Cancer Spheroids and Organoids as Novel Tools for Research and Therapy: State of the Art and Challenges to Guide Precision Medicine
Abstract
1. Introduction
2. Current Source to Generate and Culture In Vitro Tumor Spheroids and Organoids
2.1. Spheroids from Tumor Cell Lines
2.2. Organoids Derived from Primary Cells and Patients’ Biopsies
2.3. Spheroids and Organoids from Genetically Modified Cells
2.4. Cancer Organoids from Pluripotent Stem Cells
2.5. Organoids Made from Several Cell Sources
2.6. Organoids Including the TME
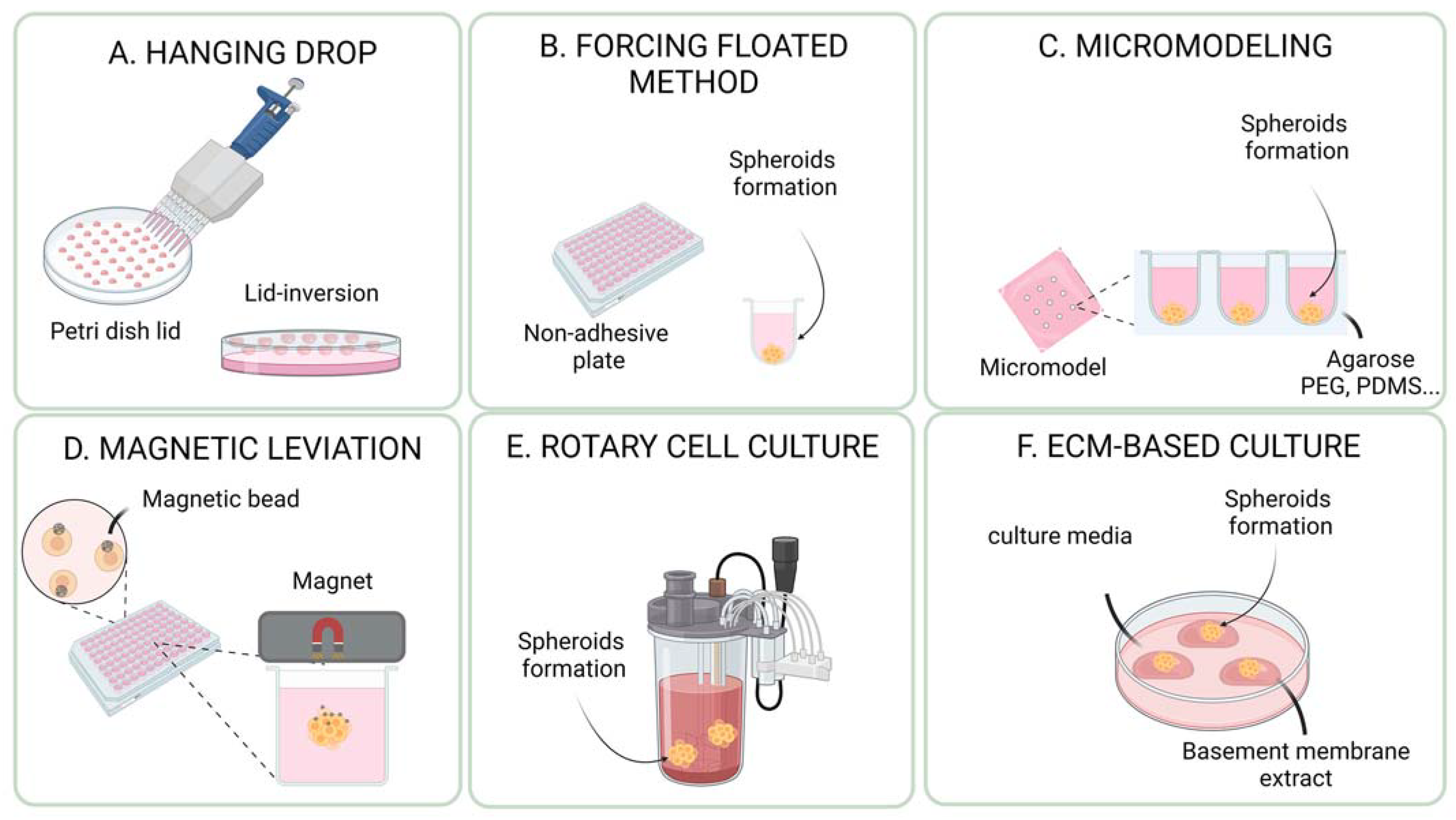
3. Methods for the Generation and Culture of Cancer Spheroids and Organoids
3.1. ECM (Extracellular Matrix)-Free Methods for Cancer Spheroids and Organoids
3.1.1. Hanging Drop Method
3.1.2. Non-Adherent Plates and Forced Floating Method
3.1.3. Forced Aggregation into Microwells
3.1.4. Magnetic Levitation
3.1.5. Rotary Cell Culture System (Bioreactor/Spinner)
3.2. ECM-Based Methods for Cancer Spheroids and Organoids
4. Methodological Options in Front of Research on Anticancer Therapy
4.1. Chemotherapy
4.2. CRISPR/Cas9
4.3. Adoptive Immune Cell Transfer Therapies
4.4. Immunotherapy
4.4.1. Immune Checkpoint Inhibitors (ICIs)
4.4.2. Monoclonal and Bispecific Antibodies (bsAbs)
4.4.3. Application of Organoids in Tumor Vaccination
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sachdeva, U.M.; Shimonosono, M.; Flashner, S.; Cruz-Acuña, R.; Gabre, J.T.; Nakagawa, H. Understanding the cellular origin and progression of esophageal cancer using esophageal organoids. Cancer Lett. 2021, 509, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.P.; Lan, H.-R.; Fang, X.-L.; Yang, X.-Y.; Jin, K.-T. Organoid Models for Precision Cancer Immunotherapy. Front. Immunol. 2022, 13, 770465. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Li, F.; He, J.; Aji, R.; Gao, D. Organoid technology in cancer precision medicine. Cancer Lett. 2019, 457, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Weeber, F.; van de Wetering, M.; Hoogstraat, M.; Dijkstra, K.K.; Krijgsman, O.; Kuilman, T.; Gadellaa-van Hooijdonk, C.G.M.; van der Velden, D.L.; Peeper, D.S.; Cuppen, E.P.J.G.; et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc. Natl. Acad. Sci. USA 2015, 112, 13308–13311. [Google Scholar] [CrossRef]
- Schutte, M.; Risch, T.; Abdavi-Azar, N.; Boehnke, K.; Schumacher, D.; Keil, M.; Yildiriman, R.; Jandrasits, C.; Borodina, T.; Amstislavskiy, V.; et al. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat. Commun. 2017, 8, 14262. [Google Scholar] [CrossRef]
- Boretto, M.; Maenhoudt, N.; Luo, X.; Hennes, A.; Boeckx, B.; Bui, B.; Heremans, R.; Perneel, L.; Kobayashi, H.; Van Zundert, I.; et al. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat. Cell Biol. 2019, 21, 1041–1051. [Google Scholar] [CrossRef]
- Takahashi, N.; Higa, A.; Hiyama, G.; Tamura, H.; Hoshi, H.; Dobashi, Y.; Katahira, K.; Ishihara, H.; Takagi, K.; Goda, K.; et al. Construction of in vitro patient-derived tumor models to evaluate anticancer agents and cancer immunotherapy. Oncol. Lett. 2021, 21, 406. [Google Scholar] [CrossRef]
- Dominijanni, A.; Devarasetty, M.; Soker, S. Manipulating the Tumor Microenvironment in Tumor Organoids Induces Phenotypic Changes and Chemoresistance. iScience 2020, 23, 101851. [Google Scholar] [CrossRef]
- Devarasetty, M.; Forsythe, S.; Shelkey, E.; Soker, S. In Vitro Modeling of the Tumor Microenvironment in Tumor Organoids. Tissue Eng. Regen. Med. 2020, 17, 759–771. [Google Scholar] [CrossRef]
- Kretzschmar, K. Cancer research using organoid technology. J. Mol. Med. 2021, 99, 501–515. [Google Scholar] [CrossRef]
- Gronholm, M.; Feodoroff, M.; Antignani, G.; Martins, B.; Hamdan, F.; Cerullo, V. Patient-Derived Organoids for Precision Cancer Immunotherapy. Cancer Res. 2021, 81, 3149–3155. [Google Scholar] [CrossRef]
- De Witt Hamer, P.C.; van Tilborg, A.A.G.; Eijk, P.P.; Sminia, P.; Troost, D.; Van Noorden, C.J.F.; Ylstra, B.; Leenstra, S. The genomic profile of human malignant glioma is altered early in primary cell culture and preserved in spheroids. Oncogene 2008, 27, 2091–2096. [Google Scholar] [CrossRef]
- Weiswald, L.B.; Bellet, D.; Dangles-Marie, V. Spherical cancer models in tumor biology. Neoplasia 2015, 17, 1–15. [Google Scholar] [CrossRef]
- Liao, J.; Qian, F.; Tchabo, N.; Mhawech-Fauceglia, P.; Beck, A.; Qian, Z.; Wang, X.; Huss, W.J.; Lele, S.B.; Morrison, C.D.; et al. Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism. PLoS ONE 2014, 9, e84941. [Google Scholar] [CrossRef]
- Sutherland, R.M.; Inch, W.R.; McCredie, J.A.; Kruuv, J. A multi-component radiation survival curve using an in vitro tumour model. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1970, 18, 491–495. [Google Scholar] [CrossRef]
- Pastrana, E.; Silva-Vargas, V.; Doetsch, F. Eyes wide open: A critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 2011, 8, 486–498. [Google Scholar] [CrossRef]
- Mirabelli, P.; Coppola, L.; Salvatore, M. Cancer Cell Lines Are Useful Model Systems for Medical Research. Cancers 2019, 11, 1098. [Google Scholar] [CrossRef]
- Perche, F.; Torchilin, V.P. Cancer cell spheroids as a model to evaluate chemotherapy protocols. Cancer Biol. Ther. 2012, 13, 1205–1213. [Google Scholar] [CrossRef]
- Roy, A.; Wang, G.; Iskander, D.; O’Byrne, S.; Elliott, N.; O’Sullivan, J.; Buck, G.; Heuston, E.F.; Wen, W.X.; Meira, A.R.; et al. Transitions in lineage specification and gene regulatory networks in hematopoietic stem/progenitor cells over human development. Cell Rep. 2021, 36, 109698. [Google Scholar] [CrossRef]
- Kuo, C.T.; Wang, J.-Y.; Lin, Y.-F.; Wo, A.M.; Chen, B.P.C.; Lee, H. Three-dimensional spheroid culture targeting versatile tissue bioassays using a PDMS-based hanging drop array. Sci. Rep. 2017, 7, 4363. [Google Scholar] [CrossRef]
- Khan, H.Y.; Kamgar, M.; Aboukameel, A.; Bannoura, S.; Chung, B.Y.; Li, Y.; Al Hallak, M.N.; Philip, P.A.; Tsai, S.; Luther, S.; et al. Targeting Cellular Metabolism With CPI-613 Sensitizes Pancreatic Cancer Cells to Radiation Therapy. Adv. Radiat. Oncol. 2023, 8, 101122. [Google Scholar] [CrossRef] [PubMed]
- Yeon, S.E.; No, D.Y.; Lee, S.-H.; Nam, S.W.; Oh, I.-H.; Lee, J.; Kuh, H.-J. Application of concave microwells to pancreatic tumor spheroids enabling anticancer drug evaluation in a clinically relevant drug resistance model. PLoS ONE 2013, 8, e73345. [Google Scholar] [CrossRef] [PubMed]
- Zuchowska, A.; Jastrzebska, E.; Zukowski, K.; Chudy, M.; Dybko, A.; Brzozka, Z. A549 and MRC-5 cell aggregation in a microfluidic Lab-on-a-chip system. Biomicrofluidics 2017, 11, 024110. [Google Scholar] [CrossRef] [PubMed]
- Mosaad, E.O.; Chambers, K.F.; Futrega, K.; Clements, J.A.; Doran, M.R. The Microwell-mesh: A high-throughput 3D prostate cancer spheroid and drug-testing platform. Sci. Rep. 2018, 8, 253. [Google Scholar] [CrossRef]
- Amaral, R.L.F.; Miranda, M.; Marcato, P.D.; Swiech, K. Comparative Analysis of 3D Bladder Tumor Spheroids Obtained by Forced Floating and Hanging Drop Methods for Drug Screening. Front. Physiol. 2017, 8, 605. [Google Scholar] [CrossRef]
- Cirigliano, S.M.; Bessone, M.I.D.; Berardi, D.E.; Flumian, C.; Joffé, E.D.B.D.K.; Perea, S.E.; Farina, H.G.; Todaro, L.B.; Urtreger, A.J. The synthetic peptide CIGB-300 modulates CK2-dependent signaling pathways affecting the survival and chemoresistance of non-small cell lung cancer cell lines. Cancer Cell Int. 2017, 17, 42. [Google Scholar] [CrossRef]
- Fan, T.W.-M.; El-Amouri, S.S.; Macedo, J.K.A.; Wang, Q.J.; Song, H.; Cassel, T.; Lane, A.N. Stable Isotope-Resolved Metabolomics Shows Metabolic Resistance to Anti-Cancer Selenite in 3D Spheroids versus 2D Cell Cultures. Metabolites 2018, 8, 40. [Google Scholar] [CrossRef]
- Widder, M.; Lemke, K.; Kekeç, B.; Förster, T.; Grodrian, A.; Gastrock, G. A modified 384-well-device for versatile use in 3D cancer cell (co-)cultivation and screening for investigations of tumor biology in vitro. Eng. Life Sci. 2018, 18, 132–139. [Google Scholar] [CrossRef]
- Khan, M.I.; Czarnecka, A.M.; Lewicki, S.; Helbrecht, I.; Brodaczewska, K.; Koch, I.; Zdanowski, R.; Król, M.; Szczylik, C. Comparative Gene Expression Profiling of Primary and Metastatic Renal Cell Carcinoma Stem Cell-Like Cancer Cells. PLoS ONE 2016, 11, e0165718. [Google Scholar] [CrossRef]
- Katt, M.E.; Placone, A.L.; Wong, A.D.; Xu, Z.S.; Searson, P.C. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front. Bioeng. Biotechnol. 2016, 4, 12. [Google Scholar] [CrossRef]
- Petljak, M.; Alexandrov, L.B.; Brammeld, J.S.; Price, S.; Wedge, D.C.; Grossmann, S.; Dawson, K.J.; Ju, Y.S.; Iorio, F.; Tubio, J.M.; et al. Characterizing Mutational Signatures in Human Cancer Cell Lines Reveals Episodic APOBEC Mutagenesis. Cell 2019, 176, 1282–1294.e20. [Google Scholar] [CrossRef] [PubMed]
- Levatic, J.; Salvadores, M.; Fuster-Tormo, F.; Supek, F. Mutational signatures are markers of drug sensitivity of cancer cells. Nat. Commun. 2022, 13, 2926. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, J.F.; van Vliet, E.J.; Sachs, N.; Rosenbluth, J.M.; Kopper, O.; Rebel, H.G.; Wehrens, E.J.; Piani, C.; Visvader, J.E.; Verissimo, C.S.; et al. Long-term culture, genetic manipulation and xenotransplantation of human normal and breast cancer organoids. Nat. Protoc. 2021, 16, 1936–1965. [Google Scholar] [CrossRef]
- Shu, D.; Shen, M.; Li, K.; Han, X.; Li, H.; Tan, Z.; Wang, Y.; Peng, Y.; Tang, Z.; Qu, C.; et al. Organoids from patient biopsy samples can predict the response of BC patients to neoadjuvant chemotherapy. Ann. Med. 2022, 54, 2581–2597. [Google Scholar] [CrossRef] [PubMed]
- Sachs, N.; de Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 2018, 172, 373–386.e10. [Google Scholar] [CrossRef]
- Chen, P.; Zhang, X.; Ding, R.; Yang, L.; Lyu, X.; Zeng, J.; Lei, J.H.; Wang, L.; Bi, J.; Shao, N.; et al. Patient-Derived Organoids Can Guide Personalized-Therapies for Patients with Advanced Breast Cancer. Adv. Sci. 2021, 8, e2101176. [Google Scholar] [CrossRef]
- Hu, Y.; Sui, X.; Song, F.; Li, Y.; Li, K.; Chen, Z.; Yang, F.; Chen, X.; Zhang, Y.; Wang, X.; et al. Lung cancer organoids analyzed on microwell arrays predict drug responses of patients within a week. Nat. Commun. 2021, 12, 2581. [Google Scholar] [CrossRef]
- Bie, Y.; Wang, J.; Xiong, L.; Wang, D.; Liao, J.; Zhang, Y.; Lin, H. Lung adenocarcinoma organoids harboring EGFR 19Del and L643V double mutations respond to osimertinib and gefitinib: A case report. Medicine 2021, 100, e24793. [Google Scholar] [CrossRef]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, C.; Hu, Y.; Li, T.; Yang, G.; Wang, G.; Zhu, J.; Shao, C.; Hou, H.; Zhou, N.; et al. Anlotinib Combined with Toripalimab as Second-Line Therapy for Advanced, Relapsed Gastric or Gastroesophageal Junction Carcinoma. Oncologist 2022, 27, e856–e869. [Google Scholar] [CrossRef]
- Lai Benjamin, F.L.; Lu, R.X.Z.; Hu, Y.; Huyer, L.D.; Dou, W.; Wang, E.Y.; Radulovich, N.; Tsao, M.S.; Sun, Y.; Radisic, M. Recapitulating pancreatic tumor microenvironment through synergistic use of patient organoids and organ-on-a-chip vasculature. Adv. Funct. Mater. 2020, 30, 2000545. [Google Scholar]
- Kopper, O.; de Witte, C.J.; Lõhmussaar, K.; Valle-Inclan, J.E.; Hami, N.; Kester, L.; Balgobind, A.V.; Korving, J.; Proost, N.; Begthel, H.; et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 2019, 25, 838–849. [Google Scholar] [CrossRef]
- Servant, R.; Garioni, M.; Vlajnic, T.; Blind, M.; Pueschel, H.; Müller, D.C.; Zellweger, T.; Templeton, A.J.; Garofoli, A.; Maletti, S.; et al. Prostate cancer patient-derived organoids: Detailed outcome from a prospective cohort of 81 clinical specimens. J. Pathol. 2021, 254, 543–555. [Google Scholar] [CrossRef]
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A Three-Dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found In Vivo. Cancer Res. 2016, 76, 2465–2477. [Google Scholar] [CrossRef]
- Broutier, L.; Mastrogiovanni, G.; Verstegen, M.M.A.; Francies, H.E.; Gavarró, L.M.; Bradshaw, C.R.; Allen, G.E.; Arnes-Benito, R.; Sidorova, O.; Gaspersz, M.P.; et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017, 23, 1424–1435. [Google Scholar] [CrossRef]
- Zeng, L.; Liao, Q.; Zhao, H.; Jiang, S.; Yang, X.; Tang, H.; He, Q.; Yang, X.; Fang, S.; He, J.; et al. 2Raltitrexed as a synergistic hyperthermia chemotherapy drug screened in patient-derived colorectal cancer organoids. Cancer Biol. Med. 2021, 18, 750–762. [Google Scholar] [CrossRef]
- Saengwimol, D.; Rojanaporn, D.; Chaitankar, V.; Chittavanich, P.; Aroonroch, R.; Boontawon, T.; Thammachote, W.; Jinawath, N.; Hongeng, S.; Kaewkhaw, R. A three-dimensional organoid model recapitulates tumorigenic aspects and drug responses of advanced human retinoblastoma. Sci. Rep. 2018, 8, 15664. [Google Scholar] [CrossRef]
- Lee, S.H.; Hu, W.; Matulay, J.T.; Silva, M.V.; Owczarek, T.B.; Kim, K.; Chua, C.W.; Barlow, L.M.J.; Kandoth, C.; Williams, A.B.; et al. Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell 2018, 173, 515–528.e17. [Google Scholar] [CrossRef]
- Driehuis, E.; van Hoeck, A.; Moore, K.; Kolders, S.; Francies, H.E.; Gulersonmez, M.C.; Stigter, E.C.A.; Burgering, B.; Geurts, V.; Gracanin, A.; et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl. Acad. Sci. USA 2019, 116, 26580–26590. [Google Scholar] [CrossRef]
- Phifer, C.J.; Bergdorf, K.N.; Bechard, M.E.; Vilgelm, A.; Baregamian, N.; McDonald, O.G.; Lee, E.; Weiss, V.L. Obtaining patient-derived cancer organoid cultures via fine-needle aspiration. STAR Protoc. 2021, 2, 100220. [Google Scholar] [CrossRef] [PubMed]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Böttinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019, 38, e100300. [Google Scholar] [CrossRef] [PubMed]
- Schutgens, F.; Rookmaaker, M.B.; Margaritis, T.; Rios, A.; Ammerlaan, C.; Jansen, J.; Gijzen, L.; Vormann, M.; Vonk, A.; Viveen, M.; et al. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat. Biotechnol. 2019, 37, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shiratsuchi, H.; Lin, J.; Chen, G.; Reddy, R.M.; Azizi, E.; Fouladdel, S.; Chang, A.C.; Lin, L.; Jiang, H.; et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget 2014, 5, 12383–12397. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, H.; Chen, K. Living biobank-based cancer organoids: Prospects and challenges in cancer research. Cancer Biol. Med. 2022, 19, 965–982. [Google Scholar] [CrossRef]
- Liu, L.; Liu, H.; Chen, K. Patient-derived organoid (PDO) platforms to facilitate clinical decision making. J. Transl. Med. 2021, 19, 40. [Google Scholar] [CrossRef]
- Pfohl, U.; Pflaume, A.; Regenbrecht, M.; Finkler, S.; Adelmann, Q.G.; Reinhard, C.; Regenbrecht, C.; Wedeken, L. Precision Oncology beyond Genomics: The Future Is Here-It Is Just Not Evenly Distributed. Cells 2021, 10, 928. [Google Scholar] [CrossRef]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity. Cell 2020, 180, 188–204.e22. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, N.; Liu, Y.C. An organoids biobank for recapitulating tumor heterogeneity and personalized medicine. Chin. J. Cancer Res. 2020, 32, 408–413. [Google Scholar] [CrossRef]
- Prasetyanti, P.R.; Medema, J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 2017, 16, 41. [Google Scholar] [CrossRef]
- Gambara, G.; Gaebler, M.; Keilholz, U.; Regenbrecht, C.R.A.; Silvestri, A. From Chemotherapy to Combined Targeted Therapeutics: In Vitro and In Vivo Models to Decipher Intra-Tumor Heterogeneity. Front. Pharmacol. 2018, 9, 77. [Google Scholar] [CrossRef]
- Xia, T.; Du, W.; Chen, X.; Zhang, Y. Organoid models of the tumor microenvironment and their applications. J. Cell. Mol. Med. 2021, 25, 5829–5841. [Google Scholar] [CrossRef]
- Tiriac, H.; Belleau, P.; Engle, D.D.; Plenker, D.; Deschenes, A.; Somerville, T.D.D.; Froeling, F.E.M.; Burkhart, R.A.; Denroche, R.E.; Jang, G.H.; et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018, 8, 1112–1129. [Google Scholar] [CrossRef]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, G.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.-H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e16. [Google Scholar] [CrossRef]
- Fujii, M.; Shimokawa, M.; Date, S.; Takano, A.; Matano, M.; Nanki, K.; Ohta, Y.; Toshimitsu, K.; Nakazato, Y.; Kawasaki, K.; et al. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell 2016, 18, 827–838. [Google Scholar] [CrossRef]
- Ko, J.W.; Sukhova, N.; Thacker, D.; Chen, P.; Flockhart, D.A. Evaluation of omeprazole and lansoprazole as inhibitors of cytochrome P450 isoforms. Drug Metab. Dispos. 1997, 25, 853–862. [Google Scholar]
- Xie, X.; Li, X.; Song, W. Tumor organoid biobank-new platform for medical research. Sci. Rep. 2023, 13, 1819. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Schwank, G.; Koo, B.-K.; Sasselli, V.; Dekkers, J.F.; Heo, I.; Demircan, T.; Sasaki, N.; Boymans, S.; Cuppen, E.; van der Ent, C.K.; et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013, 13, 653–658. [Google Scholar] [CrossRef]
- Fujii, M.; Clevers, H.; Sato, T. Modeling Human Digestive Diseases With CRISPR-Cas9-Modified Organoids. Gastroenterology 2019, 156, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Oost, K.C.; Liberali, P. Engineering human knock-in organoids. Nat. Cell Biol. 2020, 22, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, D.; Clevers, H.; Artegiani, B. CRISPR-Cas Tools and Their Application in Genetic Engineering of Human Stem Cells and Organoids. Cell Stem Cell 2020, 27, 705–731. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Clevers, H.; Sato, T. Molecular subtyping of colorectal cancer: Recent progress, new challenges and emerging opportunities. Semin. Cancer Biol. 2019, 55, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, D.; Artegiani, B.; Hu, H.; Lopes, S.C.D.S.; Clevers, H. Establishment of human fetal hepatocyte organoids and CRISPR-Cas9-based gene knockin and knockout in organoid cultures from human liver. Nat. Protoc. 2021, 16, 182–217. [Google Scholar] [CrossRef]
- Tuveson, D.; Clevers, H. Cancer modeling meets human organoid technology. Science 2019, 364, 952–955. [Google Scholar] [CrossRef]
- Artegiani, B.; van Voorthuijsen, L.; Lindeboom, R.G.; Seinstra, D.; Heo, I.; Tapia, P.; López-Iglesias, C.; Postrach, D.; Dayton, T.; Oka, R.; et al. Probing the Tumor Suppressor Function of BAP1 in CRISPR-Engineered Human Liver Organoids. Cell Stem Cell 2019, 24, 927–943.e6. [Google Scholar] [CrossRef]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef]
- Drost, J.; Van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; Van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef]
- Lo, Y.H.; Kolahi, K.S.; Du, Y.; Chang, C.-Y.; Krokhotin, A.; Nair, A.; Sobba, W.D.; Karlsson, K.; Jones, S.J.; Longacre, T.A.; et al. A CRISPR/Cas9-Engineered ARID1A-Deficient Human Gastric Cancer Organoid Model Reveals Essential and Nonessential Modes of Oncogenic Transformation. Cancer Discov. 2021, 11, 1562–1581. [Google Scholar] [CrossRef]
- Zhao, H.; Cheng, Y.; Kalra, A.; Ma, K.; Zheng, Y.; Ziman, B.; Tressler, C.; Glunde, K.; Shin, E.J.; Ngamruengphong, S.; et al. Generation and multiomic profiling of a TP53/CDKN2A double-knockout gastroesophageal junction organoid model. Sci. Transl. Med. 2022, 14, eabq6146. [Google Scholar] [CrossRef]
- Takeda, H.; Kataoka, S.; Nakayama, M.; Ali, M.A.E.; Oshima, H.; Yamamoto, D.; Park, J.-W.; Takegami, Y.; An, T.; Jenkins, N.A.; et al. CRISPR-Cas9-mediated gene knockout in intestinal tumor organoids provides functional validation for colorectal cancer driver genes. Proc. Natl. Acad. Sci. USA 2019, 116, 15635–15644. [Google Scholar] [CrossRef]
- Verissimo, C.S.; Overmeer, R.M.; Ponsioen, B.; Drost, J.; Mertens, S.; Verlaan-Klink, I.; van Gerwen, B.; van der Ven, M.; van de Wetering, M.; Egan, D.A. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. eLife 2016, 5, e18489. [Google Scholar] [CrossRef]
- Zhang, S.; Iyer, S.; Ran, H.; Dolgalev, I.; Gu, S.; Wei, W.; Foster, C.J.; Loomis, C.A.; Olvera, N.; Dao, F.; et al. Genetically Defined, Syngeneic Organoid Platform for Developing Combination Therapies for Ovarian Cancer. Cancer Discov. 2021, 11, 362–383. [Google Scholar] [CrossRef]
- Escalona, R.M.; Chu, S.; Kadife, E.; Kelly, J.K.; Kannourakis, G.; Findlay, J.K.; Ahmed, N. Knock down of TIMP-2 by siRNA and CRISPR/Cas9 mediates diverse cellular reprogramming of metastasis and chemosensitivity in ovarian cancer. Cancer Cell Int. 2022, 22, 422. [Google Scholar] [CrossRef]
- Kim, D.; Choi, B.-H.; Ryoo, I.-G.; Kwak, M.-K. High NRF2 level mediates cancer stem cell-like properties of aldehyde dehydrogenase (ALDH)-high ovarian cancer cells: Inhibitory role of all-trans retinoic acid in ALDH/NRF2 signaling. Cell Death Dis. 2018, 9, 896. [Google Scholar] [CrossRef]
- Bou-Nader, M.; Caruso, S.; Donne, R.; Celton-Morizur, S.; Calderaro, J.; Gentric, G.; Cadoux, M.; L’Hermitte, A.; Klein, C.; Guilbert, T.; et al. Polyploidy spectrum: A new marker in HCC classification. Gut 2020, 69, 355–364. [Google Scholar] [CrossRef]
- Zhang, M.; Vandana, J.J.; Lacko, L.; Chen, S. Modeling cancer progression using human pluripotent stem cell-derived cells and organoids. Stem Cell Res. 2020, 49, 102063. [Google Scholar] [CrossRef]
- Bian, S.; Repic, M.; Guo, Z.; Kavirayani, A.; Burkard, T.; Bagley, J.A.; Krauditsch, C.; Knoblich, J.A. Genetically engineered cerebral organoids model brain tumor formation. Nat. Methods 2018, 15, 631–639. [Google Scholar] [CrossRef]
- Ogawa, J.; Pao, G.M.; Shokhirev, M.N.; Verma, I.M. Glioblastoma Model Using Human Cerebral Organoids. Cell Rep. 2018, 23, 1220–1229. [Google Scholar] [CrossRef]
- Breunig, M.; Merkle, J.; Wagner, M.; Melzer, M.K.; Barth, T.F.E.; Engleitner, T.; Krumm, J.; Wiedenmann, S.; Cohrs, C.M.; Perkhofer, L.; et al. Modeling plasticity and dysplasia of pancreatic ductal organoids derived from human pluripotent stem cells. Cell Stem Cell 2021, 28, 1105–1124.e19. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Desai, R.; Conrad, D.N.; Leite, N.C.; Akshinthala, D.; Lim, C.M.; Gonzalez, R.; Muthuswamy, L.B.; Gartner, Z.; Muthuswamy, S.K. Commitment and oncogene-induced plasticity of human stem cell-derived pancreatic acinar and ductal organoids. Cell Stem Cell 2021, 28, 1090–1104.e6. [Google Scholar] [CrossRef] [PubMed]
- Dost, A.F.M.; Moye, A.L.; Vedaie, M.; Tran, L.M.; Fung, E.; Heinze, D.; Villacorta-Martin, C.; Huang, J.; Hekman, R.; Kwan, J.H.; et al. Organoids Model Transcriptional Hallmarks of Oncogenic KRAS Activation in Lung Epithelial Progenitor Cells. Cell Stem Cell 2020, 27, 663–678.e8. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Sander, J.D.; Reyon, D.; Cascio, V.M.; Joung, J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014, 32, 279–284. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Idris, M.; Alves, M.M.; Hofstra, R.M.; Mahe, M.M.; Melotte, V. Intestinal multicellular organoids to study colorectal cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188586. [Google Scholar] [CrossRef]
- Yang, L.; Güell, M.; Byrne, S.; Yang, J.L.; Angeles, A.D.L.; Mali, P.; Aach, J.; Kim-Kiselak, C.; Briggs, A.W.; Rios, X.; et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013, 41, 9049–9061. [Google Scholar] [CrossRef]
- Song, Y.; Kim, S.; Heo, J.; Shum, D.; Lee, S.-Y.; Lee, M.; Kim, A.-R.; Seo, H.R. Identification of hepatic fibrosis inhibitors through morphometry analysis of a hepatic multicellular spheroids model. Sci. Rep. 2021, 11, 10931. [Google Scholar] [CrossRef]
- Bessy, T.; Itkin, T.; Passaro, D. Bioengineering the Bone Marrow Vascular Niche. Front. Cell Dev. Biol. 2021, 9, 645496. [Google Scholar] [CrossRef]
- Janagama, D.; Hui, S.K. 3-D Cell Culture Systems in Bone Marrow Tissue and Organoid Engineering, and BM Phantoms as In Vitro Models of Hematological Cancer Therapeutics—A Review. Materials 2020, 13, 5609. [Google Scholar] [CrossRef]
- Balandran, J.C.; Dávila-Velderrain, J.; Sandoval-Cabrera, A.; Zamora-Herrera, G.; Terán-Cerqueda, V.; García-Stivalet, L.A.; Limón-Flores, J.A.; Armenta-Castro, E.; Rodríguez-Martínez, A.; Leon-Chavez, B.A.; et al. Patient-Derived Bone Marrow Spheroids Reveal Leukemia-Initiating Cells Supported by Mesenchymal Hypoxic Niches in Pediatric B-ALL. Front. Immunol. 2021, 12, 746492. [Google Scholar] [CrossRef]
- Khan, A.O.; Rodriguez-Romera, A.; Reyat, J.S.; Olijnik, A.-A.; Colombo, M.; Wang, G.; Wen, W.X.; Sousos, N.; Murphy, L.C.; Grygielska, B.; et al. Human Bone Marrow Organoids for Disease Modeling, Discovery, and Validation of Therapeutic Targets in Hematologic Malignancies. Cancer Discov. 2023, 13, 364–385. [Google Scholar] [CrossRef]
- Giger, S.; Hofer, M.; Miljkovic-Licina, M.; Hoehnel, S.; Brandenberg, N.; Guiet, R.; Ehrbar, M.; Kleiner, E.; Gegenschatz-Schmid, K.; Matthes, T.; et al. Microarrayed human bone marrow organoids for modeling blood stem cell dynamics. APL Bioeng. 2022, 6, 036101. [Google Scholar] [CrossRef]
- Zhao, H.; Jiang, E.; Shang, Z. 3D Co-culture of Cancer-Associated Fibroblast with Oral Cancer Organoids. J. Dent. Res. 2021, 100, 201–208. [Google Scholar] [CrossRef]
- Vis, M.A.M.; Ito, K.; Hofmann, S. Impact of Culture Medium on Cellular Interactions in In Vitro Co-Culture Systems. Front. Bioeng. Biotechnol. 2020, 8, 911. [Google Scholar] [CrossRef]
- Cheung, D.W. Atriopeptin III depresses the excitability of sympathetic neurones. Biochem. Biophys. Res. Commun. 1988, 154, 411–416. [Google Scholar] [CrossRef]
- Smith, R.C.; Tabar, V. Constructing and Deconstructing Cancers Using Human Pluripotent Stem Cells and Organoids. Cell Stem Cell 2019, 24, 12–24. [Google Scholar] [CrossRef]
- Ho, T.; Msallam, R. Tissues and Tumor Microenvironment (TME) in 3D: Models to Shed Light on Immunosuppression in Cancer. Cells 2021, 10, 831. [Google Scholar] [CrossRef]
- Titova, V.N. Methodological and diagnostic aspects of the determination of calcium contents. Klin. Lab. Diagn. 1996, 2, 23–26. [Google Scholar]
- Koeck, S.; Kern, J.; Zwierzina, M.; Gamerith, G.; Lorenz, E.; Sopper, S.; Zwierzina, H.; Amann, A. The influence of stromal cells and tumor-microenvironment-derived cytokines and chemokines on CD3+CD8+ tumor infiltrating lymphocyte subpopulations. Oncoimmunology 2017, 6, e1323617. [Google Scholar] [CrossRef]
- Foty, R. A simple hanging drop cell culture protocol for generation of 3D spheroids. J. Vis. Exp. 2011, 51, e2720. [Google Scholar]
- Bahar, E.; Kim, J.-Y.; Kim, D.-C.; Kim, H.-S.; Yoon, H. Combination of Niraparib, Cisplatin and Twist Knockdown in Cisplatin-Resistant Ovarian Cancer Cells Potentially Enhances Synthetic Lethality through ER-Stress Mediated Mitochondrial Apoptosis Pathway. Int. J. Mol. Sci. 2021, 22, 3916. [Google Scholar] [CrossRef] [PubMed]
- Gheytanchi, E.; Naseri, M.; Karimi-Busheri, F.; Atyabi, F.; Mirsharif, E.S.; Bozorgmehr, M.; Ghods, R.; Madjd, Z. Morphological and molecular characteristics of spheroid formation in HT-29 and Caco-2 colorectal cancer cell lines. Cancer Cell Int. 2021, 21, 204. [Google Scholar] [CrossRef] [PubMed]
- Sims, L.B.; Huss, M.K.; Frieboes, H.B.; Steinbach-Rankins, J.M. Distribution of PLGA-modified nanoparticles in 3D cell culture models of hypo-vascularized tumor tissue. J. Nanobiotechnol. 2017, 15, 67. [Google Scholar] [CrossRef]
- Audoin, M.; Sogaard, M.T.; Jauffred, L. Tumor spheroids accelerate persistently invading cancer cells. Sci. Rep. 2022, 12, 14713. [Google Scholar] [CrossRef]
- Velasco, V.; Shariati, S.A.; Esfandyarpour, R. Esfandyarpour, Microtechnology-based methods for organoid models. Microsyst. Nanoeng. 2020, 6, 76. [Google Scholar] [CrossRef]
- Benien, P.; Swami, A. 3D tumor models: History, advances and future perspectives. Future Oncol. 2014, 10, 1311–1327. [Google Scholar] [CrossRef]
- Razian, G.; Yu, Y.; Ungrin, M. Production of large numbers of size-controlled tumor spheroids using microwell plates. J. Vis. Exp. 2013, 81, e50665. [Google Scholar]
- Nishikawa, T.; Tanaka, Y.; Nishikawa, M.; Ogino, Y.; Kusamori, K.; Mizuno, N.; Mizukami, Y.; Shimizu, K.; Konishi, S.; Takahashi, Y.; et al. Optimization of Albumin Secretion and Metabolic Activity of Cytochrome P450 1A1 of Human Hepatoblastoma HepG2 Cells in Multicellular Spheroids by Controlling Spheroid Size. Biol. Pharm. Bull. 2017, 40, 334–338. [Google Scholar] [CrossRef]
- Olofsson, K.; Carannante, V.; Takai, M.; Önfelt, B.; Wiklund, M. Ultrasound-Based Scaffold-Free Core-Shell Multicellular Tumor Spheroid Formation. Micromachines 2021, 12, 329. [Google Scholar] [CrossRef]
- El Harane, S.; Durual, S.; Braschler, T.; André-Lévigne, D.; Brembilla, N.; Krause, K.; Modarressi, A.; Preynat-Seauve, O. Adipose-derived stem cell spheroids are superior to single-cell suspensions to improve fat autograft long-term survival. J. Cell. Mol. Med. 2022, 26, 1421–1433. [Google Scholar] [CrossRef]
- Jaganathan, H.; Gage, J.; Leonard, F.; Srinivasan, S.; Souza, G.R.; Dave, B.; Godin, B. Three-dimensional in vitro co-culture model of breast tumor using magnetic levitation. Sci. Rep. 2014, 4, 6468. [Google Scholar] [CrossRef]
- Leonard, F.; Godin, B. 3D In Vitro Model for Breast Cancer Research Using Magnetic Levitation and Bioprinting Method. Methods Mol. Biol. 2016, 1406, 239–251. [Google Scholar]
- Redden, R.A.; Doolin, E.J. Microgravity assay of neuroblastoma: In vitro aggregation kinetics and organoid morphology correlate with MYCN expression. In Vitro Cell. Dev. Biol. Anim. 2011, 47, 312–317. [Google Scholar] [CrossRef]
- Kaur, P.; Ward, B.; Saha, B.; Young, L.; Groshen, S.; Techy, G.; Lu, Y.; Atkinson, R.; Taylor, C.R.; Ingram, M.; et al. Human breast cancer histoid: An in vitro 3-dimensional co-culture model that mimics breast cancer tissue. J. Histochem. Cytochem. 2011, 59, 1087–1100. [Google Scholar] [CrossRef]
- Massai, D.; Isu, G.; Madeddu, D.; Cerino, G.; Falco, A.; Frati, C.; Gallo, D.; Deriu, M.A.; Labate, G.F.D.; Quaini, F.; et al. A Versatile Bioreactor for Dynamic Suspension Cell Culture. Application to the Culture of Cancer Cell Spheroids. PLoS ONE 2016, 11, e0154610. [Google Scholar] [CrossRef]
- Stampar, M.; Frandsen, H.S.; Rogowska-Wrzesinska, A.; Wrzesinski, K.; Filipič, M.; Žegura, B. Hepatocellular carcinoma (HepG2/C3A) cell-based 3D model for genotoxicity testing of chemicals. Sci. Total Environ. 2021, 755 Pt 2, 143255. [Google Scholar] [CrossRef]
- Phelan, M.A.; Lelkes, P.I.; Swaroop, A. Mini and customized low-cost bioreactors for optimized high-throughput generation of tissue organoids. Stem Cell Investig. 2018, 5, 33. [Google Scholar] [CrossRef]
- Ferreira, L.P.; Gaspar, V.M.; Mano, J.F. Decellularized Extracellular Matrix for Bioengineering Physiomimetic 3D In Vitro Tumor Models. Trends Biotechnol. 2020, 38, 1397–1414. [Google Scholar] [CrossRef]
- Hume, R.D.; Pensa, S.; Brown, E.J.; Kreuzaler, P.A.; Hitchcock, J.; Husmann, A.; Campbell, J.J.; Lloyd-Thomas, A.O.; Watson, C.J. Tumour cell invasiveness and response to chemotherapeutics in adipocyte invested 3D engineered anisotropic collagen scaffolds. Sci. Rep. 2018, 8, 12658. [Google Scholar] [CrossRef]
- Taubenberger, A.V.; Bray, L.J.; Haller, B.; Shaposhnykov, A.; Binner, M.; Freudenberg, U.; Guck, J.; Werner, C. 3D extracellular matrix interactions modulate tumour cell growth, invasion and angiogenesis in engineered tumour microenvironments. Acta Biomater. 2016, 36, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; Van Es, J.H.; Van Den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- LeSavage, B.L.; Suhar, R.A.; Broguiere, N.; Lutolf, M.P.; Heilshorn, S.C. Next-generation cancer organoids. Nat. Mater. 2022, 21, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Kleinman, H.K.; Martin, G.R. Matrigel: Basement membrane matrix with biological activity. Semin. Cancer Biol. 2005, 15, 378–386. [Google Scholar] [CrossRef]
- Badea, M.A.; Balas, M.; Hermenean, A.; Ciceu, A.; Herman, H.; Ionita, D.; Dinischiotu, A. Influence of Matrigel on Single- and Multiple-Spheroid Cultures in Breast Cancer Research. SLAS Discov. 2019, 24, 563–578. [Google Scholar] [CrossRef]
- Swaminathan, S.; Hamid, Q.; Sun, W.; Clyne, A.M. Bioprinting of 3D breast epithelial spheroids for human cancer models. Biofabrication 2019, 11, 025003. [Google Scholar] [CrossRef]
- Kim, S.; Min, S.; Choi, Y.S.; Jo, S.-H.; Jung, J.H.; Han, K.; Kim, J.; An, S.; Ji, Y.W.; Kim, Y.-G.; et al. Tissue extracellular matrix hydrogels as alternatives to Matrigel for culturing gastrointestinal organoids. Nat. Commun. 2022, 13, 1692. [Google Scholar] [CrossRef]
- Giobbe, G.G.; Crowley, C.; Luni, C.; Campinoti, S.; Khedr, M.; Kretzschmar, K.; De Santis, M.M.; Zambaiti, E.; Michielin, F.; Meran, L.; et al. Extracellular matrix hydrogel derived from decellularized tissues enables endodermal organoid culture. Nat. Commun. 2019, 10, 5658. [Google Scholar] [CrossRef]
- Heo, J.H.; Kang, D.; Seo, S.J.; Jin, Y. Engineering the Extracellular Matrix for Organoid Culture. Int. J. Stem Cells 2022, 15, 60–69. [Google Scholar] [CrossRef]
- Lee, H.J.; Mun, S.; Pham, D.M.; Kim, P. Extracellular Matrix-Based Hydrogels to Tailoring Tumor Organoids. ACS Biomater. Sci. Eng. 2021, 7, 4128–4135. [Google Scholar] [CrossRef]
- Rezakhani, S.; Gjorevski, N.; Lutolf, M. Extracellular matrix requirements for gastrointestinal organoid cultures. Biomaterials 2021, 276, 121020. [Google Scholar] [CrossRef]
- Mo, S.; Tang, P.; Luo, W.; Zhang, L.; Li, Y.; Hu, X.; Ma, X.; Chen, Y.; Bao, Y.; He, X.; et al. Patient-Derived Organoids from Colorectal Cancer with Paired Liver Metastasis Reveal Tumor Heterogeneity and Predict Response to Chemotherapy. Adv. Sci. 2022, 9, e2204097. [Google Scholar] [CrossRef]
- Votanopoulos, K.I.; Mazzocchi, A.; Sivakumar, H.; Forsythe, S.; Aleman, J.; Levine, E.A.; Skardal, A. Appendiceal Cancer Patient-Specific Tumor Organoid Model for Predicting Chemotherapy Efficacy Prior to Initiation of Treatment: A Feasibility Study. Ann. Surg. Oncol. 2019, 26, 139–147. [Google Scholar] [CrossRef]
- Ubink, I.; Bolhaqueiro, A.C.F.; Elias, S.G.; Raats, D.A.E.; Constantinides, A.; Peters, N.A.; Wassenaar, E.C.E.; de Hingh, I.H.J.T.; Rovers, K.P.; van Grevenstein, W.M.U.; et al. Organoids from colorectal peritoneal metastases as a platform for improving hyperthermic intraperitoneal chemotherapy. Br. J. Surg. 2019, 106, 1404–1414. [Google Scholar] [CrossRef] [PubMed]
- Woo, D.H.; Chen, Q.; Yang, T.-L.B.; Glineburg, M.R.; Hoge, C.; Leu, N.A.; Johnson, F.B.; Lengner, C.J. Enhancing a Wnt-Telomere Feedback Loop Restores Intestinal Stem Cell Function in a Human Organotypic Model of Dyskeratosis Congenita. Cell Stem Cell 2016, 19, 397–405. [Google Scholar] [CrossRef]
- Koikawa, K.; Kibe, S.; Suizu, F.; Sekino, N.; Kim, N.; Manz, T.D.; Pinch, B.J.; Akshinthala, D.; Verma, A.; Gaglia, G.; et al. Targeting Pin1 renders pancreatic cancer eradicable by synergizing with immunochemotherapy. Cell 2021, 184, 4753–4771.e27. [Google Scholar] [CrossRef] [PubMed]
- Sayed, S.; Sidorova, O.A.; Hennig, A.; Augsburg, M.; Vesga, C.P.C.; Abohawya, M.; Schmitt, L.T.; Sürün, D.; Stange, D.E.; Mircetic, J.; et al. Efficient Correction of Oncogenic KRAS and TP53 Mutations through CRISPR Base Editing. Cancer Res. 2022, 82, 3002–3015. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; Van De Haar, J.; Fanchi, L.F.; Slagter, M.; Van Der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-Culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598.e12. [Google Scholar] [CrossRef]
- Dekkers, J.F.; Alieva, M.; Cleven, A.; Keramati, F.; Wezenaar, A.K.L.; van Vliet, E.J.; Puschhof, J.; Brazda, P.; Johanna, I.; Meringa, A.D.; et al. Uncovering the mode of action of engineered T cells in patient cancer organoids. Nat. Biotechnol. 2023, 41, 60–69. [Google Scholar] [CrossRef]
- Ando, Y.; Siegler, E.L.; Ta, H.P.; Cinay, G.E.; Zhou, H.; Gorrell, K.A.; Au, H.; Jarvis, B.M.; Wang, P.; Shen, K. Evaluating CAR-T Cell Therapy in a Hypoxic 3D Tumor Model. Adv. Healthc. Mater. 2019, 8, e1900001. [Google Scholar] [CrossRef]
- Yu, L.; Li, Z.; Mei, H.; Li, W.; Chen, D.; Liu, L.; Zhang, Z.; Sun, Y.; Song, F.; Chen, W.; et al. Patient-derived organoids of bladder cancer recapitulate antigen expression profiles and serve as a personal evaluation model for CAR-T cells in vitro. Clin. Transl. Immunol. 2021, 10, e1248. [Google Scholar] [CrossRef]
- Breznik, B.; Ko, M.-W.; Tse, C.; Chen, P.-C.; Senjor, E.; Majc, B.; Habič, A.; Angelillis, N.; Novak, M.; Župunski, V.; et al. Infiltrating natural killer cells bind, lyse and increase chemotherapy efficacy in glioblastoma stem-like tumorospheres. Commun. Biol. 2022, 5, 436. [Google Scholar] [CrossRef]
- Voabil, P.; de Bruijn, M.; Roelofsen, L.M.; Hendriks, S.H.; Brokamp, S.; Braber, M.V.D.; Broeks, A.; Sanders, J.; Herzig, P.; Zippelius, A.; et al. An ex vivo tumor fragment platform to dissect response to PD-1 blockade in cancer. Nat. Med. 2021, 27, 1250–1261. [Google Scholar] [CrossRef]
- Martin, S.Z.; Wagner, D.C.; Hörner, N.; Horst, D.; Lang, H.; Tagscherer, K.E.; Roth, W. Ex vivo tissue slice culture system to measure drug-response rates of hepatic metastatic colorectal cancer. BMC Cancer 2019, 19, 1030. [Google Scholar] [CrossRef]
- Kong, J.C.H.; Guerra, G.R.; Millen, R.M.; Roth, S.; Xu, H.; Neeson, P.J.; Darcy, P.K.; Kershaw, M.H.; Sampurno, S.; Malaterre, J.; et al. Tumor-Infiltrating Lymphocyte Function Predicts Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. JCO Precis. Oncol. 2018, 2, 1–15. [Google Scholar] [CrossRef]
- Koh, V.; Chakrabarti, J.; Torvund, M.; Steele, N.; Hawkins, J.A.; Ito, Y.; Wang, J.; Helmrath, M.A.; Merchant, J.L.; Ahmed, S.A.; et al. Hedgehog transcriptional effector GLI mediates mTOR-Induced PD-L1 expression in gastric cancer organoids. Cancer Lett. 2021, 518, 59–71. [Google Scholar] [CrossRef]
- Fan, T.W.; Higashi, R.M.; Song, H.; Daneshmandi, S.; Mahan, A.L.; Purdom, M.S.; Bocklage, T.J.; Pittman, T.A.; He, D.; Wang, C.; et al. Innate immune activation by checkpoint inhibition in human patient-derived lung cancer tissues. eLife 2021, 10, e69578. [Google Scholar] [CrossRef]
- Gonzalez-Exposito, R.; Semiannikova, M.; Griffiths, B.; Khan, K.; Barber, L.J.; Woolston, A.; Spain, G.; Von Loga, K.; Challoner, B.; Patel, R.; et al. CEA expression heterogeneity and plasticity confer resistance to the CEA-targeting bispecific immunotherapy antibody cibisatamab (CEA-TCB) in patient-derived colorectal cancer organoids. J. Immunother. Cancer 2019, 7, 101. [Google Scholar] [CrossRef]
- Olivera, I.; Sanz-Pamplona, R.; Bolaños, E.; Rodriguez, I.; Etxeberria, I.; Cirella, A.; Egea, J.; Garasa, S.; Migueliz, I.; Eguren-Santamaria, I.; et al. A Therapeutically Actionable Protumoral Axis of Cytokines Involving IL-8, TNFalpha, and IL-1beta. Cancer Discov. 2022, 12, 2140–2157. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, F.; Wang, L.; Diao, Y.; Chen, D. TNF-alpha promotes the malignant transformation of intestinal stem cells through the NF-kappaB and Wnt/beta-catenin signaling pathways. Oncol. Rep. 2020, 44, 577–588. [Google Scholar] [CrossRef]
- Lucarini, V.; Buccione, C.; Ziccheddu, G.; Peschiaroli, F.; Sestili, P.; Puglisi, R.; Mattia, G.; Zanetti, C.; Parolini, I.; Bracci, L.; et al. Combining Type I Interferons and 5-Aza-2′-Deoxycitidine to Improve Anti-Tumor Response against Melanoma. J. Investig. Dermatol. 2017, 137, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Herpers, B.; Eppink, B.; James, M.I.; Cortina, C.; Cañellas-Socias, A.; Boj, S.F.; Hernando-Momblona, X.; Glodzik, D.; Roovers, R.C.; van de Wetering, M.; et al. Functional patient-derived organoid screenings identify MCLA-158 as a therapeutic EGFR x LGR5 bispecific antibody with efficacy in epithelial tumors. Nat. Cancer 2022, 3, 418–436. [Google Scholar] [CrossRef] [PubMed]
- Rau, A.; Janssen, N.; Kühl, L.; Sell, T.; Kalmykova, S.; Mürdter, T.E.; Dahlke, M.-H.; Sers, C.; Morkel, M.; Schwab, M.; et al. Triple Targeting of HER Receptors Overcomes Heregulin-Mediated Resistance to EGFR Blockade in Colorectal Cancer. Mol. Cancer Ther. 2022, 21, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.; Keany, M.P.; Dong, H.; Al-Alem, L.F.; Pandya, U.M.; Lazo, S.; Boehnke, K.; Lynch, K.N.; Xu, R.; Zarrella, D.T.; et al. Enhanced Efficacy of Simultaneous PD-1 and PD-L1 Immune Checkpoint Blockade in High-Grade Serous Ovarian Cancer. Cancer Res. 2021, 81, 158–173. [Google Scholar] [CrossRef]
- Huang, L.; Rong, Y.; Tang, X.; Yi, K.; Qi, P.; Hou, J.; Liu, W.; He, Y.; Gao, X.; Yuan, C.; et al. Engineered exosomes as an in situ DC-primed vaccine to boost antitumor immunity in breast cancer. Mol. Cancer 2022, 21, 45. [Google Scholar] [CrossRef]
- Wang, W.; Yuan, T.; Ma, L.; Zhu, Y.; Bao, J.; Zhao, X.; Zhao, Y.; Zong, Y.; Zhang, Y.; Yang, S.; et al. Hepatobiliary Tumor Organoids Reveal HLA Class I Neoantigen Landscape and Antitumoral Activity of Neoantigen Peptide Enhanced with Immune Checkpoint Inhibitors. Adv. Sci. 2022, 9, e2105810. [Google Scholar] [CrossRef]
- Izadpanah, A.; Delirezh, N.; Mahmodlou, R. Ex Vivo Optimization of Glucose-Regulated Protein 94/Glycoprotein 96 Expressions in Mammospheres; Implication for Breast Cancer Immunotherapy. Cell J. 2022, 24, 261–266. [Google Scholar]
- Trofimova, O.; Korotkaja, K.; Skrastina, D.; Jansons, J.; Spunde, K.; Isaguliants, M.; Zajakina, A. Alphavirus-Driven Interferon Gamma (IFNg) Expression Inhibits Tumor Growth in Orthotopic 4T1 Breast Cancer Model. Vaccines 2021, 9, 1247. [Google Scholar] [CrossRef]
- Yao, X.; Dong, Z.; Zhang, Q.; Wang, Q.; Lai, D. Epithelial ovarian cancer stem-like cells expressing alpha-gal epitopes increase the immunogenicity of tumor associated antigens. BMC Cancer 2015, 15, 956. [Google Scholar] [CrossRef]
- Ligtenberg, M.A.; Witt, K.; Galvez-Cancino, F.; Sette, A.; Lundqvist, A.; Lladser, A.; Kiessling, R. Cripto-1 vaccination elicits protective immunity against metastatic melanoma. Oncoimmunology 2016, 5, e1128613. [Google Scholar] [CrossRef]
- Witt, K.; Ligtenberg, M.A.; Conti, L.; Lanzardo, S.; Ruiu, R.; Wallmann, T.; Tufvesson-Stiller, H.; Chambers, B.J.; Rolny, C.; Lladser, A.; et al. Cripto-1 Plasmid DNA Vaccination Targets Metastasis and Cancer Stem Cells in Murine Mammary Carcinoma. Cancer Immunol. Res. 2018, 6, 1417–1425. [Google Scholar] [CrossRef]
- Carter, M.E.; Hartkopf, A.D.; Wagner, A.; Volmer, L.L.; Brucker, S.Y.; Berchtold, S.; Lauer, U.M.; Koch, A. A Three-Dimensional Organoid Model of Primary Breast Cancer to Investigate the Effects of Oncolytic Virotherapy. Front. Mol. Biosci. 2022, 9, 826302. [Google Scholar] [CrossRef]
- Harryvan, T.J.; Golo, M.; Dam, N.; Schoonderwoerd, M.J.A.; Farshadi, E.A.; Hornsveld, M.; Hoeben, R.C.; Hawinkels, L.J.A.C.; Kemp, V. Gastrointestinal cancer-associated fibroblasts expressing Junctional Adhesion Molecule-A are amenable to infection by oncolytic reovirus. Cancer Gene Ther. 2022, 29, 1918–1929. [Google Scholar] [CrossRef]
- Zhu, Z.; Mesci, P.; Bernatchez, J.A.; Gimple, R.C.; Wang, X.; Schafer, S.T.; Wettersten, H.I.; Beck, S.; Clarck, S.T.; Wu, Q.; et al. Zika Virus Targets Glioblastoma Stem Cells through a SOX2-Integrin αvβ5 Axis. Cell Stem Cell 2020, 26, 187–204.e10. [Google Scholar] [CrossRef]
- Engel, R.M.; Chan, W.H.; Nickless, D.; Hlavca, S.; Richards, E.; Kerr, G.; Oliva, K.; McMurrick, P.J.; Jardé, T.; Abud, H.E. Patient-Derived Colorectal Cancer Organoids Upregulate Revival Stem Cell Marker Genes following Chemotherapeutic Treatment. J. Clin. Med. 2020, 9, 128. [Google Scholar] [CrossRef]
- Narasimhan, V.; Wright, J.A.; Churchill, M.; Wang, T.; Rosati, R.; Lannagan, T.R.M.; Vrbanac, L.; Richardson, A.B.; Kobayashi, H.; Price, T.; et al. Medium-Throughput Drug Screening of Patient-Derived Organoids from Colorectal Peritoneal Metastases to Direct Personalized Therapy. Clin. Cancer Res. 2020, 26, 3662–3670. [Google Scholar] [CrossRef]
- de Witte, C.J.; Valle-Inclan, J.E.; Hami, N.; Lõhmussaar, K.; Kopper, O.; Vreuls, C.P.H.; Jonges, G.N.; van Diest, P.; Nguyen, L.; Clevers, H.; et al. Patient-Derived Ovarian Cancer Organoids Mimic Clinical Response and Exhibit Heterogeneous Inter- and Intrapatient Drug Responses. Cell Rep. 2020, 31, 107762. [Google Scholar] [CrossRef]
- Tsuyoshi, H.; Inoue, D.; Kurokawa, T.; Yoshida, Y. Hyperthermic intraperitoneal chemotherapy (HIPEC) for gynecological cancer. J. Obstet. Gynaecol. Res. 2020, 46, 1661–1671. [Google Scholar] [CrossRef]
- Leiting, J.L.; Cloyd, J.M.; Ahmed, A.; Fournier, K.; Lee, A.J.; Dessureault, S.; Felder, S.; Veerapong, J.; Baumgartner, J.M.; Clarke, C.; et al. Comparison of open and closed hyperthermic intraperitoneal chemotherapy: Results from the United States hyperthermic intraperitoneal chemotherapy collaborative. World J. Gastrointest. Oncol. 2020, 12, 756–767. [Google Scholar] [CrossRef]
- June, C.H.; O’Connor, R.S.; Kawalekar, O.U.; Ghassemi, S.; Milone, M.C. CAR T cell immunotherapy for human cancer. Science 2018, 359, 1361–1365. [Google Scholar] [CrossRef]
- Upadhaya, S.; Hubbard-Lucey, V.M.; Yu, J.X. Immuno-oncology drug development forges on despite COVID-19. Nat. Rev. Drug Discov. 2020, 19, 751–752. [Google Scholar] [CrossRef] [PubMed]
- Zumwalde, N.A.; Haag, J.D.; Sharma, D.; Mirrielees, J.A.; Wilke, L.G.; Gould, M.N.; Gumperz, J.E. Analysis of Immune Cells from Human Mammary Ductal Epithelial Organoids Reveals Vdelta2+ T Cells that Efficiently Target Breast Carcinoma Cells in the Presence of Bisphosphonate. Cancer Prev. Res. 2016, 9, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Ayuso, J.M.; Truttschel, R.; Gong, M.M.; Humayun, M.; Virumbrales-Munoz, M.; Vitek, R.; Felder, M.; Gillies, S.D.; Sondel, P.; Wisinski, K.B.; et al. Evaluating natural killer cell cytotoxicity against solid tumors using a microfluidic model. Oncoimmunology 2019, 8, 1553477. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Dong, H.; Liang, Y.; Ham, J.D.; Rizwan, R.; Chen, J. CAR-NK cells: A promising cellular immunotherapy for cancer. EBioMedicine 2020, 59, 102975. [Google Scholar] [CrossRef]
- Schnalzger, T.E.; De Groot, M.H.; Zhang, C.; Mosa, M.H.; Michels, B.E.; Röder, J.; Darvishi, T.; Wels, W.S.; Farin, H.F. 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 2019, 38, e100928. [Google Scholar] [CrossRef]
- Wu, T.; Dai, Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017, 387, 61–68. [Google Scholar] [CrossRef]
- Kirkwood, J.M.; Butterfield, L.H.; Tarhini, A.A.; Zarour, H.; Kalinski, P.; Ferrone, S. Immunotherapy of cancer in 2012. CA Cancer J. Clin. 2012, 62, 309–335. [Google Scholar] [CrossRef]
- Yuki, K.; Cheng, N.; Nakano, M.; Kuo, C.J. Organoid Models of Tumor Immunology. Trends Immunol. 2020, 41, 652–664. [Google Scholar] [CrossRef]
- Scognamiglio, G.; De Chiara, A.; Parafioriti, A.; Armiraglio, E.; Fazioli, F.; Gallo, M.; Aversa, L.; Camerlingo, R.; Cacciatore, F.; Colella, G. Patient-derived organoids as a potential model to predict response to PD-1/PD-L1 checkpoint inhibitors. Br. J. Cancer 2019, 121, 979–982. [Google Scholar] [CrossRef]
- Saleh, R.; Toor, S.M.; Al-Ali, D.; Nair, V.S.; Elkord, E. Blockade of PD-1, PD-L1, and TIM-3 Altered Distinct Immune- and Cancer-Related Signaling Pathways in the Transcriptome of Human Breast Cancer Explants. Genes 2020, 11, 703. [Google Scholar] [CrossRef]
- Courau, T.; Bonnereau, J.; Chicoteau, J.; Bottois, H.; Remark, R.; Miranda, L.A.; Toubert, A.; Blery, M.; Aparicio, T.; Allez, M.; et al. Cocultures of human colorectal tumor spheroids with immune cells reveal the therapeutic potential of MICA/B and NKG2A targeting for cancer treatment. J. Immunother. Cancer 2019, 7, 74. [Google Scholar] [CrossRef]
- Votanopoulos, K.I.; Forsythe, S.; Msc, H.S.; Mazzocchi, A.; Aleman, J.; Miller, L.; Levine, E.; Triozzi, P.; Skardal, A. Model of Patient-Specific Immune-Enhanced Organoids for Immunotherapy Screening: Feasibility Study. Ann. Surg. Oncol. 2020, 27, 1956–1967. [Google Scholar] [CrossRef]
- Jenkins, R.W.; Aref, A.R.; Lizotte, P.H.; Ivanova, E.; Stinson, S.; Zhou, C.W.; Bowden, M.; Deng, J.; Liu, H.; Miao, D.; et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov. 2018, 8, 196–215. [Google Scholar] [CrossRef]
- Aref, A.R.; Campisi, M.; Ivanova, E.; Portell, A.; Larios, D.; Piel, B.P.; Mathur, N.; Zhou, C.; Coakley, R.V.; Bartels, A.; et al. 3D microfluidic ex vivo culture of organotypic tumor spheroids to model immune checkpoint blockade. Lab Chip 2018, 18, 3129–3143. [Google Scholar] [CrossRef]
- Sun, W.; Luo, Z.; Lee, J.; Kim, H.-J.; Lee, K.; Tebon, P.; Feng, Y.; Dokmeci, M.R.; Sengupta, S.; Khademhosseini, A. Organ-On-A-Chip for Cancer and Immune Organs Modeling. Adv. Healthc. Mater. 2019, 8, e1801363. [Google Scholar] [CrossRef]
- Cui, X.; Ma, C.; Vasudevaraja, V.; Serrano, J.; Tong, J.; Peng, Y.; Shen, G.; Frenster, J.; Morales, R.-T.T.; Qian, W.; et al. Dissecting the immunosuppressive tumor microenvironments in Glioblastoma-on-a-Chip for optimized PD-1 immunotherapy. eLife 2020, 9, e52253. [Google Scholar] [CrossRef]
- Moore, N.; Doty, D.; Zielstorff, M.; Kariv, I.; Moy, L.Y.; Gimbel, A.; Chevillet, J.R.; Lowry, N.; Santos, J.; Mott, V.; et al. A multiplexed microfluidic system for evaluation of dynamics of immune-tumor interactions. Lab Chip 2018, 18, 1844–1858. [Google Scholar] [CrossRef]
- Zhou, G.; Lieshout, R.; van Tienderen, G.S.; de Ruiter, V.; van Royen, M.E.; Boor, P.P.C.; Magré, L.; Desai, J.; Köten, K.; Kan, Y.Y.; et al. Modelling immune cytotoxicity for cholangiocarcinoma with tumour-derived organoids and effector T cells. Br. J. Cancer 2022, 127, 649–660. [Google Scholar] [CrossRef]
- Kang, S.; Tanaka, T.; Narazaki, M.; Kishimoto, T. Targeting Interleukin-6 Signaling in Clinic. Immunity 2019, 50, 1007–1023. [Google Scholar] [CrossRef]
- Hollingsworth, R.E.; Jansen, K. Turning the corner on therapeutic cancer vaccines. NPJ Vaccines 2019, 4, 7. [Google Scholar] [CrossRef]
- O’Keeffe, M.; Mok, W.H.; Radford, K.J. Human dendritic cell subsets and function in health and disease. Cell Mol. Life Sci. 2015, 72, 4309–4325. [Google Scholar] [CrossRef] [PubMed]
- Caushi, J.X.; Zhang, J.; Ji, Z.; Vaghasia, A.; Zhang, B.; Hsiue, E.H.-C.; Mog, B.J.; Hou, W.; Justesen, S.; Blosser, R.; et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Naturie 2021, 596, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Demmers, L.C.; Kretzschmar, K.; Van Hoeck, A.; Bar-Epraïm, Y.E.; Toorn, H.W.P.V.D.; Koomen, M.; van Son, G.; van Gorp, J.; Pronk, A.; Smakman, N.; et al. Single-cell derived tumor organoids display diversity in HLA class I peptide presentation. Nat. Commun. 2020, 11, 5338. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Fan, H.; Li, Y.; Zheng, H.; Li, X.; Li, C.; Chen, L.; Ju, Y.; Meng, S. CD133 epitope vaccine with gp96 as adjuvant elicits an antitumor T cell response against leukemia. Sheng Wu Gong Cheng Xue Bao 2017, 33, 1006–1017. [Google Scholar]
- Raimondi, G.; Mato-Berciano, A.; Pascual-Sabater, S.; Rovira-Rigau, M.; Cuatrecasas, M.; Fondevila, C.; Sánchez-Cabús, S.; Begthel, H.; Boj, S.F.; Clevers, H.; et al. Patient-derived pancreatic tumour organoids identify therapeutic responses to oncolytic adenoviruses. EBioMedicine 2020, 56, 102786. [Google Scholar] [CrossRef]
- Pascual-Sabater, S.; Raimondi, G.; Mato-Berciano, A.; Vaquero, E.C.; Ausania, F.; Fillat, C. Preclinical testing of oncolytic adenovirus sensitivity in patient-derived tumor organoids. STAR Protoc. 2021, 2, 101017. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Grebenyuk, S.; Ranga, A. Engineering Organoid Vascularization. Front. Bioeng. Biotechnol. 2019, 7, 39. [Google Scholar] [CrossRef]
- Miller, J.S.; Stevens, K.R.; Yang, M.T.; Baker, B.M.; Nguyen, D.-H.T.; Cohen, D.M.; Toro, E.; Chen, A.A.; Galie, P.A.; Yu, X.; et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 2012, 11, 768–774. [Google Scholar] [CrossRef]
- Huber, B.; Engelhardt, S.; Meyer, W.; Krüger, H.; Wenz, A.; Schönhaar, V.; Tovar, G.E.M.; Kluger, P.J.; Borchers, K. Blood-Vessel Mimicking Structures by Stereolithographic Fabrication of Small Porous Tubes Using Cytocompatible Polyacrylate Elastomers, Biofunctionalization and Endothelialization. J. Funct. Biomater. 2016, 7, 11. [Google Scholar] [CrossRef]
- Pittman, R.N. Oxygen transport in the microcirculation and its regulation. Microcirculation 2013, 20, 117–137. [Google Scholar] [CrossRef]
- Badalamenti, G.; Fanale, D.; Incorvaia, L.; Barraco, N.; Listi, A.; Maragliano, R.; Vicenzi, B.; Calo, V.; Iovanna, J.L.; Bazan, V.; et al. Role of tumor-infiltrating lymphocytes in patients with solid tumors: Can a drop dig a stone? Cell. Immunol 2019, 343, 103753. [Google Scholar] [CrossRef]
- Nashimoto, Y.; Hayashi, T.; Kunita, I.; Nakamasu, A.; Torisawa, Y.-S.; Nakayama, M.; Takigawa-Imamura, H.; Kotera, H.; Nishiyama, K.; Miura, T.; et al. Integrating perfusable vascular networks with a three-dimensional tissue in a microfluidic device. Integr. Biol. 2017, 9, 506–518. [Google Scholar] [CrossRef]


| Cell Sources | Advantages | Inconvenience |
|---|---|---|
| Cancer cell lines |
|
|
| Primary cells |
|
|
| Genetically modified cells |
|
|
| Pluripotent stem cells |
|
|
| Mixture of different cell sources |
|
|
| Organoids including immune cells |
|
|
| Method or Technology | Advantages | Inconvenient |
|---|---|---|
| Hanging drop method |
|
|
| Low-attachment plate method |
|
|
| Microwell method |
|
|
| Magnetic levitation |
|
May require additional steps to remove the beads after the cells have formed spheroids or organoids |
| Rotatory systems (Bioreactors and spinner flask) |
|
|
| Extracellular-matrix-based method |
|
|
| Application | Cell Source | Culture System | Cancer Type | References |
|---|---|---|---|---|
| Chemotherapy | Primary biopsy | Embedded or resuspended in Matrigel, BME Type 2, ECM-mimicking HA/collagen-based Hydrogel | Colorectal, pancreatic cancer, metastatic colorectal cancer, breast cancer, ovarian cancer and appendiceal cancer | [35,62,63,119,142,143,144] |
| CRISPR/Cas9 therapy | HEK293T OVCAR5 cells HCT116 cells iPSCs Human and mice biopsies | Embedded in Matrigel, ultra- low-attachment 6 well culture plates, Drops of Matrigel | Mice tubo-ovarian carcinoma, ovarian cancer cells, colon cancer, murine and human small intestine and pancreatic ductal adenocarcinoma | [70,84,85,86,145,146,147] |
| Adoptive cell transfer therapy | Tumors tissues Biobank PDOs SKOV3 human epithelial ovarian cancer cells biopsies | Embedded in Geltrex Basement membrane extract (BME, Cultrex), PDMS fluidic chamber, ultra-low attachment 6-well culture plates, Matrigel, agar-coated plates, embedded in growth factor reduced Matrigel | Colorectal cancer, non-small cancer lung cells, breast cancer and normal breast, Human ovarian cancer cells, glioblastoma and bladder cancer | [57,148,149,150,151,152] |
| Immune checkpoint inhibitors | Biopsies | ALI, Special cell-culture inserts, embedding in Geltrex, BME, multiwell plates, embedded in 3% low-melting agarose | Melanoma, colorectal cancer, non-small cancer lung cells, breast cancer, ovarian cancer or renal cell carcinoma, colorectal carcinoma, gastric cancer and lung cancer | [64,148,153,154,155,156,157] |
| Monoclonal and bispecific antibodies | Biopsies, colon epithelial cell line NCM460, A375 human melanoma cells, Prospect C and Prospect R trials | Resuspended in 100% Matrigel, ultra-low attachment culture dishes, embedded in growth factor reduced Matrigel | Human colorectal cancer, colon cancer line, melanoma cancer and metastatic colorectal cancers | [158,159,160,161,162,163,164] |
| Cancer vaccines | MDA-MB-231, MCF7, MDA-MB-435S, SKBR3 cancer cells, 4T1/eGFP cells, SKOV3 cells, B16F10 cells TUBO cells, 4T1 cells, HT29 colorectal cancer cells, biopsies | Embedded in 4% agarose, BME; cultrex PC BME RGF type 2, 24-well plates a thin layer of 1.5% agarose, ultralow attachment plates, cancer cells grow as non-adherent spheroid cells, ultra-low attachment plates, Matrigel | Breast cancer, hepatobiliary tumor, colorectal cancers, ovarian cancer, metastatic melanoma, murine mammary carcinoma and glioblastoma. | [165,166,167,168,169,170,171,172,173,174] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Harane, S.; Zidi, B.; El Harane, N.; Krause, K.-H.; Matthes, T.; Preynat-Seauve, O. Cancer Spheroids and Organoids as Novel Tools for Research and Therapy: State of the Art and Challenges to Guide Precision Medicine. Cells 2023, 12, 1001. https://doi.org/10.3390/cells12071001
El Harane S, Zidi B, El Harane N, Krause K-H, Matthes T, Preynat-Seauve O. Cancer Spheroids and Organoids as Novel Tools for Research and Therapy: State of the Art and Challenges to Guide Precision Medicine. Cells. 2023; 12(7):1001. https://doi.org/10.3390/cells12071001
Chicago/Turabian StyleEl Harane, Sanae, Bochra Zidi, Nadia El Harane, Karl-Heinz Krause, Thomas Matthes, and Olivier Preynat-Seauve. 2023. "Cancer Spheroids and Organoids as Novel Tools for Research and Therapy: State of the Art and Challenges to Guide Precision Medicine" Cells 12, no. 7: 1001. https://doi.org/10.3390/cells12071001
APA StyleEl Harane, S., Zidi, B., El Harane, N., Krause, K.-H., Matthes, T., & Preynat-Seauve, O. (2023). Cancer Spheroids and Organoids as Novel Tools for Research and Therapy: State of the Art and Challenges to Guide Precision Medicine. Cells, 12(7), 1001. https://doi.org/10.3390/cells12071001






