Animal Model to a Double-Blind, Randomized, Placebo-Controlled Trial" Cells 12, no. 5: 768. https://doi.org/10.3390/cells12050768
APA StyleLin, E.-K., Chang, W.-W., Jhong, J.-H., Tsai, W.-H., Chou, C.-H., & Wang, I.-J. (2023). Lacticaseibacillus paracasei GM-080 Ameliorates Allergic Airway Inflammation in Children with Allergic Rhinitis: From an Animal Model to a Double-Blind, Randomized, Placebo-Controlled Trial. Cells, 12(5), 768. https://doi.org/10.3390/cells12050768