B7H4 Expression Is More Frequent in MSS Status Colorectal Cancer and Is Negatively Associated with Tumour Infiltrating Lymphocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Characteristics of the Patient Group
2.2. Evaluation of the B7H4 Expression by ELISA
2.3. Evaluation of the B7H4 Expression by IHC
2.4. Assessment of the MSI/MSS Status
2.5. Assessment of the Tumour-Infiltrating CD8+ T Cells
2.6. Assessment of the TILs and Budding
2.7. Assessment of the Cytokines Screening Panel
2.8. Exploration of Biological Characteristics of B7H4
2.9. Statistical Analysis
3. Results
3.1. The Expression of B7H4 Is Upregulated in Tumour Tissues
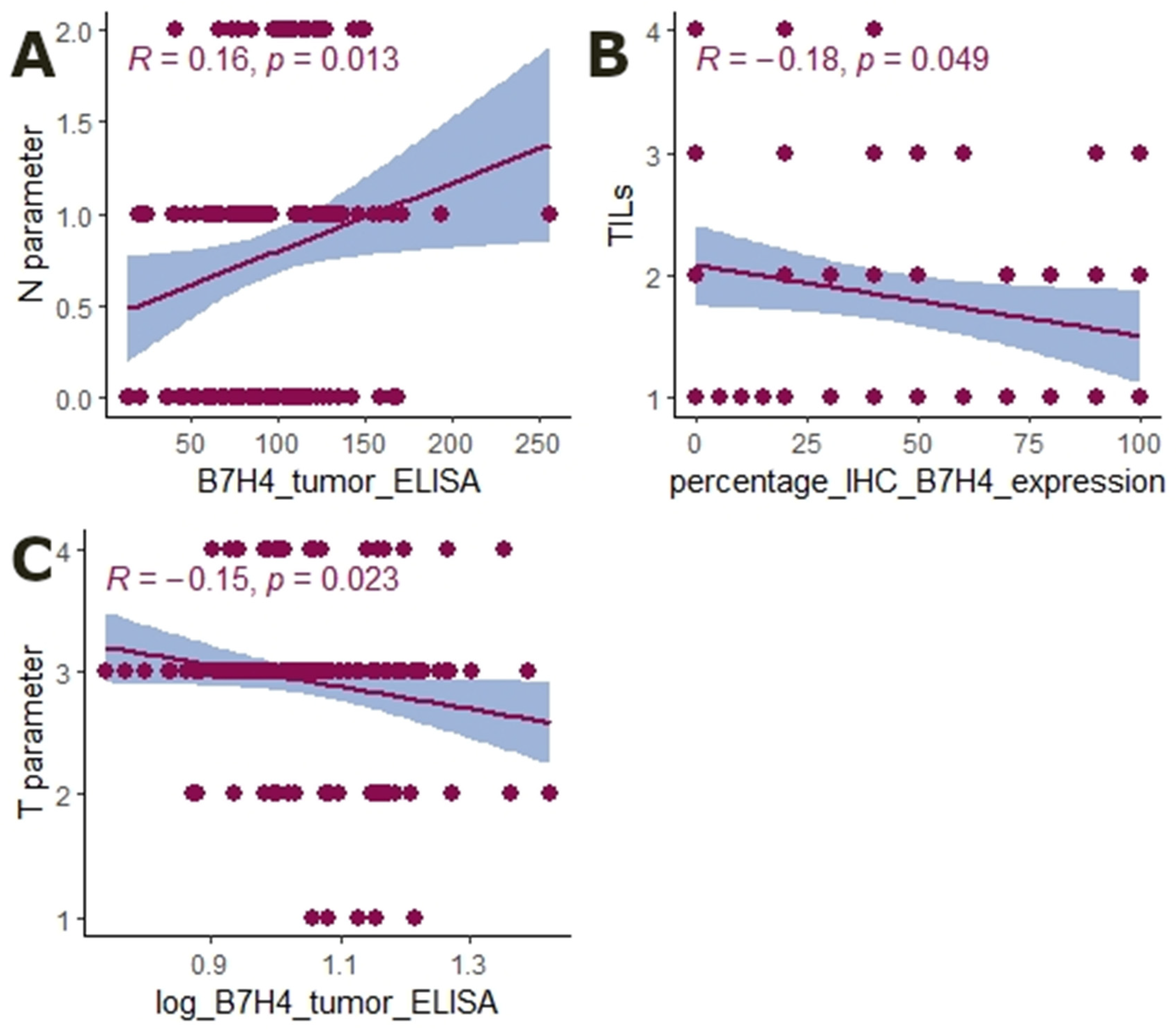
3.2. The Expression of B7H4 Is Associated with the MSS Status of the Tumour
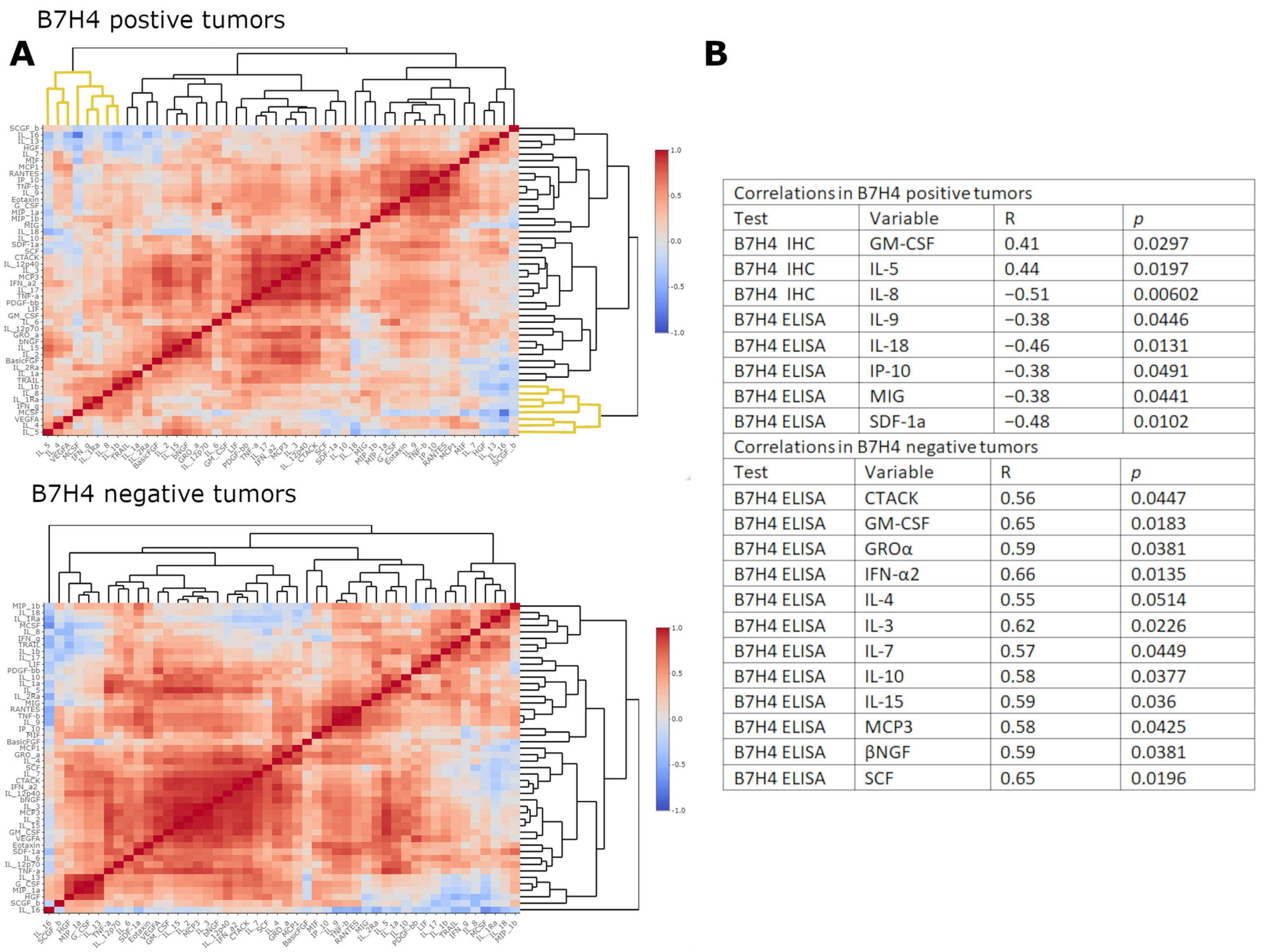
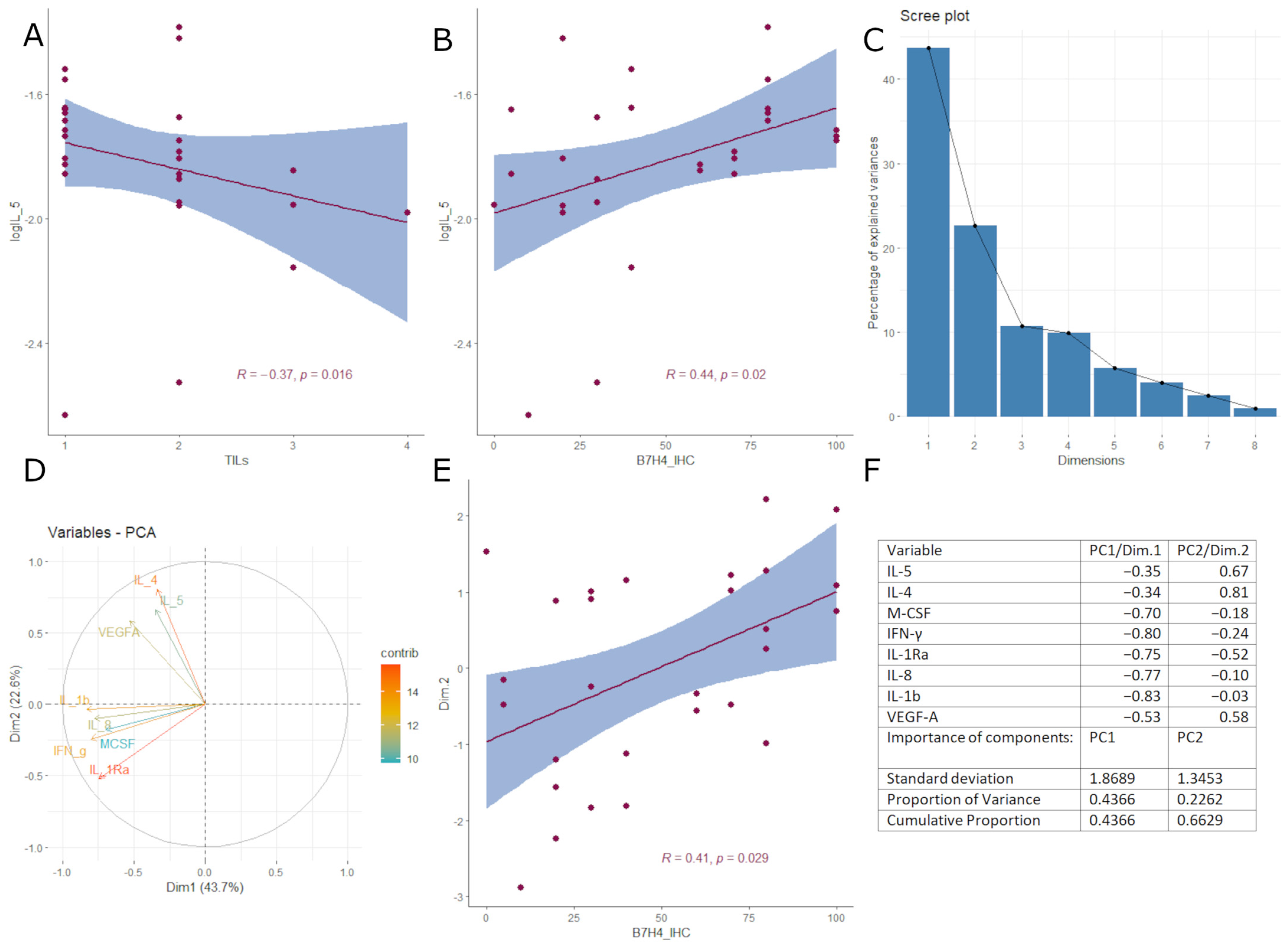
3.3. B7H4 Expression Is Negatively Associated with Pro-Inflammatory Cytokines
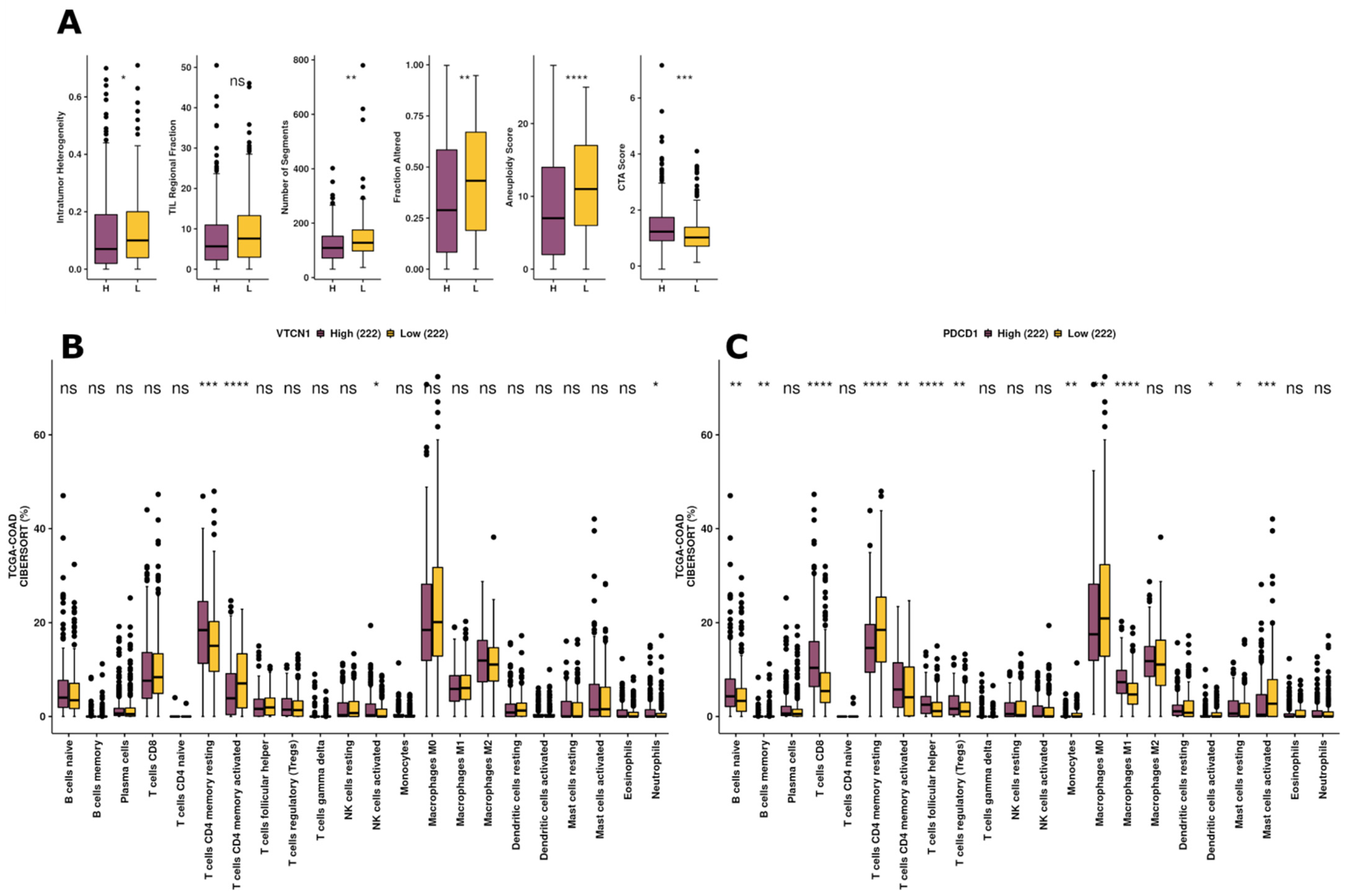
3.4. Exploration of Immunogenicity and Immune Infiltration Landscape of B7H4 Based on TCGA-COAD Data
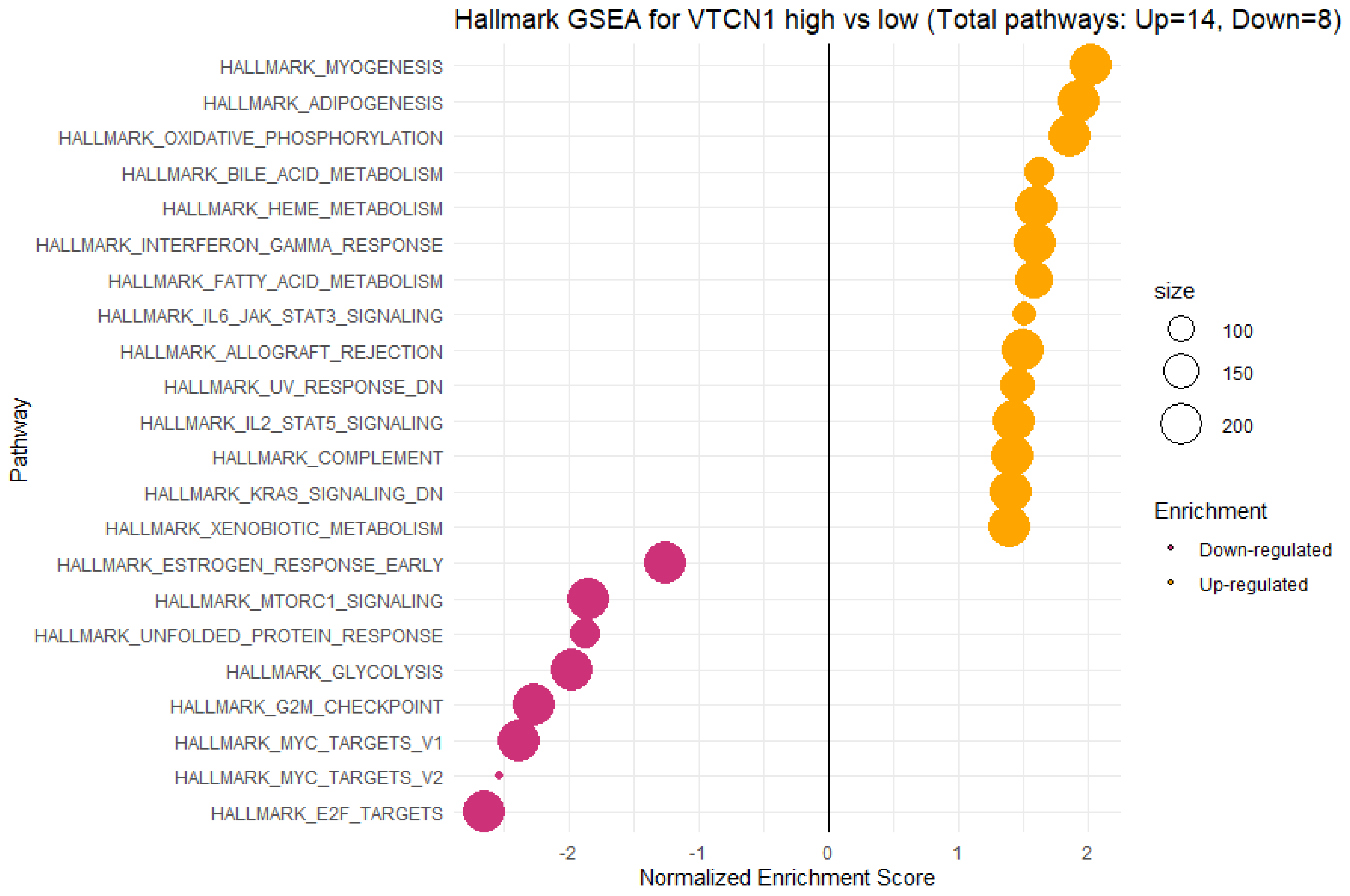
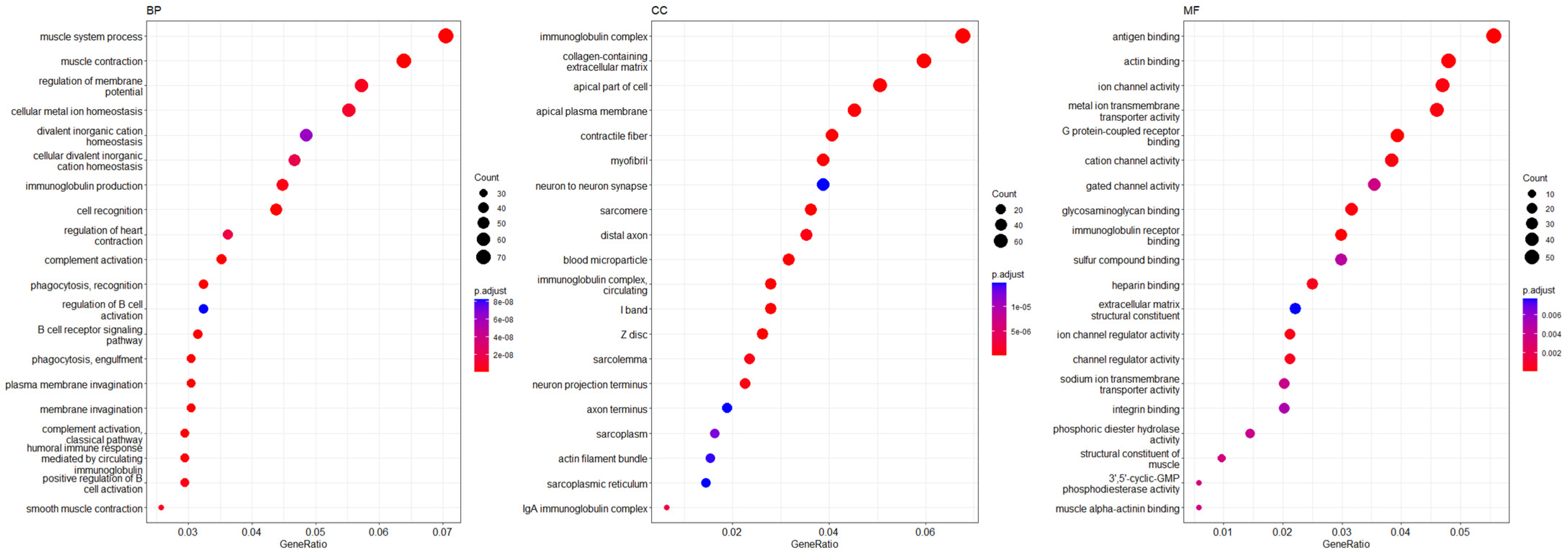
3.5. Functional Annotations and Predicted Signalling Pathways for VTCN1 (B7H4) Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sica, G.L.; Choi, I.H.; Zhu, G.; Tamada, K.; Wang, S.D.; Tamura, H.; Chapoval, A.I.; Flies, D.B.; Bajorath, J.; Chen, L. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity 2003, 18, 849–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaishnav, J.; Khan, F.; Yadav, M.; Parmar, N.; Buch, H.; Jadeja, S.D.; Dwivedi, M.; Begum, R. V-set domain containing T-cell activation inhibitor-1 (VTCN1): A potential target for the treatment of autoimmune diseases. Immunobiology 2022, 227, 152274. [Google Scholar] [CrossRef] [PubMed]
- Schildberg, F.A.; Klein, S.R.; Freeman, G.J.; Sharpe, A.H. Coinhibitory Pathways in the B7-CD28 Ligand-Receptor Family. Immunity 2016, 44, 955–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janakiram, M.; Shah, U.A.; Liu, W.; Zhao, A.; Schoenberg, M.P.; Zang, X. The third group of the B7-CD28 immune checkpoint family: HHLA2, TMIGD2, B7x, and B7-H3. Immunol. Rev. 2017, 276, 26–39. [Google Scholar] [CrossRef] [Green Version]
- Carreno, B.M.; Collins, M. The B7 family of ligands and its receptors: New pathways for costimulation and inhibition of immune responses. Annu. Rev. Immunol. 2002, 20, 29–53. [Google Scholar] [CrossRef]
- Collins, M.; Ling, V.; Carreno, B.M. The B7 family of immune-regulatory ligands. Genome Biol. 2005, 6, 223. [Google Scholar] [CrossRef] [Green Version]
- Ni, L.; Dong, C. New B7 Family Checkpoints in Human Cancers. Mol. Cancer Ther. 2017, 16, 1203–1211. [Google Scholar] [CrossRef] [Green Version]
- Wojtukiewicz, M.Z.; Rek, M.M.; Karpowicz, K.; Górska, M.; Polityńska, B.; Wojtukiewicz, A.M.; Moniuszko, M.; Radziwon, P.; Tucker, S.C.; Honn, K.V. Inhibitors of immune checkpoints-PD-1, PD-L1, CTLA-4-new opportunities for cancer patients and a new challenge for internists and general practitioners. Cancer Metastasis Rev. 2021, 40, 949–982. [Google Scholar] [CrossRef]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087.e3. [Google Scholar] [CrossRef]
- Pyo, J.; Park, H.-J. Treatment Efficacy of Immune Checkpoint Inhibitors for Patients with Advanced or Metastatic Colorectal Cancer: A Systematic Review and Meta-Analysis. J. Clin. Med. 2021, 10, 3599. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.-J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Overman, M.J.; Lonardi, S.; Wong, K.Y.M.; Lenz, H.-J.; Gelsomino, F.; Aglietta, M.; Morse, M.A.; van Cutsem, E.; McDermott, R.; Hill, A.; et al. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. J. Clin. Oncol. 2018, 36, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Sillo, T.O.; Beggs, A.D.; Morton, D.G.; Middleton, G. Mechanisms of immunogenicity in colorectal cancer. Br. J. Surg. 2019, 106, 1283–1297. [Google Scholar] [CrossRef] [Green Version]
- Llosa, N.J.; Cruise, M.; Tam, A.; Wicks, E.C.; Hechenbleikner, E.M.; Taube, J.M.; Blosser, R.L.; Fan, H.; Wang, H.; Luber, B.S.; et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015, 5, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Arrichiello, G.; Poliero, L.; Borrelli, C.; Paragliola, F.; Nacca, V.; Napolitano, S.; Della Corte, C.M.; Martini, G.; Martinelli, E. Immunotherapy in colorectal cancer: Is the long-awaited revolution finally happening? Cancer Treat. Res. Commun. 2021, 28, 100442. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef] [Green Version]
- Schürch, C.M.; Bhate, S.S.; Barlow, G.L.; Phillips, D.J.; Noti, L.; Zlobec, I.; Chu, P.; Black, S.; Demeter, J.; McIlwain, D.R.; et al. Coordinated Cellular Neighborhoods Orchestrate Antitumoral Immunity at the Colorectal Cancer Invasive Front. Cell 2020, 182, 1341–1359.e19. [Google Scholar] [CrossRef]
- Wong, H.S.-C.; Chang, C.-M.; Liu, X.; Huang, W.-C.; Chang, W.-C. Characterization of cytokinome landscape for clinical responses in human cancers. Oncoimmunology 2016, 5, e1214789. [Google Scholar] [CrossRef] [Green Version]
- Czajka-Francuz, P.; Cisoń-Jurek, S.; Czajka, A.; Kozaczka, M.; Wojnar, J.; Chudek, J.; Francuz, T. Systemic Interleukins’ Profile in Early and Advanced Colorectal Cancer. Int. J. Mol. Sci. 2021, 23, 124. [Google Scholar] [CrossRef]
- Kula, A.; Dawidowicz, M.; Mielcarska, S.; Kiczmer, P.; Chrabańska, M.; Rynkiewicz, M.; Świętochowska, E.; Waniczek, D. Periostin in Angiogenesis and Inflammation in CRC-A Preliminary Observational Study. Medicina 2022, 58, 96. [Google Scholar] [CrossRef]
- Olave, M.C.; Graham, R.P. Mismatch repair deficiency: The what, how and why it is important. Genes Chromosom. Cancer 2022, 61, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Grudzińska, E.; Grzegorczyn, S.; Czuba, Z.P. Chemokines and Growth Factors Produced by Lymphocytes in the Incompetent Great Saphenous Vein. Mediat. Inflamm. 2019, 2019, 7057303. [Google Scholar] [CrossRef] [Green Version]
- Idzik, M.; Poloczek, J.; Skrzep-Poloczek, B.; Dróżdż, E.; Chełmecka, E.; Czuba, Z.; Jochem, J.; Stygar, D. The Effects of 21-Day General Rehabilitation after Hip or Knee Surgical Implantation on Plasma Levels of Selected Interleukins, VEGF, TNF-α, PDGF-BB, and Eotaxin-1. Biomolecules 2022, 12, 605. [Google Scholar] [CrossRef]
- Dampier, C.H.; Devall, M.; Jennelle, L.T.; Díez-Obrero, V.; Plummer, S.J.; Moreno, V.; Casey, G. Oncogenic Features in Histologically Normal Mucosa: Novel Insights Into Field Effect From a Mega-Analysis of Colorectal Transcriptomes. Clin. Transl. Gastroenterol. 2020, 11, e00210. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Li, J.; Chen, Y.; Que, Z.; Du, J.; Zhang, J. B7 Family Members in Pancreatic Ductal Adenocarcinoma: Attractive Targets for Cancer Immunotherapy. Int. J. Mol. Sci. 2022, 23, 15005. [Google Scholar] [CrossRef]
- Smith, J.B.; Lanitis, E.; Dangaj, D.; Buza, E.; Poussin, M.; Stashwick, C.; Scholler, N.; Powell, D.J., Jr. Tumor Regression and Delayed Onset Toxicity Following B7-H4 CAR T Cell Therapy. Mol. Ther. 2016, 24, 1987–1999. [Google Scholar] [CrossRef] [Green Version]
- John, P.; Wei, Y.; Liu, W.; Du, M.; Guan, F.; Zang, X. The B7x Immune Checkpoint Pathway: From Discovery to Clinical Trial. Trends Pharmacol. Sci. 2019, 40, 883–896. [Google Scholar] [CrossRef]
- Yan, X.; Hong, B.; Feng, J.; Jin, Y.; Chen, M.; Li, F.; Qian, Y. B7-H4 is a potential diagnostic and prognostic biomarker in colorectal cancer and correlates with the epithelial-mesenchymal transition. BMC Cancer 2022, 22, 1053. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Shi, L.; Yang, J.; Wang, H.; Yang, H.; Wang, Q. B7 family member H4 induces epithelial-mesenchymal transition and promotes the proliferation, migration and invasion of colorectal cancer cells. Bioengineered 2022, 13, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Lv, X.; Liu, Z.; Zhan, S.; Xu, Y.; Zhang, X.; Liu, C.; Cao, L. Overexpression of B7-H4 is associated with infiltrating immune cells and poor prognosis in metastatic colorectal cancer. Int. Immunopharmacol. 2021, 90, 107144. [Google Scholar] [CrossRef]
- Li, L.; Ouyang, Y.; Wang, W.; Hou, D.; Zhu, Y. The landscape and prognostic value of tumor-infiltrating immune cells in gastric cancer. PeerJ 2019, 7, e7993. [Google Scholar] [CrossRef]
- Detoc, M.; Bruel, S.; Frappe, P.; Tardy, B.; Botelho-Nevers, E.; Gagneux-Brunon, A. Intention to participate in a COVID-19 vaccine clinical trial and to get vaccinated against COVID-19 in France during the pandemic. Vaccine 2020, 38, 7002–7006. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Guo, H.; Wang, Y.; Liu, B.; Zhou, H. Profiles of tumor-infiltrating immune cells and prognostic genes associated with the microenvironment of bladder cancer. Int. Immunopharmacol. 2020, 85, 106641. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Yu, W.; He, J.; Liu, W.; Yang, J.; Lin, X.; Zhang, Y.; Wang, X.; Jiang, W.; Luo, J.; et al. Reprogramming immunosuppressive myeloid cells facilitates immunotherapy for colorectal cancer. EMBO Mol. Med. 2021, 13, e12798. [Google Scholar] [CrossRef]
- Cheng, H.; Borczuk, A.; Janakiram, M.; Ren, X.; Lin, J.; Assal, A.; Halmos, B.; Perez-Soler, R.; Zang, X. Wide Expression and Significance of Alternative Immune Checkpoint Molecules, B7x and HHLA2, in PD-L1-Negative Human Lung Cancers. Clin. Cancer Res. 2018, 24, 1954–1964. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Cao, Y.; Zhang, S.; Gao, F. Association between low expression levels of interleukin-9 and colon cancer progression. Exp. Ther. Med. 2015, 10, 942–946. [Google Scholar] [CrossRef] [Green Version]
- Noelle, R.J.; Nowak, E.C. Cellular sources and immune functions of interleukin-9. Nat. Rev. Immunol. 2010, 10, 683–687. [Google Scholar] [CrossRef] [Green Version]
- Nonomura, Y.; Otsuka, A.; Nakashima, C.; Seidel, J.A.; Kitoh, A.; Dainichi, T.; Nakajima, S.; Sawada, Y.; Matsushita, S.; Aoki, M.; et al. Peripheral blood Th9 cells are a possible pharmacodynamic biomarker of nivolumab treatment efficacy in metastatic melanoma patients. Oncoimmunology 2016, 5, e1248327. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, M.; Zhao, H.; Huang, Y.; Li, D.; Mao, D.; Zhang, Z.; Zhu, X.; Dong, X.; Zhao, X. IL-9 Exerts Antitumor Effects in Colon Cancer and Transforms the Tumor Microenvironment In Vivo. Technol. Cancer Res. Treat. 2019, 18, 1533033819857737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Zhang, Z.; Sun, P.; Song, G.; Wang, L.; Sun, Z.; Yuan, N.; Wang, Q.; Lun, L. Interleukin-18 Is a Prognostic Marker and Plays a Tumor Suppressive Role in Colon Cancer. Dis. Mrk. 2020, 2020, 6439614. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Yin, H.; Han, C.; Mao, Q.; Tang, J.; Ji, Z.; Yan, X.; Wang, L.; Liu, S.; Ai, C. The role of CXCL10 in prognosis of patients with colon cancer and tumor microenvironment remodeling. Medicine 2021, 100, e27224. [Google Scholar] [CrossRef] [PubMed]
- Goïta, A.A.; Guenot, D. Colorectal Cancer: The Contribution of CXCL12 and Its Receptors CXCR4 and CXCR7. Cancers 2022, 14, 1810. [Google Scholar] [CrossRef]
- Pelaia, C.; Paoletti, G.; Puggioni, F.; Racca, F.; Pelaia, G.; Canonica, G.W.; Heffler, E. Interleukin-5 in the Pathophysiology of Severe Asthma. Front. Physiol. 2019, 10, 1514. [Google Scholar] [CrossRef]
- Blomberg, O.S.; Spagnuolo, L.; Garner, H.; Voorwerk, L.; Isaeva, O.I.; van Dyk, E.; Bakker, N.; Chalabi, M.; Klaver, C.; Duijst, M.; et al. IL-5-producing CD4(+) T cells and eosinophils cooperate to enhance response to immune checkpoint blockade in breast cancer. Cancer Cell 2023, 41, 106–123.e10. [Google Scholar] [CrossRef]
- Zhou, Q.; Peng, R.-Q.; Wu, X.-J.; Xia, Q.; Hou, J.-H.; Ding, Y.; Zhou, Q.-M.; Zhang, X.; Pang, Z.-Z.; Wan, D.-S.; et al. The density of macrophages in the invasive front is inversely correlated to liver metastasis in colon cancer. J. Transl. Med. 2010, 8, 13. [Google Scholar] [CrossRef] [Green Version]
- Lin, A.; Zhang, J.; Luo, P. Crosstalk Between the MSI Status and Tumor Microenvironment in Colorectal Cancer. Front. Immunol. 2020, 11, 2039. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Sobral, D.; Martins, M.; Kaplan, S.; Golkaram, M.; Salmans, M.; Khan, N.; Vijayaraghavan, R.; Casimiro, S.; Fernandes, A.; Borralho, P.; et al. Genetic and microenvironmental intra-tumor heterogeneity impacts colorectal cancer evolution and metastatic development. Commun. Biol. 2022, 5, 937. [Google Scholar] [CrossRef] [PubMed]
- Zang, X.; Loke, P.n.; Kim, J.; Murphy, K.; Waitz, R.; Allison, J.P. B7x: A widely expressed B7 family member that inhibits T cell activation. Proc. Natl. Acad. Sci. USA 2003, 100, 10388–10392. [Google Scholar] [CrossRef] [Green Version]
- Prasad, D.V.R.; Richards, S.; Mai, X.M.; Dong, C. B7S1, a novel B7 family member that negatively regulates T cell activation. Immunity 2003, 18, 863–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Lee, Y.; Li, Y.; Jiang, Y.; Lu, H.; Zang, W.; Zhao, X.; Liu, L.; Chen, Y.; Tan, H.; et al. Co-inhibitory Molecule B7 Superfamily Member 1 Expressed by Tumor-Infiltrating Myeloid Cells Induces Dysfunction of Anti-tumor CD8(+) T Cells. Immunity 2018, 48, 773–786.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abadi, Y.M.; Jeon, H.; Ohaegbulam, K.C.; Scandiuzzi, L.; Ghosh, K.; Hofmeyer, K.A.; Lee, J.S.; Ray, A.; Gravekamp, C.; Zang, X. Host b7x promotes pulmonary metastasis of breast cancer. J. Immunol. 2013, 190, 3806–3814. [Google Scholar] [CrossRef] [Green Version]
- Ohaegbulam, K.C.; Liu, W.; Jeon, H.; Almo, S.C.; Zang, X. Tumor-expressed immune checkpoint B7x promotes cancer progression and antigen-specific CD8 T cell exhaustion and suppressive innate immune cells. Oncotarget 2017, 8, 82740–82753. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; He, Z.; Ye, J.; Liu, Z.; She, X.; Gao, X.; Liang, R. Progress in Understanding the IL-6/STAT3 Pathway in Colorectal Cancer. Onco. Targets Ther. 2020, 13, 13023–13032. [Google Scholar] [CrossRef]
- Halim, C.E.; Deng, S.; Ong, M.S.; Yap, C.T. Involvement of STAT5 in Oncogenesis. Biomedicines 2020, 8, 316. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Wang, F.; Wang, C.; Jin, L. Phenotypic and Functional Analyses of B7S1 in Ovarian Cancer. Front. Mol. Biosci. 2021, 8, 686803. [Google Scholar] [CrossRef]
- Kent, L.N.; Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef]
- Shirley Polager; Doron Ginsberg. E2F Mediates Sustained G2 Arrest and Down-regulation of Stathmin and AIM-1 Expression in Response to Genotoxic Stress*. J. Biol. Chem. 2003, 278, 1443–1449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leo, M.; Muccillo, L.; Pranzini, E.; Barisciano, G.; Parri, M.; Lopatriello, G.; Carlomagno, M.; Santi, A.; Taddei, M.L.; Sabatino, L. Transcriptomic Analysis of Colorectal Cancer Cells Treated with Oil Production Waste Products (OPWPs) Reveals Enrichment of Pathways of Mitochondrial Functionality. Cells 2022, 11, 3992. [Google Scholar] [CrossRef] [PubMed]




| Female | Male | All Cases | |
|---|---|---|---|
| 76 (45.51%) | 91 (54.49%) | 167 (100%) | |
| Age | 66.63 ± 9.48 | 63.75 ± 9.46 | 65.09 ± 9.55 |
| Tumour localization | |||
| Left-sided | 49 (66.22%) | 65 (73.86%) | 114 (70.37%) |
| Right-sided | 25 (33.78%) | 23 (26.13%) | 48 (29.63%) |
| T parameter | |||
| T1 | 1 (1.33%) | 7 (8.05%) | 8 (4.94%) |
| T2 | 16 (21.33%) | 12 (13.79%) | 28 (22.22%) |
| T3 | 47 (62.67%) | 54 (62.07%) | 101 (62.35%) |
| T4 | 11 (14.67%) | 14 (16.09%) | 25 (15.43%) |
| N parameter | |||
| N0 | 32 (42.67%) | 37 (42.05%) | 69 (42.33%) |
| N1 | 30 (40.00%) | 37 (42.05%) | 67 (41.10%) |
| N2 | 13 (17.33%) | 14 (15.91%) | 27 (16.56%) |
| M parameter | |||
| M0 | 66 (88.00%) | 69 (79.31%) | 135 (83.33%) |
| M1 | 9 (12.00%) | 18 (20.69%) | 27 (16.67%) |
| TNM stage | |||
| I | 13 (17.33%) | 13 (14.77%) | 26 (15.95%) |
| II | 19 (25.33%) | 21 (28.86%) | 40 (24.54%) |
| III | 34 (45.33%) | 37 (42.05%) | 71 (43.55%) |
| IV | 9 (12.00%) | 17 (19.32%) | 26 (15.95%) |
| Grading | |||
| Low | 63 (85.14%) | 75 (85.23%) | 138 (85.16%) |
| High | 11 (14.86%) | 13 (14.777%) | 24 (14.81%) |
| Adjuvant treatment | |||
| Yes | 7 (9.21%) | 14 (15.38%) | 21 (12.35%) |
| No | 69 (90.79%) | 77 (84.62%) | 149 (87.65%) |
| Characteristics | B7H4 Tumour Expression | ||
|---|---|---|---|
| Positive | Negative | p | |
| Age | 63.82 | 67.88 | 0.13 |
| Tumour localization | 0.18 | ||
| Left-sided | 48 (81.36%) | 11 (18.64%) | |
| Right-sided | 12 (66.67%) | 6 (33.33%) | |
| T parameter | 0.67 | ||
| T1 | 4 (100%) | 0 (0%) | |
| T2 | 8 (80%) | 2 (20%) | |
| T3 | 41 (78.85%) | 11 (21.15%) | |
| T4 | 7 (70%) | 3 (30%) | |
| N parameter | 0.14 | ||
| N0 | 25 (69.44%) | 11 (30.56%) | |
| N1 | 25 (89.29%) | 3 (10.71%) | |
| N2 | 10 (83.33%) | 2 (16.67%) | |
| M parameter | 0.91 | ||
| M0 | 51 (78.46%) | 14 (21.54%) | |
| M1 | 8 (80%) | 2 (20%) | |
| TNM stage | 0.20 | ||
| I | 10 (83.33%) | 2 (16.67%) | |
| II | 14 (63.64%) | 8 (36.36%) | |
| III | 28 (87.50%) | 4 (12.50%) | |
| IV | 8 (80%) | 2 (20%) | |
| Grading | 0.91 | ||
| Low | 51 (78.46%) | 14 (21.54%) | |
| High | 8 (80%) | 2 (20%) | |
| MSS/MSI status | 0.005 | ||
| MSS tumours | 56 (83.58%) | 11 (16.42%) | |
| MSI tumours | 4 (40%) | 6 (60%) | |
| TILs | 0.42 | ||
| 0–5% | 30 (83.33%) | 6 (16.67%) | |
| 6–25% | 18 (85.71%) | 3 (14.29%) | |
| 26–50% | 10 (62.50%) | 6 (37.50%) | |
| 51–75% | 2 (66.67%) | 1 (33.33%) | |
| 76–100% | 0 (0%) | 0 (0%) | |
| CD8 Lymphocytes | 0.54 | ||
| 0–5% | 27 (79.41%) | 7 (20.59%) | |
| 6–25% | 17 (80.95%) | 4 (19.05%) | |
| 26–50% | 10 (83.33%) | 2 (16.67%) | |
| 51–75% | 3 (50%) | 3 (50%) | |
| 76–100% | 3 (75%) | 1 (25%) | |
| Budding | 0.93 | ||
| 0–4 | 32 (80%) | 8 (20%) | |
| 5–9 | 18 (78.26%) | 5 (21.74%) | |
| >9 | 9 (75%) | 3 (25%) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dawidowicz, M.; Kula, A.; Mielcarska, S.; Kiczmer, P.; Skiba, H.; Krygier, M.; Chrabańska, M.; Piecuch, J.; Szrot, M.; Robotycka, J.; et al. B7H4 Expression Is More Frequent in MSS Status Colorectal Cancer and Is Negatively Associated with Tumour Infiltrating Lymphocytes. Cells 2023, 12, 861. https://doi.org/10.3390/cells12060861
Dawidowicz M, Kula A, Mielcarska S, Kiczmer P, Skiba H, Krygier M, Chrabańska M, Piecuch J, Szrot M, Robotycka J, et al. B7H4 Expression Is More Frequent in MSS Status Colorectal Cancer and Is Negatively Associated with Tumour Infiltrating Lymphocytes. Cells. 2023; 12(6):861. https://doi.org/10.3390/cells12060861
Chicago/Turabian StyleDawidowicz, Miriam, Agnieszka Kula, Sylwia Mielcarska, Paweł Kiczmer, Hanna Skiba, Małgorzata Krygier, Magdalena Chrabańska, Jerzy Piecuch, Monika Szrot, Julia Robotycka, and et al. 2023. "B7H4 Expression Is More Frequent in MSS Status Colorectal Cancer and Is Negatively Associated with Tumour Infiltrating Lymphocytes" Cells 12, no. 6: 861. https://doi.org/10.3390/cells12060861






