Large-Scale Polymorphism Analysis of Dog Leukocyte Antigen Class I and Class II Genes (DLA-88, DLA-12/88L and DLA-DRB1) and Comparison of the Haplotype Diversity between Breeds in Japan
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA and DNA Samples
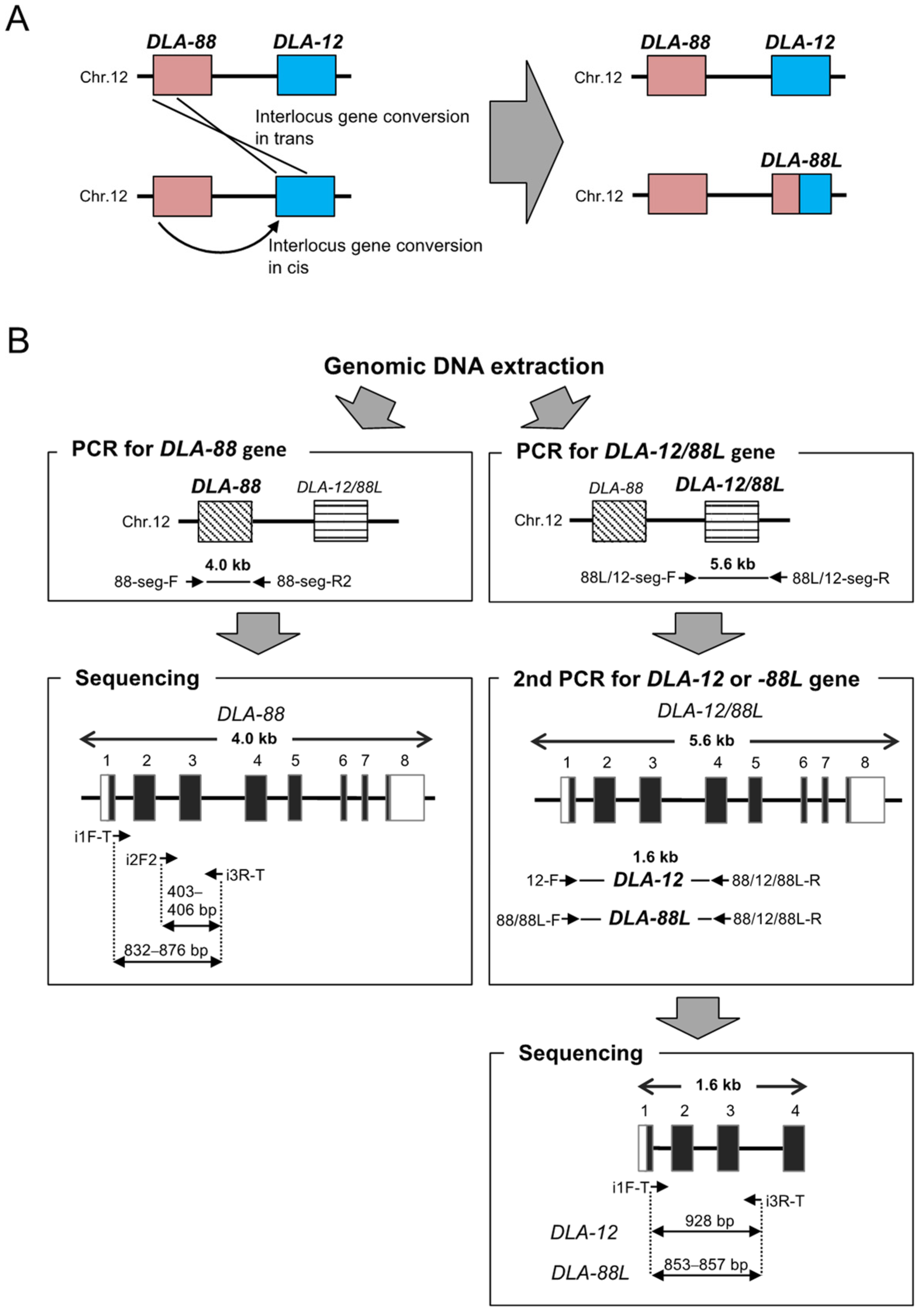
2.2. PCR Amplification of DLA-88 and DLA-12/88L Genes
2.3. PCR Amplification of DLA-DRB1 Gene
2.4. Sanger-Sequencing
2.5. Allele Assignment and Confirmation of Novel DLA Alleles
2.6. Nomenclature of Novel DLA Alleles
2.7. Estimation of DLA-88–DLA-12/88L–DLA-DRB1 (88-12/88L-DRB1) Haplotypes
2.8. Data Analysis
3. Results
3.1. Allele Number and Frequency of DLA-88, DLA-88L, DLA-12, and DLA-DRB1
3.2. Phylogenetic Relationships of the DLA-88, DLA-88L, and DLA-12 Alleles
3.3. Evaluation of DLA-DRB1 Polymorphisms between Same Dog Breeds in Japan and the United Kingdom
3.4. Frequency of the 88-12/88L-DRB1 Haplotypes
3.5. Comparison of Genetic Diversity between Dog Breeds by Haplotype Numbers and Heterozygosity
3.6. Characteristics of Genetic Relationship of the 88-12/88L-DRB1 Haplotypes by Principal Component Analysis
3.7. Number of Potential Recipients for 88-12/88L-DRB1-Matched Transplantation, Assuming Homozygous-Derived Somatic Stem Cells as Donors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Nomenclature
| Haplotype(s) | A set of closely linked alleles on a chromosome carrying the three-gene loci combination of 88-12-DRB1 or 88-88L-DRB1 and represented by 88-12/88L-DRB1 where 12/88L could be either 12 or 88L. |
| Sub-haplotype(s) | A single locus (88, 12, 88L, 12/88L, DRB1) or two-locus subtype (88-12/88L, 88/DRB1, 12/88L-DRB1) of the three locus-haplotype 88-12/88L-DRB1. |
| Homozygous haplotypes | Two identical haplotypes in diploid cells. |
| Heterozygous haplotypes | Two different haplotypes in diploid cells. |
| Mongrels | Mixtures of different pure dog breeds or interbreeds. |
References
- König, R.; Huang, L.-Y.; Germain, R.N. MHC class II interaction with CD4 mediated by a region analogous to the MHC class I binding site for CD8. Nature 1992, 356, 796–798. [Google Scholar] [CrossRef] [PubMed]
- Garcia, K.C.; Scott, C.A.; Brunmark, A.; Carbone, F.R.; Peterson, P.A.; Wilson, I.A.; Teyton, L. CD8 enhances formation of stable T-cell receptor/MHC class I molecule complexes. Nature 1996, 384, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Shiina, T.; Inoko, H.; Kulski, J. An update of the HLA genomic region, locus information and disease associations: 2004. Tissue Antigens 2004, 64, 631–649. [Google Scholar] [CrossRef]
- Shiina, T.; Hosomichi, K.; Inoko, H.; Kulski, J.K. The HLA genomic loci map: Expression, interaction, diversity and disease. J. Hum. Genet. 2009, 54, 15–39. [Google Scholar] [CrossRef]
- Matzaraki, V.; Kumar, V.; Wijmenga, C.; Zhernakova, A. The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol. 2017, 18, 76. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.A.; Tatapudi, V.S.; Leffell, M.S.; Zachary, A.A. HLA in transplantation. Nat. Rev. Nephrol. 2018, 14, 558–570. [Google Scholar] [CrossRef]
- Eapen, M.; Klein, J.P.; Sanz, G.F.; Spellman, S.; Ruggeri, A.; Anasetti, C.; Brown, M.; Champlin, R.E.; Garcia-Lopez, J.; Hattersely, G. Effect of donor–recipient HLA matching at HLA A, B, C, and DRB1 on outcomes after umbilical-cord blood transplantation for leukaemia and myelodysplastic syndrome: A retrospective analysis. Lancet Oncol. 2011, 12, 1214–1221. [Google Scholar] [CrossRef]
- Zachary, A.A.; Leffell, M.S. HLA mismatching strategies for solid organ transplantation–a balancing act. Front. Immunol. 2016, 7, 575. [Google Scholar] [CrossRef]
- Sugita, S.; Iwasaki, Y.; Makabe, K.; Kimura, T.; Futagami, T.; Suegami, S.; Takahashi, M. Lack of T cell response to iPSC-derived retinal pigment epithelial cells from HLA homozygous donors. Stem Cell Rep. 2016, 7, 619–634. [Google Scholar] [CrossRef]
- Schoenebeck, J.J.; Ostrander, E.A. Insights into morphology and disease from the dog genome project. Annu. Rev. Cell Dev. Biol. 2014, 30, 535–560. [Google Scholar] [CrossRef] [PubMed]
- Graves, S.S.; Storb, R. Developments and translational relevance for the canine haematopoietic cell transplantation preclinical model. Vet. Comp. Oncol. 2020, 18, 471–483. [Google Scholar] [CrossRef]
- Kirkness, E.F.; Bafna, V.; Halpern, A.L.; Levy, S.; Remington, K.; Rusch, D.B.; Delcher, A.L.; Pop, M.; Wang, W.; Fraser, C.M. The dog genome: Survey sequencing and comparative analysis. Science 2003, 301, 1898–1903. [Google Scholar] [CrossRef] [PubMed]
- Lindblad-Toh, K.; Wade, C.M.; Mikkelsen, T.S.; Karlsson, E.K.; Jaffe, D.B.; Kamal, M.; Clamp, M.; Chang, J.L.; Kulbokas, E.J.; Zody, M.C. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 2005, 438, 803–819. [Google Scholar] [CrossRef] [PubMed]
- Graumann, M.; DeRose, S.; Ostrander, E.; Storb, R. Polymorphism analysis of four canine MHC class I genes. Tissue Antigens 1998, 51, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.; Buntzman, A.S.; Vincent, B.G.; Grover, E.N.; Gojanovich, G.S.; Collins, E.J.; Frelinger, J.A.; Hess, P.R. Allelic diversity at the DLA-88 locus in Golden Retriever and Boxer breeds is limited. Tissue Antigens 2012, 80, 175–183. [Google Scholar] [CrossRef]
- Venkataraman, G.M.; Geraghty, D.; Fox, J.; Graves, S.S.; Zellmer, E.; Storer, B.E.; Torok-Storb, B.J.; Storb, R. Canine DLA-79 gene: An improved typing method, identification of new alleles and its role in graft rejection and graft-versus-host disease. Tissue Antigens 2013, 81, 204–211. [Google Scholar] [CrossRef]
- Venkataraman, G.M.; Kennedy, L.J.; Little, M.T.; Graves, S.S.; Harkey, M.A.; Torok-Storb, B.J.; Storb, R. Thirteen novel canine dog leukocyte antigen-88 alleles identified by sequence-based typing. Hla 2017, 90, 165–170. [Google Scholar] [CrossRef]
- Miyamae, J.; Suzuki, S.; Katakura, F.; Uno, S.; Tanaka, M.; Okano, M.; Matsumoto, T.; Kulski, J.K.; Moritomo, T.; Shiina, T. Identification of novel polymorphisms and two distinct haplotype structures in dog leukocyte antigen class I genes: DLA-88, DLA-12 and DLA-64. Immunogenetics 2018, 70, 237–255. [Google Scholar] [CrossRef]
- Wagner, J.; DeRose, S.; Burnett, R.; Storb, R. Nucleotide sequence and polymorphism analysis of canine DRA cDNA clones. Tissue Antigens 1995, 45, 284–287. [Google Scholar] [CrossRef]
- Kennedy, L.J.; Barnes, A.; Happ, G.; Quinnell, R.; Bennett, D.; Angles, J.; Day, M.; Carmichael, N.; Innes, J.; Isherwood, D. Extensive interbreed, but minimal intrabreed, variation of DLA class II alleles and haplotypes in dogs. Tissue Antigens 2002, 59, 194–204. [Google Scholar] [CrossRef]
- Kennedy, L.; Barnes, A.; Short, A.; Brown, J.; Lester, S.; Seddon, J.; Fleeman, L.; Francino, O.; Brkljacic, M.; Knyazev, S. Canine DLA diversity: 1. New alleles and haplotypes. Tissue Antigens 2007, 69, 272–288. [Google Scholar] [CrossRef]
- Kennedy, L.J.; Ollier, W.E.; Marti, E.; Wagner, J.L.; Storb, R.F. Canine immunogenetics. In The Genetics of the Dog; CABI: Wallingford, UK, 2012; pp. 91–135. [Google Scholar]
- Tsai, K.L.; Starr-Moss, A.N.; Venkataraman, G.M.; Robinson, C.; Kennedy, L.J.; Steiner, J.M.; Clark, L.A. Alleles of the major histocompatibility complex play a role in the pathogenesis of pancreatic acinar atrophy in dogs. Immunogenetics 2013, 65, 501–509. [Google Scholar] [CrossRef]
- Hardt, C.; Ferencik, S.; Tak, R.; Hoogerbrugge, P.; Wagner, V.; Grosse-Wilde, H. Sequence-based typing reveals a novel DLA-88 allele, DLA-88* 04501, in a beagle family. Tissue Antigens 2006, 67, 163–165. [Google Scholar] [CrossRef]
- Ollier, W.E.; Kennedy, L.J.; Thomson, W.; Barnes, A.N.; Bell, S.C.; Bennett, D.; Angles, J.M.; Innes, J.F.; Carter, S.D. Dog MHC alleles containing the human RA shared epitope confer susceptibility to canine rheumatoid arthritis. Immunogenetics 2001, 53, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, L.; Barnes, A.; Short, A.; Brown, J.; Seddon, J.; Fleeman, L.; Brkljacic, M.; Happ, G.; Catchpole, B.; Ollier, W. Canine DLA diversity: 3. Disease studies. Tissue Antigens 2007, 69, 292–296. [Google Scholar] [CrossRef]
- Denyer, A.; Massey, J.; Davison, L.; Ollier, W.; Catchpole, B.; Kennedy, L. Dog leucocyte antigen (DLA) class II haplotypes and risk of canine diabetes mellitus in specific dog breeds. Canine Med. Genet. 2020, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, M.; Miyamae, J.; Okano, M.; Kanemoto, H.; Katakura, F.; Shiina, T.; Ohno, K.; Tsujimoto, H.; Moritomo, T.; Watari, T. Dog leukocyte antigen (DLA) class II genotypes associated with chronic enteropathy in French bulldogs and miniature dachshunds. Vet. Immunol. Immunopathol. 2021, 237, 110271. [Google Scholar] [CrossRef]
- Friedenberg, S.G.; Buhrman, G.; Chdid, L.; Olby, N.J.; Olivry, T.; Guillaumin, J.; O’Toole, T.; Goggs, R.; Kennedy, L.J.; Rose, R.B. Evaluation of a DLA-79 allele associated with multiple immune-mediated diseases in dogs. Immunogenetics 2016, 68, 205–217. [Google Scholar] [CrossRef]
- Miyamae, J.; Okano, M.; Nishiya, K.; Katakura, F.; Kulski, J.K.; Moritomo, T.; Shiina, T. Haplotype structures and polymorphisms of dog leukocyte antigen (DLA) class I loci shaped by intralocus and interlocus recombination events. Immunogenetics 2022, 74, 245–259. [Google Scholar] [CrossRef] [PubMed]
- Miyamae, J.; Yagi, H.; Sato, K.; Okano, M.; Nishiya, K.; Katakura, F.; Sakai, M.; Nakayama, T.; Moritomo, T.; Shiina, T. Evaluation of alloreactive T cells based on the degree of MHC incompatibility using flow cytometric mixed lymphocyte reaction assay in dogs. Immunogenetics 2019, 71, 635–645. [Google Scholar] [CrossRef]
- Wagner, J.; Burnett, R.; Works, J.; Storb, R. Molecular analysis of DLA-DRBB1 polymorphism. Tissue Antigens 1996, 48, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, L.J.; Angles, J.; Barnes, A.; Carter, S.; Francino, O.; Gerlach, J.; Happ, G.; Ollier, W.; Thomson, W.; Wagner, J. Nomenclature for factors of the dog major histocompatibility system (DLA), 2000: Second report of the ISAG DLA Nomenclature Committee. Anim. Genet. 2001, 32, 193–199. [Google Scholar] [CrossRef]
- Stephens, M.; Smith, N.J.; Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ohtsubo, Y.; Ikeda-Ohtsubo, W.; Nagata, Y.; Tsuda, M. GenomeMatcher: A graphical user interface for DNA sequence comparison. BMC Bioinform. 2008, 9, 376. [Google Scholar] [CrossRef]
- Quignon, P.; Herbin, L.; Cadieu, E.; Kirkness, E.F.; Hédan, B.; Mosher, D.S.; Galibert, F.; André, C.; Ostrander, E.A.; Hitte, C. Canine population structure: Assessment and impact of intra-breed stratification on SNP-based association studies. PLoS ONE 2007, 2, e1324. [Google Scholar] [CrossRef] [PubMed]
- Parker, H.G.; Dreger, D.L.; Rimbault, M.; Davis, B.W.; Mullen, A.B.; Carpintero-Ramirez, G.; Ostrander, E.A. Genomic analyses reveal the influence of geographic origin, migration, and hybridization on modern dog breed development. Cell Rep. 2017, 19, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Lampi, S.; Donner, J.; Anderson, H.; Pohjoismäki, J. Variation in breeding practices and geographic isolation drive subpopulation differentiation, contributing to the loss of genetic diversity within dog breed lineages. Canine Med. Genet. 2020, 7, 5. [Google Scholar] [CrossRef]
- Runstadler, J.; Angles, J.; Pedersen, N.C. Dog leucocyte antigen class II diversity and relationships among indigenous dogs of the island nations of Indonesia (Bali), Australia and New Guinea. Tissue Antigens 2006, 68, 418–426. [Google Scholar] [CrossRef]
- Kang, M.; Ahn, B.; Youk, S.; Cho, H.-s.; Choi, M.; Hong, K.; Do, J.T.; Song, H.; Jiang, H.; Kennedy, L.J. High Allelic Diversity of Dog Leukocyte Antigen Class II in East Asian Dogs: Identification of New Alleles and Haplotypes. J. Mamm. Evol. 2021, 28, 773–784. [Google Scholar] [CrossRef]
- Niskanen, A.; Hagström, E.; Lohi, H.; Ruokonen, M.; Esparza-Salas, R.; Aspi, J.; Savolainen, P. MHC variability supports dog domestication from a large number of wolves: High diversity in Asia. Heredity 2013, 110, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Gojobori, J.; Arakawa, N.; Xiayire, X.; Matsumoto, Y.; Matsumura, S.; Hongo, H.; Ishiguro, N.; Terai, Y. The Japanese wolf is most closely related to modern dogs and its ancestral genome has been widely inherited by dogs throughout East Eurasia. bioRxiv 2021, 2021, 463851. [Google Scholar]
- Niskanen, A.; Kennedy, L.; Ruokonen, M.; Kojola, I.; Lohi, H.; Isomursu, M.; Jansson, E.; Pyhäjärvi, T.; Aspi, J. Balancing selection and heterozygote advantage in major histocompatibility complex loci of the bottlenecked Finnish wolf population. Mol. Ecol. 2014, 23, 875–889. [Google Scholar] [CrossRef]
- Arbanasić, H.; Huber, Đ.; Kusak, J.; Gomerčić, T.; Hrenović, J.; Galov, A. Extensive polymorphism and evidence of selection pressure on major histocompatibility complex DLA-DRB1, DQA1 and DQB1 class II genes in Croatian grey wolves. Tissue Antigens 2013, 81, 19–27. [Google Scholar] [CrossRef]
- Kennedy, L.J.; Angles, J.M.; Barnes, A.; Carmichael, L.E.; Radford, A.D.; Ollier, W.E.; Happ, G.M. DLA-DRB1, DQA1, and DQB1 alleles and haplotypes in North American gray wolves. J. Hered. 2007, 98, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Marsden, C.D.; Ortega-Del Vecchyo, D.; O’Brien, D.P.; Taylor, J.F.; Ramirez, O.; Vilà, C.; Marques-Bonet, T.; Schnabel, R.D.; Wayne, R.K.; Lohmueller, K.E. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc. Natl. Acad. Sci. USA 2016, 113, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C.; Liu, H.; Leonard, A.; Griffioen, L. A search for genetic diversity among Italian Greyhounds from Continental Europe and the USA and the effect of inbreeding on susceptibility to autoimmune disease. Canine Genet. Epidemiol. 2015, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Osborne, A.J.; Pearson, J.; Negro, S.S.; Chilvers, B.L.; Kennedy, M.A.; Gemmell, N.J. Heterozygote advantage at MHC DRB may influence response to infectious disease epizootics. Mol. Ecol. 2015, 24, 1419–1432. [Google Scholar] [CrossRef]
- Burger, D.; Thomas, S.; Aepli, H.; Dreyer, M.; Fabre, G.; Marti, E.; Sieme, H.; Robinson, M.R.; Wedekind, C. Major histocompatibility complex-linked social signalling affects female fertility. Proc. R. Soc. B Biol. Sci. 2017, 284, 20171824. [Google Scholar] [CrossRef]
- Nakatsuji, N.; Nakajima, F.; Tokunaga, K. HLA-haplotype banking and iPS cells. Nat. Biotechnol. 2008, 26, 739–740. [Google Scholar] [CrossRef]
- Taylor, C.J.; Bolton, E.M.; Pocock, S.; Sharples, L.D.; Pedersen, R.A.; Bradley, J.A. Banking on human embryonic stem cells: Estimating the number of donor cell lines needed for HLA matching. Lancet 2005, 366, 2019–2025. [Google Scholar] [CrossRef]
- Lin, G.; Xie, Y.; OuYang, Q.; Qian, X.; Xie, P.; Zhou, X.; Xiong, B.; Tan, Y.; Li, W.; Deng, L. HLA-matching potential of an established human embryonic stem cell bank in China. Cell Stem Cell 2009, 5, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Palomo, B.; García-Martinez, I.; Gayoso, J.; Raya, A.; Veiga, A.; Abad, M.L.; Eiras, A.; Guzmán-Fulgencio, M.; Luis-Hidalgo, M.; Eguizabal, C. Evaluation of the Spanish population coverage of a prospective HLA haplobank of induced pluripotent stem cells. Stem Cell Res. Ther. 2021, 12, 233. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Tsukamoto, M.; Tanaka, M.; Kuwamura, M.; Ohtaka, M.; Nishimura, K.; Nakanishi, M.; Sugiura, K.; Hatoya, S. Efficient reprogramming of canine peripheral blood mononuclear cells into induced pluripotent stem cells. Stem Cells Dev. 2021, 30, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Yoshimatsu, S.; Edamura, K.; Yoshii, Y.; Iguchi, A.; Kondo, H.; Shibuya, H.; Sato, T.; Shiozawa, S.; Okano, H. Non-viral derivation of a transgene-free induced pluripotent stem cell line from a male beagle dog. Stem Cell Res. 2021, 53, 102375. [Google Scholar] [CrossRef]
- Scarfone, R.A.; Pena, S.M.; Russell, K.A.; Betts, D.H.; Koch, T.G. The use of induced pluripotent stem cells in domestic animals: A narrative review. BMC Vet. Res. 2020, 16, 477. [Google Scholar] [CrossRef]
- Angles, J.; Kennedy, L.; Pedersen, N.C. Frequency and distribution of alleles of canine MHC-II DLA-DQB1, DLA-DQA1 and DLA-DRB1 in 25 representative American Kennel Club breeds. Tissue Antigens 2005, 66, 173–184. [Google Scholar] [CrossRef]
- Gershony, L.C.; Belanger, J.M.; Short, A.D.; Le, M.; Hytönen, M.K.; Lohi, H.; Famula, T.R.; Kennedy, L.J.; Oberbauer, A.M. DLA class II risk haplotypes for autoimmune diseases in the bearded collie offer insight to autoimmunity signatures across dog breeds. Canine Genet. Epidemiol. 2019, 6, 2. [Google Scholar] [CrossRef]
- DeVos, J.M.; Gaber, A.O.; Knight, R.J.; Land, G.A.; Suki, W.N.; Gaber, L.W.; Patel, S.J. Donor-specific HLA-DQ antibodies may contribute to poor graft outcome after renal transplantation. Kidney Int. 2012, 82, 598–604. [Google Scholar] [CrossRef]
- Tikkanen, J.M.; Singer, L.G.; Kim, S.J.; Li, Y.; Binnie, M.; Chaparro, C.; Chow, C.-W.; Martinu, T.; Azad, S.; Keshavjee, S.; et al. De novo DQ donor-specific antibodies are associated with chronic lung allograft dysfunction after lung transplantation. Am. J. Respir. Crit. Care Med. 2016, 194, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Greer, K.; Wong, A.; Liu, H.; Famula, T.; Pedersen, N.C.; Ruhe, A.; Wallace, M.; Neff, M. Necrotizing meningoencephalitis of Pug dogs associates with dog leukocyte antigen class II and resembles acute variant forms of multiple sclerosis. Tissue Antigens 2010, 76, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Stromberg, S.J.; Thomasy, S.M.; Marangakis, A.D.; Kim, S.; Cooper, A.E.; Brown, E.A.; Maggs, D.J.; Bannasch, D.L. Evaluation of the major histocompatibility complex (MHC) class II as a candidate for sudden acquired retinal degeneration syndrome (SARDS) in Dachshunds. Vet. Ophthalmol. 2019, 22, 751–759. [Google Scholar] [CrossRef] [PubMed]






| Breed | Number of Dogs | a Rank of the Number of Dogs Registered in Japan | Breed | Number of Dogs | a Rank of the Number of Dogs Registered in Japan |
|---|---|---|---|---|---|
| Miniature Dachshund | 49 | 3 | Kaninchen Dachshund | 5 | |
| Toy Poodle | 44 | 1 | Bichon frize | 4 | 16 |
| Golden Retriever | 41 | 11 | Cairn Terrier | 4 | |
| Yorkshire Terrier | 41 | 8 | Japanese Spitz | 4 | 28 |
| Beagle | 39 | 24 | Pekingese | 4 | 19 |
| Chihuahua | 39 | 2 | Standard Poodle | 4 | |
| Labrador Retriever | 39 | 13 | Chin | 3 | |
| French Bulldog | 38 | 6 | Chinese Crested Dog | 3 | |
| Shiba | 37 | 9 | Doberman | 3 | |
| Shetland Sheepdog | 35 | 23 | Great Pyrenees | 3 | |
| Miniature Schnauzer | 33 | 5 | Kooikerhondje | 3 | |
| Welsh Corgi | 33 | 14 | Saint Bernard | 3 | |
| Pomeranian | 29 | 4 | Dalmatian | 2 | |
| Shih Tzu | 28 | 10 | Norfolk Terrier | 2 | |
| Maltese | 26 | 7 | Weimaraner | 2 | |
| Papillon | 24 | 15 | Basenji | 1 | |
| Pug | 21 | 12 | Brussels Griffon | 1 | |
| American Cocker Spaniel | 20 | 26 | Rough Collie | 1 | |
| Border Collie | 19 | 18 | Irish Setter | 1 | |
| Cavalier King Charles Spaniel | 18 | 22 | Lakeland Terrier | 1 | |
| Husky | 12 | 27 | Leonberger | 1 | |
| Miniature Pinsher | 12 | 21 | Miniature Bull Terrier | 1 | |
| Bernese Mountain Dog | 11 | 29 | Rottweiler | 1 | |
| Bulldog | 10 | 30 | Saluki | 1 | |
| Akita | 9 | Shikoku | 1 | ||
| Boston Terrier | 6 | 25 | Staffordshire Bull Terrier | 1 | |
| Italian Grey Hound | 6 | 20 | Toy Manchester Terrier | 1 | |
| Jack Russell Terrier | 6 | 17 | Wippet | 1 | |
| English Cocker Spaniel | 5 | mongrel | 27 | ||
| Flat-Coated Retriever | 5 | ||||
| German Shepherd | 5 | Total | 829 |
| Locus | DLA-88 | DLA-12/88L | DLA-DRB1 | |
|---|---|---|---|---|
| DLA-12 | DLA-88L | |||
| Number of Nucleotide Sequences Detected in This Study | 89 | 25 | 18 | 61 |
| Previously published sequences | 72 | 20 | 11 | 55 |
| Novel sequences | 17 (8) | 5 (2) | 7 (3) | 6 (4) |
| a Number of nucleotide sequences not detected in this study | 56 | 0 | - | 108 |
| Number of unique amino acid sequences | 88 | 21 | 18 | 61 |
| Haplotype | Haplotypes Detected Two or More Times | Number of Other Single Haplotypes a | Frequency (%) | Homozygote | ||
|---|---|---|---|---|---|---|
| Number of Sub-Haplotypes | Number of Haplotypes | Number of Sub-Haplotypes | Number of Dogs | |||
| DLA-88–DLA-12/88L–DRB1 | 131 | 1547 | 59 | - | 52 | 198 |
| two haplotype structures | ||||||
| DLA-88–DLA-12–DRB1 | 101 | 1228 | 36 | 79.4 | 42 | 152 |
| DLA-88–DLA-88L–DRB1 | 30 | 319 | 23 | 20.6 | 10 | 46 |
| Haplotype ID. a | DLA-88* | DLA-12/88L | DLA-DRB1* | Number of Haplotypes | Haplotypefrequency (%) | Number of Dogs with the Haplotypes | Number of Homozygous Dogs | Number of Breeds with the Haplotype Except for Mongrels c | |
|---|---|---|---|---|---|---|---|---|---|
| DLA-12* | DLA-88L b (DLA-88*) | ||||||||
| 12 | 004:02 | 001:01:01 | - | 006:01 | 68 | 4.23 | 60 | 8 | 10 |
| 20 | 003:02 | - | 017:01 | 002:01 | 64 | 3.99 | 45 | 19 | 6 (Shetland Sheepdog (73.4%)) |
| 31 | 003:02 | - | 017:01 | 009:01 | 64 | 3.99 | 59 | 5 | 11 |
| 25 | 028:01 | - | 029:01 | 015:02 | 62 | 3.86 | 52 | 10 | 8 |
| 8 | 501:01 | 001:01:01 | - | 001:01 | 53 | 3.30 | 47 | 6 | 8 |
| 21 | 508:01 | 001:01:03 | - | 012:01 | 50 | 3.11 | 42 | 8 | 3 |
| 7 | 012:01 | 001:01:01 | - | 015:01 | 48 | 2.99 | 42 | 6 | 9 |
| 23 | 013:02 | 003:01 | - | 009:01 | 46 | 2.86 | 33 | 13 | 2 (Miniature Schunauzer (95.7%)) |
| 116 | 006:01 | 001:01:01 | - | 056:01 | 41 | 2.55 | 31 | 10 | 1 (Shiba (95.1%)) |
| 73 | 051:01 | 001:01:01 | - | 012:01 | 40 | 2.49 | 33 | 7 | 2 (Golden Retriever (85.0%)) |
| 6 | 502:01 | 001:01:01 | - | 001:02 | 38 | 2.37 | 28 | 10 | 1 (Beagle (94.7%)) |
| 63 | 004:02 | 001:01:01 | - | 015:01 | 34 | 2.12 | 28 | 6 | 14 |
| 18 | 005:01 | 002:04 | - | 020:01 | 32 | 1.99 | 28 | 4 | 9 |
| 17 | 501:01 | 001:01:01 | - | 006:01 | 31 | 1.93 | 27 | 4 | 8 |
| 2 | 006:01 | 001:01:01 | - | 006:01 | 30 | 1.87 | 24 | 6 | 8 (AmericanCocker Spaniel (70.0%)) |
| 51 | 034:01 | 002:03 | - | 023:01 | 29 | 1.81 | 23 | 6 | 5 (Shetland Sheepdog (75.9%)) |
| 91 | 001:03 | 001:01:01 | - | 046:01 | 26 | 1.62 | 18 | 8 | 1 (Papillon (96.2%)) |
| 46 | 014:01:02 | 001:05 | - | 025:01 | 25 | 1.56 | 19 | 6 | 4 (Shih Tzu (72.0%)) |
| 71 | 002:01 | 001:01:01 | - | 011:01 | 23 | 1.43 | 22 | 1 | 3 |
| 52 | 006:01 | 001:01:01 | - | 002:03 | 23 | 1.43 | 21 | 2 | 2 (Dachshund (78.3%)) |
| 66 | 006:01 | 001:01:01 | - | 001:01 | 22 | 1.37 | 22 | 0 | 6 |
| 10 | 508:01 | 001:01:03 | - | 002:01 | 21 | 1.31 | 21 | 0 | 7 |
| 22 | 511:01 | 001:03 | - | 015:02 | 21 | 1.31 | 19 | 2 | 3 (Laborador Retriever (90.5%)) |
| 99 | 035:01 | nov18 | - | 006:01 | 20 | 1.25 | 15 | 5 | 2 (Shih Tzu (85.0%)) |
| 69 | 054:01 | 002:02 | - | 012:01 | 20 | 1.25 | 17 | 3 | 2 (Welsh Corgi (85.0%)) |
| 37 | 006:01 | 001:01:01 | - | 015:01 | 18 | 1.12 | 17 | 1 | 3 (Toy Poodle (77.8%)) |
| 48 | 028:05 | - | 029:01 | 015:02 | 18 | 1.12 | 18 | 0 | 6 |
| 33 | 501:01 | 001:01:01 | - | 012:01 | 16 | 1.00 | 16 | 0 | 5 |
| 117 | 511:01 | 001:03 | - | 092:01:1 | 16 | 1.00 | 15 | 1 | 1 (Shiba (87.5%)) |
| Breed | Number of Dogs a | Number of Estimated Haplotypes | Number of Homozygous Dogs | Ho | He | HWE Test | Fis | Hr b |
|---|---|---|---|---|---|---|---|---|
| Shetland Sheepdog | 35 | 3 | 24 (68.6%) | 0.314 | 0.450 | - | 0.315 | 2.13 |
| American Cocker Spaniel | 20 | 5 | 9 (45.0%) | 0.550 | 0.580 | p < 0.05 | 0.077 | 2.94 |
| Miniature Schunauzer | 32 (1) | 9 | 12 (37.5%) | 0.625 | 0.510 | - | −0.21 | 3.62 |
| Cavalier King Charles Spaniel | 18 | 6 | 5 (27.8%) | 0.722 | 0.716 | p < 0.001 | 0.020 | 3.79 |
| Shiba | 37 | 11 | 14 (37.8%) | 0.622 | 0.658 | p < 0.001 | 0.069 | 3.95 |
| Golden Retriever | 39 (2) | 11 | 11 (28.2%) | 0.718 | 0.713 | p < 0.001 | 0.006 | 4.26 |
| Papillon | 24 | 9 | 10 (41.7%) | 0.583 | 0.687 | p < 0.05 | 0.171 | 4.60 |
| Shih Tzu | 28 | 12 | 10 (35.7%) | 0.643 | 0.762 | p < 0.001 | 0.209 | 4.64 |
| Miniature Pinsher | 12 | 6 | 2 (16.7%) | 0.833 | 0.771 | - | −0.038 | 4.64 |
| Bernese Mountain Dog | 11 | 7 | 3 (27.3%) | 0.727 | 0.698 | - | 0.006 | 4.79 |
| French Bulldog | 38 | 10 | 12 (31.6%) | 0.684 | 0.751 | - | 0.102 | 4.87 |
| Husky | 12 | 8 | 5 (41.7%) | 0.583 | 0.733 | - | 0.245 | 5.06 |
| Beagle | 37 (2) | 13 | 11 (29.7%) | 0.703 | 0.730 | p < 0.05 | 0.051 | 5.10 |
| Bulldog | 10 | 7 | 2 (20.0%) | 0.800 | 0.790 | - | 0.040 | 5.20 |
| Labrador Retriever | 38 (1) | 15 | 7 (18.4%) | 0.821 | 0.807 | p < 0.001 | 0.003 | 5.50 |
| Welsh Corgi | 31 (2) | 9 | 5 (16.1%) | 0.848 | 0.837 | - | 0.015 | 5.62 |
| Pomeranian | 29 | 17 | 6 (20.7%) | 0.793 | 0.757 | - | −0.03 | 5.64 |
| Yorkshire Terrier | 41 | 13 | 3 (7.3%) | 0.927 | 0.836 | - | −0.096 | 5.85 |
| Pug | 21 | 11 | 4 (19.0%) | 0.810 | 0.840 | - | 0.061 | 5.87 |
| Maltese | 26 | 13 | 5 (19.2%) | 0.808 | 0.847 | - | 0.066 | 6.02 |
| Miniature Dachshund | 49 | 15 | 5 (10.2%) | 0.898 | 0.874 | - | −0.018 | 6.40 |
| Border Collie | 19 | 12 | 4 (21.1%) | 0.789 | 0.871 | - | 0.121 | 6.52 |
| Chihuahua | 39 | 20 | 2 (5.1%) | 0.949 | 0.917 | - | −0.022 | 7.44 |
| Toy Poodle | 44 | 27 | 4 (9.1%) | 0.909 | 0.927 | - | 0.031 | 7.79 |
| Mongrel | 27 | 34 | 1 (3.7%) | 0.963 | 0.962 | - | 0.018 | 9.16 |
| Breed | Haplotype ID. a | Most Frequent Haplotype in Each Breed | The Number of Dogs with the Haplotype within the Breed (%) | |||
|---|---|---|---|---|---|---|
| DLA-88* | DLA-12/88L | DLA-DRB1* | ||||
| DLA-12* | DLA-88L b (DLA-88*) | |||||
| Miniature Schunauzer | 23 | 013:02 | 003:01 | - | 009:01 | 32/33 (97.0) |
| Shetland Sheepdog | 20 | 003:02 | - | 017:01 | 002:01 | 29/35 (82.9) |
| American Cocker Spaniel | 31 | 003:02 | - | 017:01 | 009:01 | 16/20 (80.0) |
| Shiba | 116 | 006:01 | 001:01:01 | - | 056:01 | 29/37 (78.4) |
| Pomeranian | 12 | 004:02 | 001:01:01 | - | 006:01 | 22/29 (75.9) |
| Bernese Mountain Dog | 17 | 501:01 | 001:01:01 | - | 006:01 | 8/11 (72.7) |
| Papillon | 91 | 001:03 | 001:01:01 | - | 046:01 | 17/24 (70.8) |
| Beagle | 6 | 502:01 | 001:01:01 | - | 001:02 | 26/39 (66.7) |
| French Bulldog | 25 | 028:01 | - | 029:01 | 015:02 | 25/38 (65.8) |
| Golden Retriever | 33 | 501:01 | 001:01:01 | - | 012:01 | 26/41 (63.4) |
| Cavalier King Charles Spaniel | 71 | 002:01 | 001:01:01 | - | 011:01 | 11/18 (61.1) |
| Yorkshire Terrier | 12 | 004:02 | 001:01:01 | - | 006:01 | 24/41 (58.5) |
| Husky | 77 | 060:02 | 001:01:04 | - | 040:01 | 7/12 (58.3) |
| Labrador Retriever | 21 | 508:01 | 001:01:03 | - | 012:01 | 21/39 (53.8) |
| Bulldog | 25 | 028:01 | - | 029:01 | 015:02 | 5/10 (50.0) |
| Miniature Pinscher | 63 | 004:02 | 001:01:01 | - | 015:01 | 6/12 (50.0) |
| ShihTzu | 46 | 014:01:02 | 001:05 | - | 025:01 | 14/28 (50.0) |
| Welsh Corgi | 69 | 054:01 | 002:02 | - | 012:01 | 15/33 (45.5) |
| Maltese | 7 | 012:01 | 001:01:01 | - | 015:01 | 11/26 (42.3) |
| Miniature Dachshund | 8 | 501:01 | 001:01:01 | - | 001:01 | 20/49 (40.8) |
| Pug | 110 | 058:01 | - | 024:03 | 010:011 | 8/21 (38.1) |
| Border Collie | 24 | 028:03 | - | 029:01 | 015:02 | 6/19 (31.6) |
| Toy Poodle | 37 | 006:01 | 001:01:01 | - | 015:01 | 13/44 (29.5) |
| Chihuahua | 30 | 508:01 | 001:01:03 | - | 015:01 | 10/39 (25.6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miyamae, J.; Okano, M.; Katakura, F.; Kulski, J.K.; Moritomo, T.; Shiina, T. Large-Scale Polymorphism Analysis of Dog Leukocyte Antigen Class I and Class II Genes (DLA-88, DLA-12/88L and DLA-DRB1) and Comparison of the Haplotype Diversity between Breeds in Japan. Cells 2023, 12, 809. https://doi.org/10.3390/cells12050809
Miyamae J, Okano M, Katakura F, Kulski JK, Moritomo T, Shiina T. Large-Scale Polymorphism Analysis of Dog Leukocyte Antigen Class I and Class II Genes (DLA-88, DLA-12/88L and DLA-DRB1) and Comparison of the Haplotype Diversity between Breeds in Japan. Cells. 2023; 12(5):809. https://doi.org/10.3390/cells12050809
Chicago/Turabian StyleMiyamae, Jiro, Masaharu Okano, Fumihiko Katakura, Jerzy K. Kulski, Tadaaki Moritomo, and Takashi Shiina. 2023. "Large-Scale Polymorphism Analysis of Dog Leukocyte Antigen Class I and Class II Genes (DLA-88, DLA-12/88L and DLA-DRB1) and Comparison of the Haplotype Diversity between Breeds in Japan" Cells 12, no. 5: 809. https://doi.org/10.3390/cells12050809
APA StyleMiyamae, J., Okano, M., Katakura, F., Kulski, J. K., Moritomo, T., & Shiina, T. (2023). Large-Scale Polymorphism Analysis of Dog Leukocyte Antigen Class I and Class II Genes (DLA-88, DLA-12/88L and DLA-DRB1) and Comparison of the Haplotype Diversity between Breeds in Japan. Cells, 12(5), 809. https://doi.org/10.3390/cells12050809








