SCD5 Regulation by VHL Affects Cell Proliferation and Lipid Homeostasis in ccRCC
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Plasmids
2.2. Antibodies
2.3. Cell Culture and Transfections
2.4. shRNA Mediated Knockdown (Stable Polyclonal Cell Lines)
2.5. Quantitative Real Time PCR
2.6. Gene Expression Analysis in C. elegans
2.7. RNA Sequencing and Data Analysis
2.8. Data Availability
2.9. Cell Viability Assay
2.10. C. elegans Growth Conditions
2.11. Microscopy and Quantification of FAT-7::GFP Expression
2.12. Hypoxia Treatment
2.13. Metabolomics Analysis
2.14. Statistical Analyses
3. Results
3.1. SCD5 Expression in ccRCC
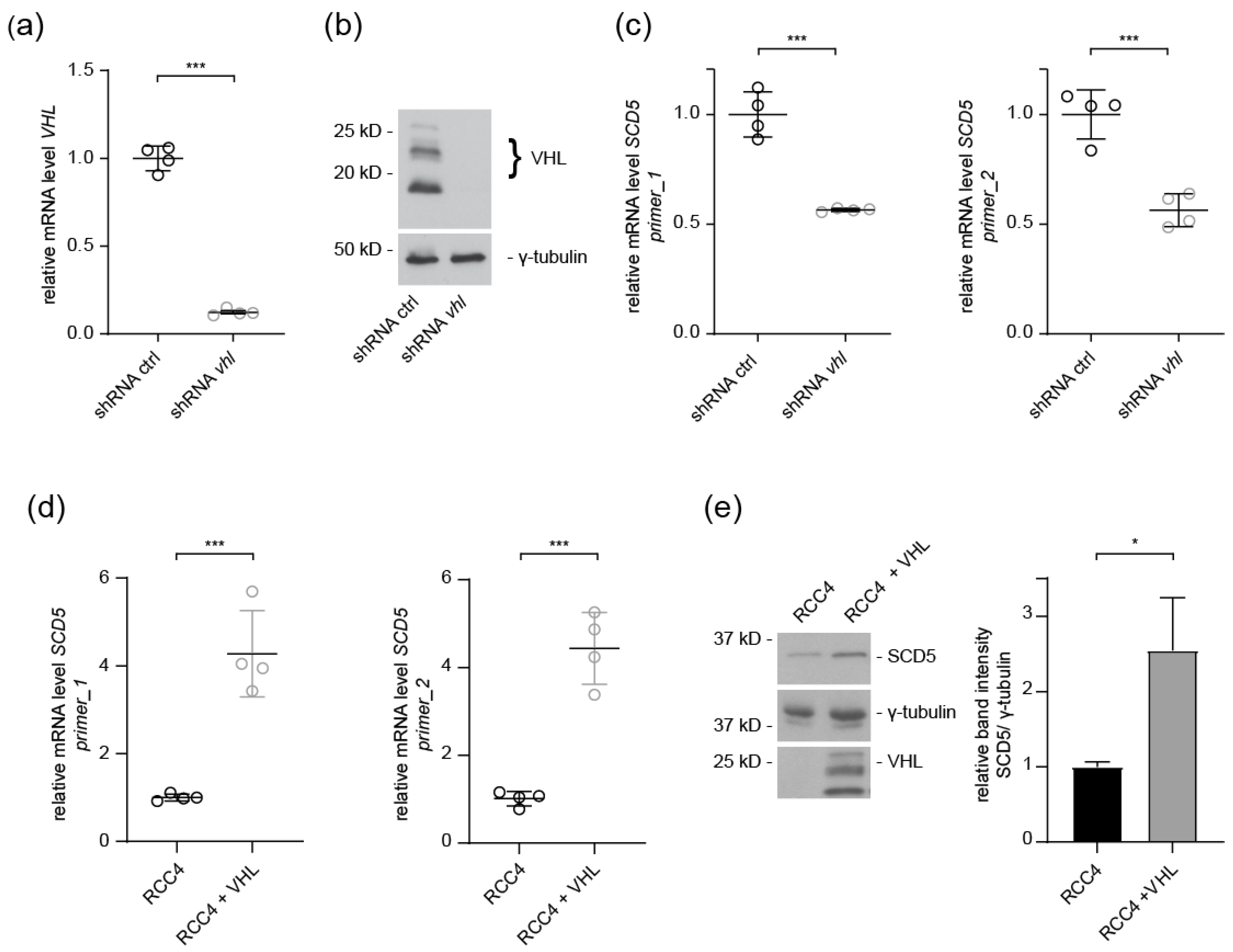
3.2. SCD5 Expression Depends on VHL
3.3. VHL Regulates SCD5 Expression in C. elegans
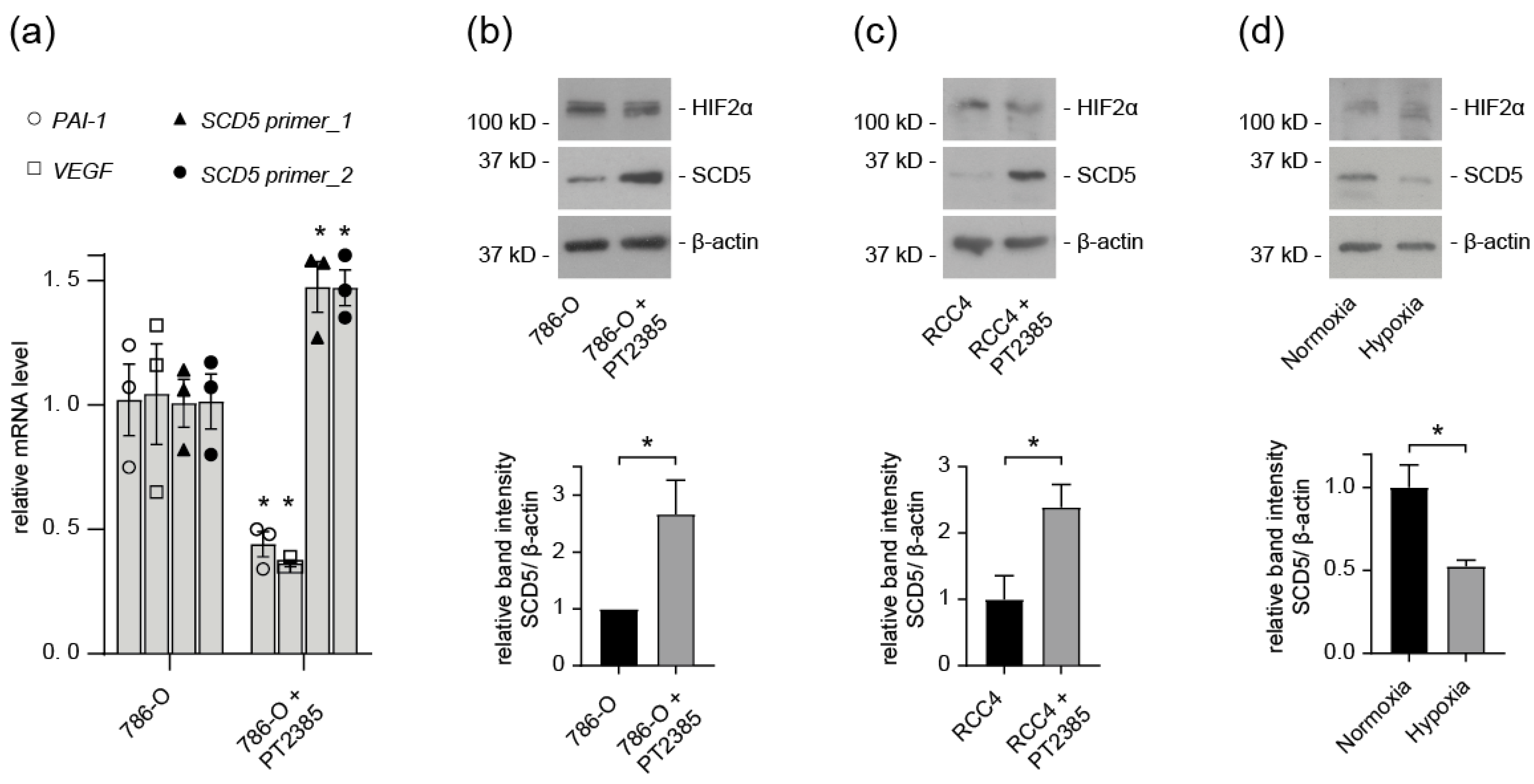
3.4. SCD5 Expression in ccRCC Cells Depends on HIF2
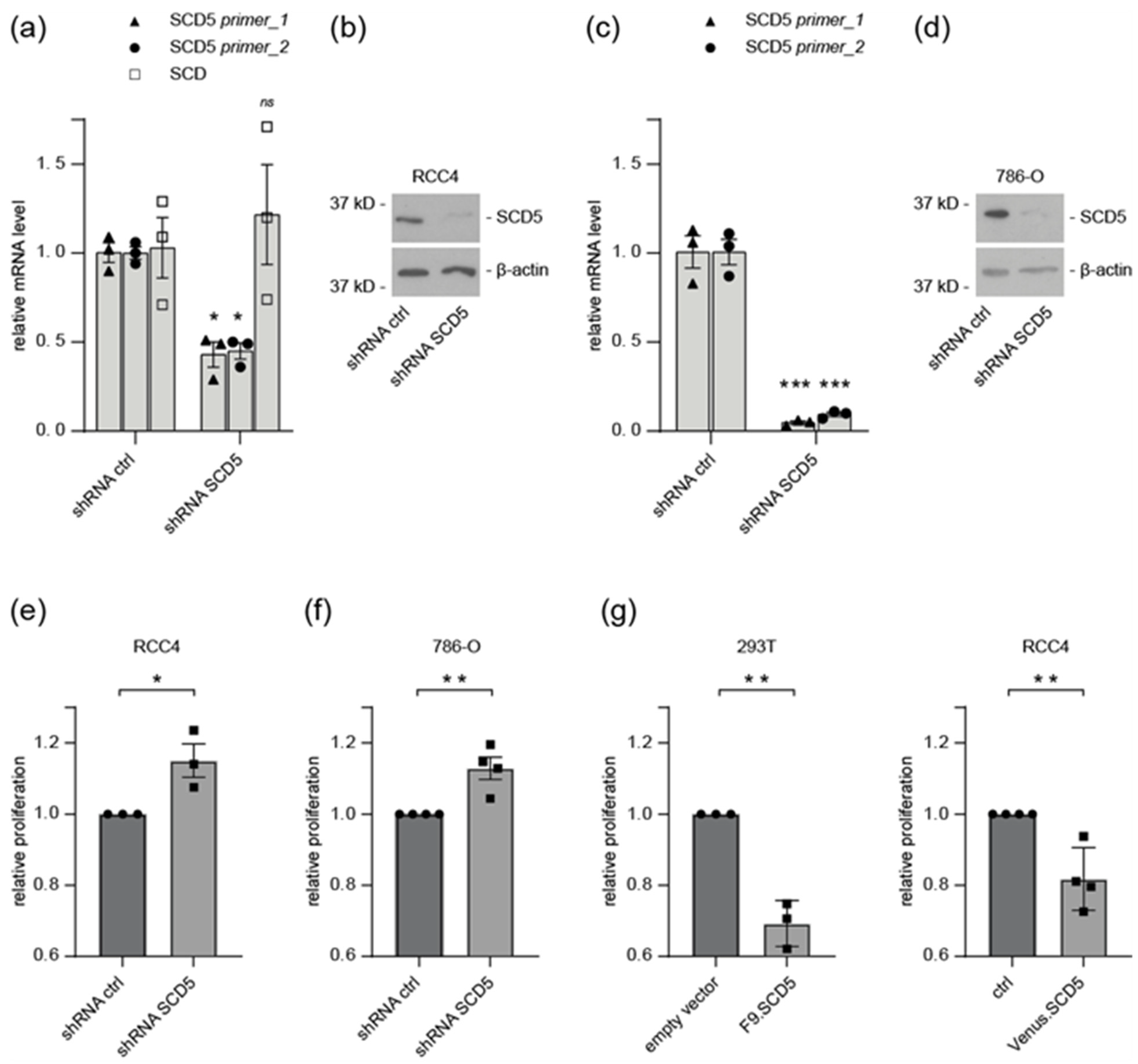
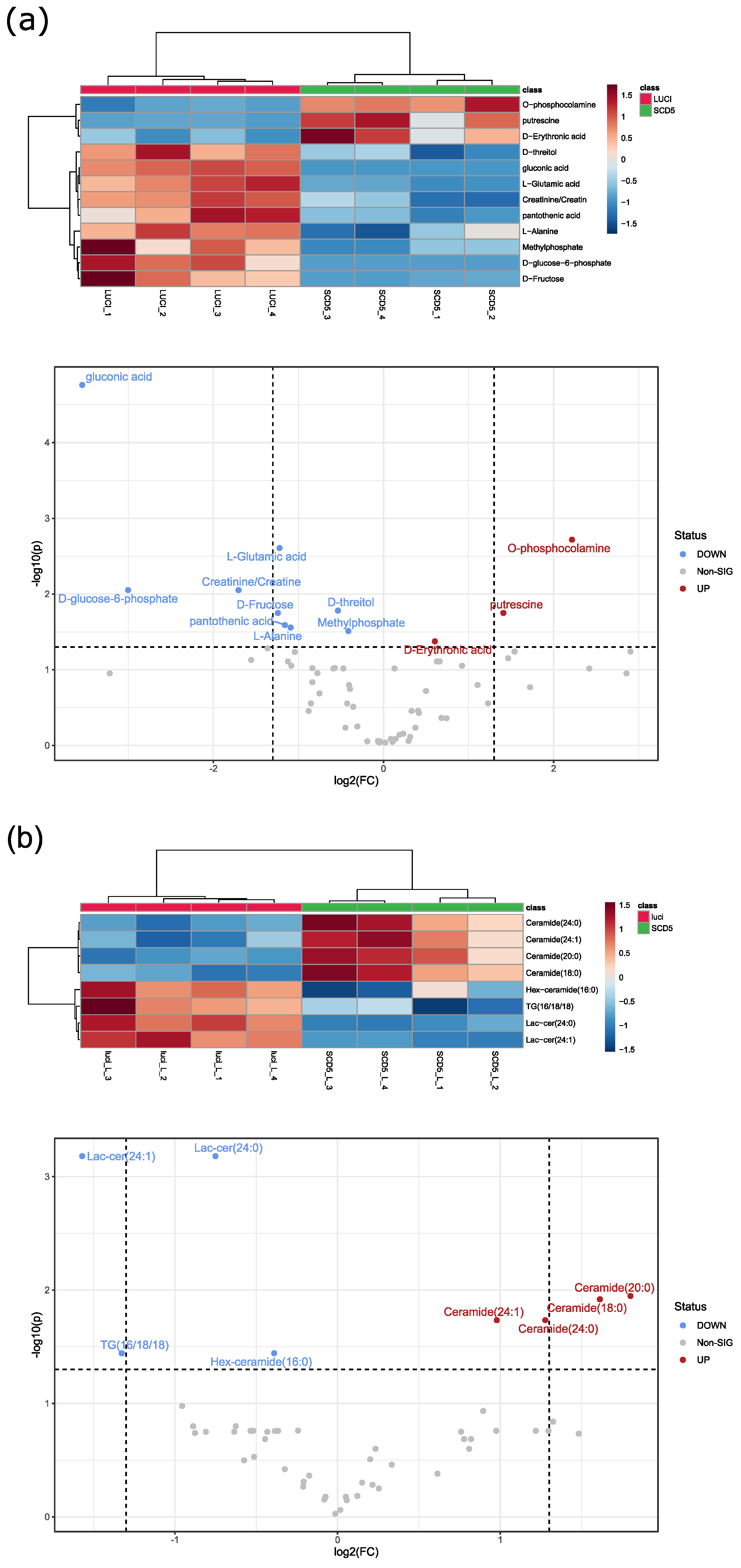
3.5. SCD5 Affects Cell Physiology and Metabolism
4. Discussion
4.1. Regulation of SCD5 Expression
4.2. Role of SCD5 in Metabolism and Cell Proliferation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Epstein, A.C.; Gleadle, J.M.; McNeill, L.A.; Hewitson, K.S.; O’Rourke, J.; Mole, D.R.; Mukherji, M.; Metzen, E.; Wilson, M.I.; Dhanda, A.; et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 2001, 107, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Frew, I.J.; Moch, H. A clearer view of the molecular complexity of clear cell renal cell carcinoma. Annu. Rev. Pathol. 2015, 10, 263–289. [Google Scholar] [CrossRef]
- Rohrig, F.; Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 2016, 16, 732–749. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Lu, Y.; Song, J.; Huang, M.; Zhang, J.; Huang, Y. The role of stearoyl-coenzyme A desaturase 1 in clear cell renal cell carcinoma. Tumour Biol. 2016, 37, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Angelucci, C.; D’Alessio, A.; Iacopino, F.; Proietti, G.; Di Leone, A.; Masetti, R.; Sica, G. Pivotal role of human stearoyl-CoA desaturases (SCD1 and 5) in breast cancer progression: Oleic acid-based effect of SCD1 on cell migration and a novel pro-cell survival role for SCD5. Oncotarget 2018, 9, 24364–24380. [Google Scholar] [CrossRef]
- Wang, J.; Xu, Y.; Zhu, L.; Zou, Y.; Kong, W.; Dong, B.; Huang, J.; Chen, Y.; Xue, W.; Huang, Y.; et al. High Expression of Stearoyl-CoA Desaturase 1 Predicts Poor Prognosis in Patients with Clear-Cell Renal Cell Carcinoma. PLoS ONE 2016, 11, e0166231. [Google Scholar] [CrossRef]
- Mason, P.; Liang, B.; Li, L.; Fremgen, T.; Murphy, E.; Quinn, A.; Madden, S.L.; Biemann, H.P.; Wang, B.; Cohen, A.; et al. SCD1 inhibition causes cancer cell death by depleting mono-unsaturated fatty acids. PLoS ONE 2012, 7, e33823. [Google Scholar] [CrossRef]
- Melana, J.P.; Mignolli, F.; Stoyanoff, T.; Aguirre, M.V.; Balboa, M.A.; Balsinde, J.; Rodriguez, J.P. The Hypoxic Microenvironment Induces Stearoyl-CoA Desaturase-1 Overexpression and Lipidomic Profile Changes in Clear Cell Renal Cell Carcinoma. Cancers 2021, 13, 2962. [Google Scholar] [CrossRef]
- Zhao, W.; Sun, L.; Li, X.; Wang, J.; Zhu, Y.; Jia, Y.; Tong, Z. SCD5 expression correlates with prognosis and response to neoadjuvant chemotherapy in breast cancer. Sci. Rep. 2021, 11, 8976. [Google Scholar] [CrossRef]
- Wang, J.; Yu, L.; Schmidt, R.E.; Su, C.; Huang, X.; Gould, K.; Cao, G. Characterization of HSCD5, a novel human stearoyl-CoA desaturase unique to primates. Biochem. Biophys. Res. Commun. 2005, 332, 735–742. [Google Scholar] [CrossRef]
- Bellenghi, M.; Puglisi, R.; Pedini, F.; De Feo, A.; Felicetti, F.; Bottero, L.; Sangaletti, S.; Errico, M.C.; Petrini, M.; Gesumundo, C.; et al. SCD5-induced oleic acid production reduces melanoma malignancy by intracellular retention of SPARC and cathepsin B. J. Pathol. 2015, 236, 315–325. [Google Scholar] [CrossRef]
- Puglisi, R.; Bellenghi, M.; Pontecorvi, G.; Gulino, A.; Petrini, M.; Felicetti, F.; Bottero, L.; Mattia, G.; Care, A. SCD5 restored expression favors differentiation and epithelial-mesenchymal reversion in advanced melanoma. Oncotarget 2018, 9, 7567–7581. [Google Scholar] [CrossRef]
- Ganner, A.; Gehrke, C.; Klein, M.; Thegtmeier, L.; Matulenski, T.; Wingendorf, L.; Wang, L.; Pilz, F.; Greidl, L.; Meid, L.; et al. VHL suppresses RAPTOR and inhibits mTORC1 signaling in clear cell renal cell carcinoma. Sci. Rep. 2021, 11, 14827. [Google Scholar] [CrossRef]
- Thoma, C.R.; Frew, I.J.; Hoerner, C.R.; Montani, M.; Moch, H.; Krek, W. pVHL and GSK3beta are components of a primary cilium-maintenance signalling network. Nat. Cell Biol. 2007, 9, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Wiznerowicz, M.; Trono, D. Conditional suppression of cellular genes: Lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 2003, 77, 8957–8961. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; van den Beek, M.; Blankenberg, D.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef] [PubMed]
- Lagies, S.; Pichler, R.; Vladimirov, G.; Gawron, J.; Bazner, F.; Schreiner, A.; Kadena, D.; Plattner, D.A.; Lienkamp, S.S.; Kammerer, B. Metabolic and Lipidomic Assessment of Kidney Cells Exposed to Nephrotoxic Vancomycin Dosages. Int. J. Mol. Sci. 2021, 22, 10111. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.E.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Chen, F.; Chandrashekar, D.S.; Varambally, S.; Creighton, C.J. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 2019, 10, 5679. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Ganner, A.; Neumann-Haefelin, E. Genetic kidney diseases: Caenorhabditis elegans as model system. Cell Tissue Res. 2017, 369, 105–118. [Google Scholar] [CrossRef]
- Gossage, L.; Eisen, T.; Maher, E.R. VHL, the story of a tumour suppressor gene. Nat. Rev. Cancer 2015, 15, 55–64. [Google Scholar] [CrossRef]
- Mlynarczyk, G.; Miklosz, A.; Suchanski, J.; Reza, S.; Romanowicz, L.; Sobolewski, K.; Chabowski, A.; Baranowski, M. Grade-dependent changes in sphingolipid metabolism in clear cell renal cell carcinoma. J. Cell. Biochem. 2022, 123, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Igal, R.A.; Sinner, D.I. Stearoyl-CoA desaturase 5 (SCD5), a Delta-9 fatty acyl desaturase in search of a function. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158840. [Google Scholar] [CrossRef]
- Keith, B.; Johnson, R.S.; Simon, M.C. HIF1alpha and HIF2alpha: Sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer 2011, 12, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Mahon, P.C.; Hirota, K.; Semenza, G.L. FIH-1: A novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001, 15, 2675–2686. [Google Scholar] [CrossRef]
- Yun, Z.; Maecker, H.L.; Johnson, R.S.; Giaccia, A.J. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: A mechanism for regulation of adipogenesis by hypoxia. Dev. Cell 2002, 2, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Igal, R.A. Stearoyl CoA desaturase-1: New insights into a central regulator of cancer metabolism. Biochim. Biophys. Acta 2016, 1861, 1865–1880. [Google Scholar] [CrossRef]
- Bellenghi, M.; Talarico, G.; Botti, L.; Puglisi, R.; Tabolacci, C.; Portararo, P.; Piva, A.; Pontecorvi, G.; Care, A.; Colombo, M.P.; et al. SCD5-dependent inhibition of SPARC secretion hampers metastatic spreading and favors host immunity in a TNBC murine model. Oncogene 2022, 41, 4055–4065. [Google Scholar] [CrossRef]
- Peck, B.; Schulze, A. Lipid desaturation—The next step in targeting lipogenesis in cancer? FEBS J. 2016, 283, 2767–2778. [Google Scholar] [CrossRef]
- Ackerman, D.; Tumanov, S.; Qiu, B.; Michalopoulou, E.; Spata, M.; Azzam, A.; Xie, H.; Simon, M.C.; Kamphorst, J.J. Triglycerides Promote Lipid Homeostasis during Hypoxic Stress by Balancing Fatty Acid Saturation. Cell Rep. 2018, 24, 2596–2605.e2595. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wei, X.; Feng, H.; Hu, B.; Liu, B.; Luan, Y.; Ruan, Y.; Liu, X.; Liu, Z.; Wang, S.; et al. An eleven metabolic gene signature-based prognostic model for clear cell renal cell carcinoma. Aging 2020, 12, 23165–23186. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Liu, Y.D.; Su, J.; Yuan, D.; Sun, F.; Zhu, J. Systematic analysis of alternative splicing signature unveils prognostic predictor for kidney renal clear cell carcinoma. J. Cell. Physiol. 2019, 234, 22753–22764. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganner, A.; Philipp, A.; Lagies, S.; Wingendorf, L.; Wang, L.; Pilz, F.; Welte, T.; Grand, K.; Lienkamp, S.S.; Klein, M.; et al. SCD5 Regulation by VHL Affects Cell Proliferation and Lipid Homeostasis in ccRCC. Cells 2023, 12, 835. https://doi.org/10.3390/cells12060835
Ganner A, Philipp A, Lagies S, Wingendorf L, Wang L, Pilz F, Welte T, Grand K, Lienkamp SS, Klein M, et al. SCD5 Regulation by VHL Affects Cell Proliferation and Lipid Homeostasis in ccRCC. Cells. 2023; 12(6):835. https://doi.org/10.3390/cells12060835
Chicago/Turabian StyleGanner, Athina, Antonia Philipp, Simon Lagies, Laura Wingendorf, Lu Wang, Felicitas Pilz, Thomas Welte, Kelli Grand, Soeren S. Lienkamp, Marinella Klein, and et al. 2023. "SCD5 Regulation by VHL Affects Cell Proliferation and Lipid Homeostasis in ccRCC" Cells 12, no. 6: 835. https://doi.org/10.3390/cells12060835
APA StyleGanner, A., Philipp, A., Lagies, S., Wingendorf, L., Wang, L., Pilz, F., Welte, T., Grand, K., Lienkamp, S. S., Klein, M., Kammerer, B., Frew, I. J., Walz, G., & Neumann-Haefelin, E. (2023). SCD5 Regulation by VHL Affects Cell Proliferation and Lipid Homeostasis in ccRCC. Cells, 12(6), 835. https://doi.org/10.3390/cells12060835





