The RUNX Family of Proteins, DNA Repair, and Cancer
Abstract
:1. Introduction
2. RUNX1 Leukemic Fusions in Hematopoietic Malignancies and Genomic Instability
2.1. RUNX1-ETO and Genomic Instability
2.2. ETV6-RUNX1 and Genomic Instability
2.3. RUNX1-EVI1 and Genomic Instability
3. RUNX1 Mutations in Cancers and Genomic Instability
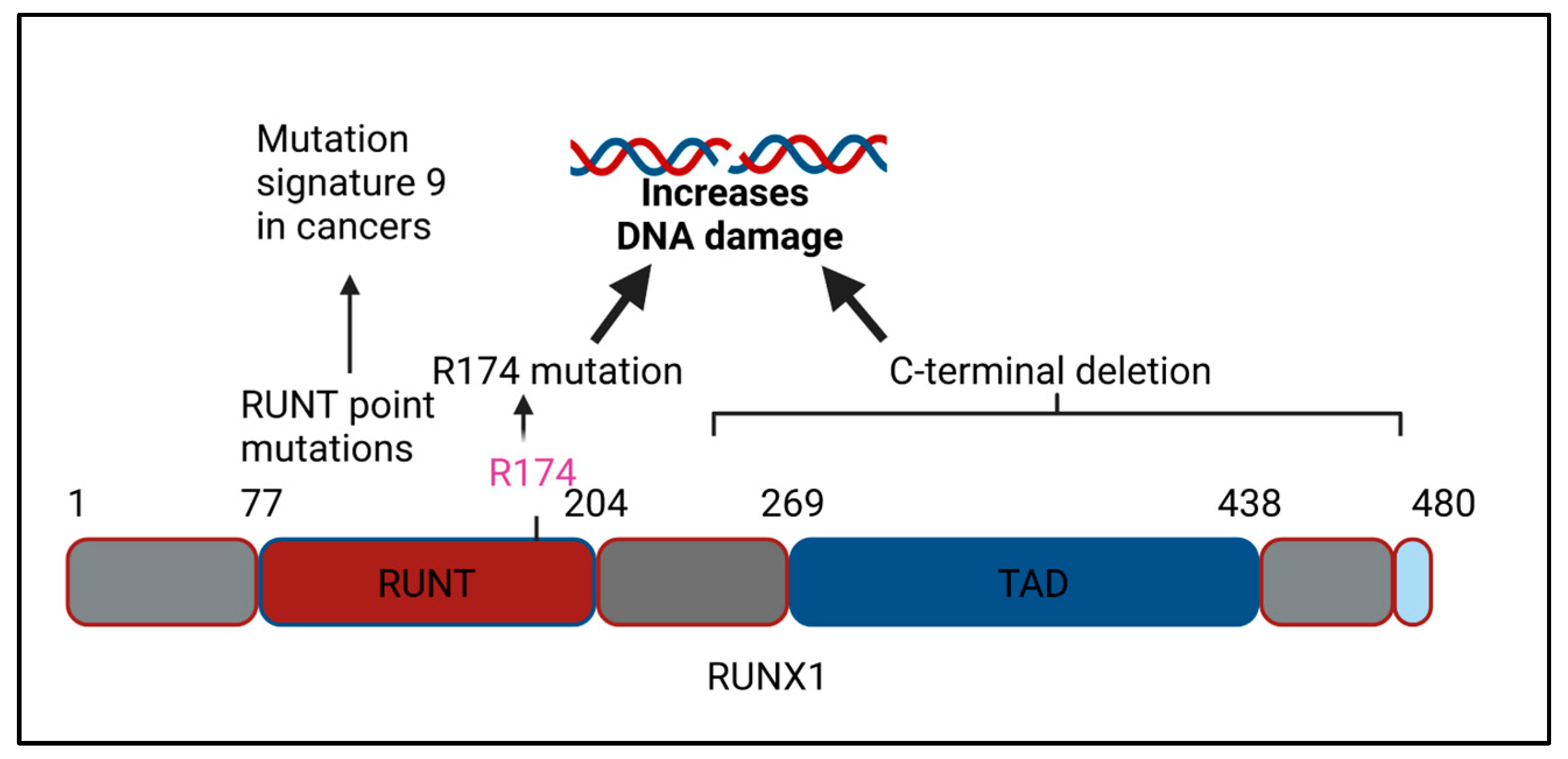
3.1. RUNX1 C-Terminal Deletions and Genomic Instability
3.2. RUNX1 RUNT Domain Mutations and Genomic Instability
3.3. RUNX1 CML Blast Crisis Mutations and Genomic Instability
3.4. RUNX1 MDS Mutations and Genomic Instability
4. RUNX3 Defects in Human Cancers and Genomic Instability
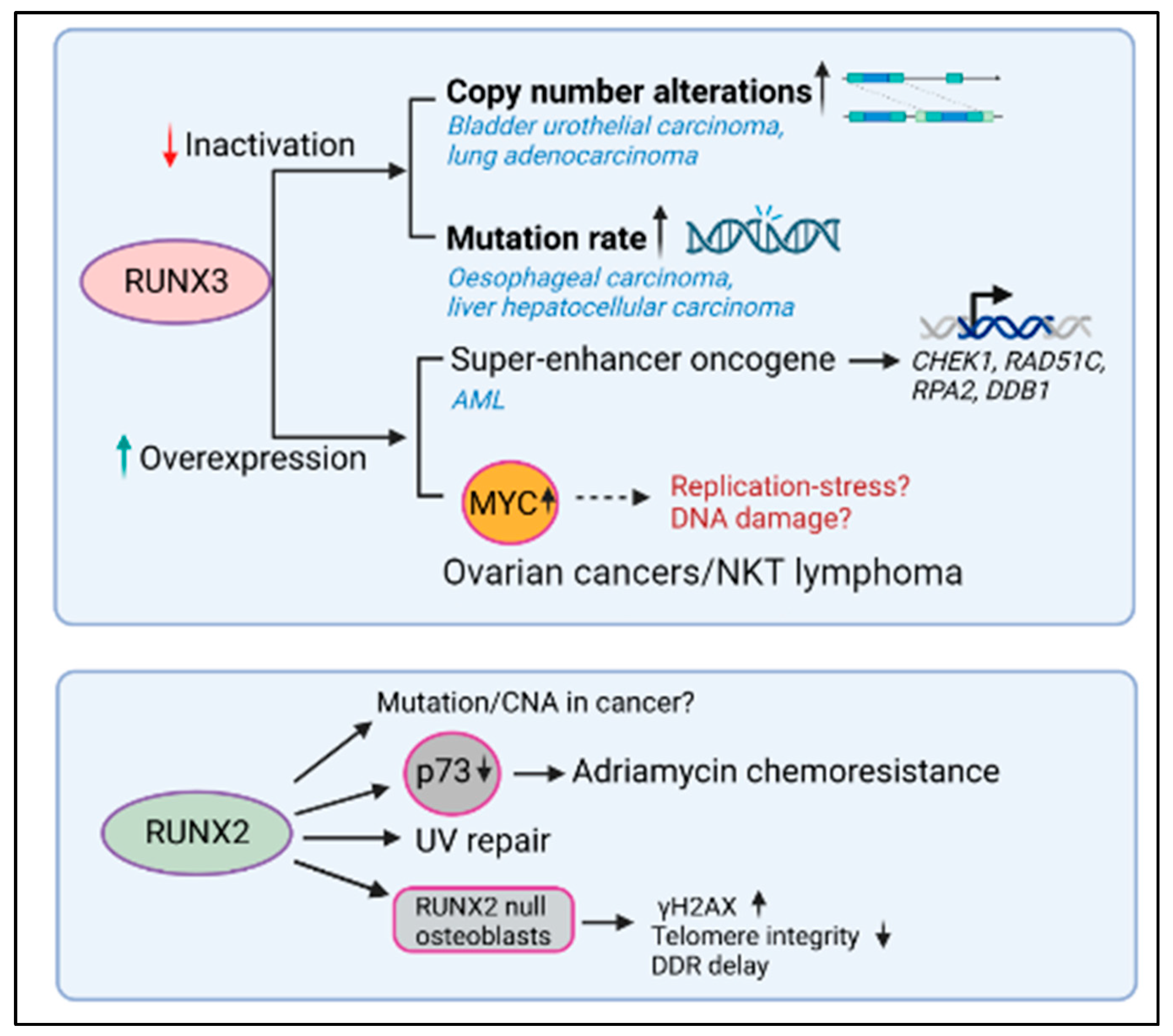
4.1. RUNX3 Inactivation and Genomic Instability
4.2. RUNX3 Activation and Genomic Instability
5. RUNX2 Defects in Cancers and Genomic Instability
RUNX2 Dysregulation and Genomic Instability
6. Molecular Mechanisms Underlying RUNX Dysregulation and Genomic Instability in Cancer
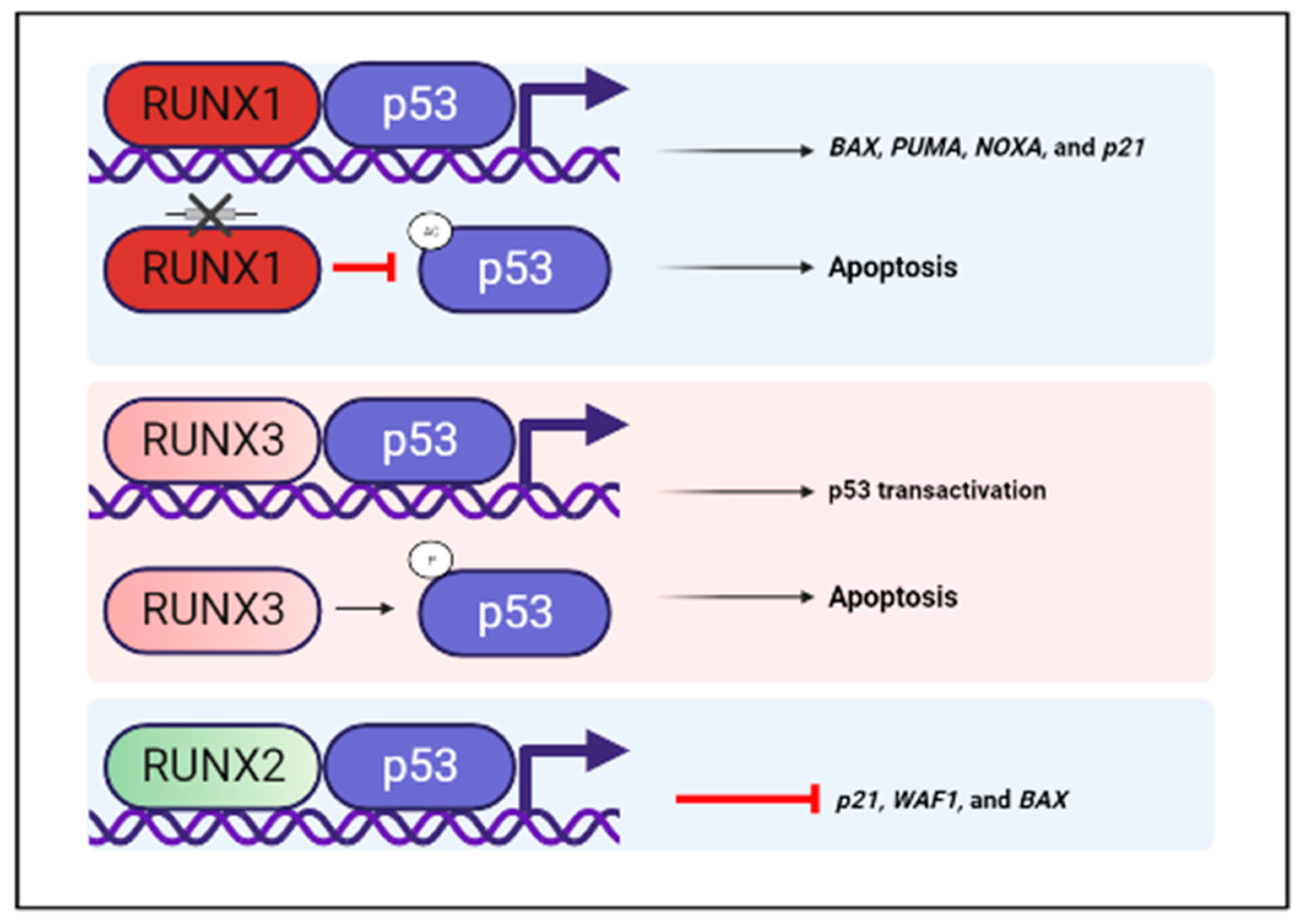
6.1. RUNX and p53-A Crosstalk between Two Tumor Suppressors
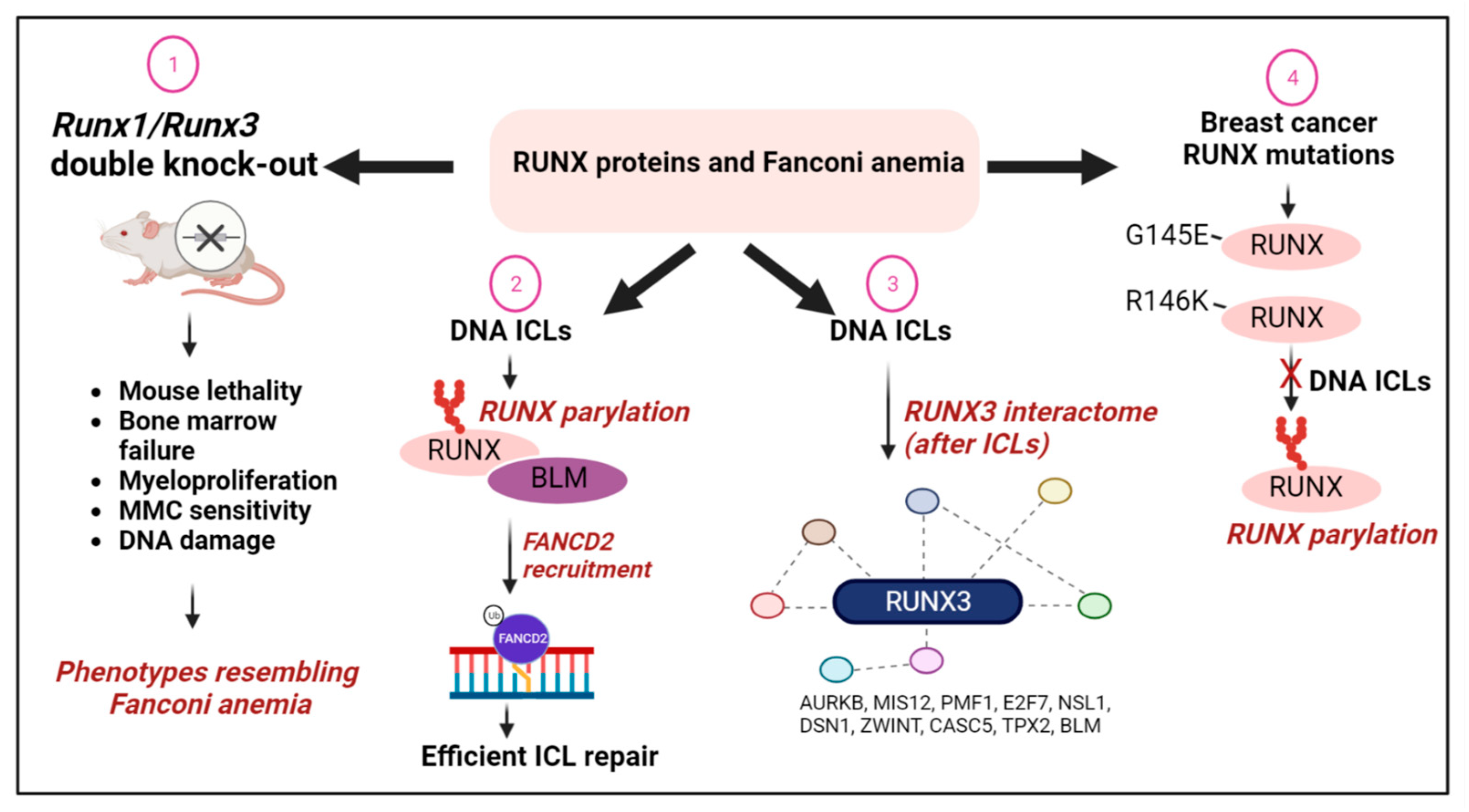
6.2. RUNX Proteins and the Fanconi Anemia Pathway of DNA Repair
6.3. The Association of RUNX Proteins with DNA Repair Complexes
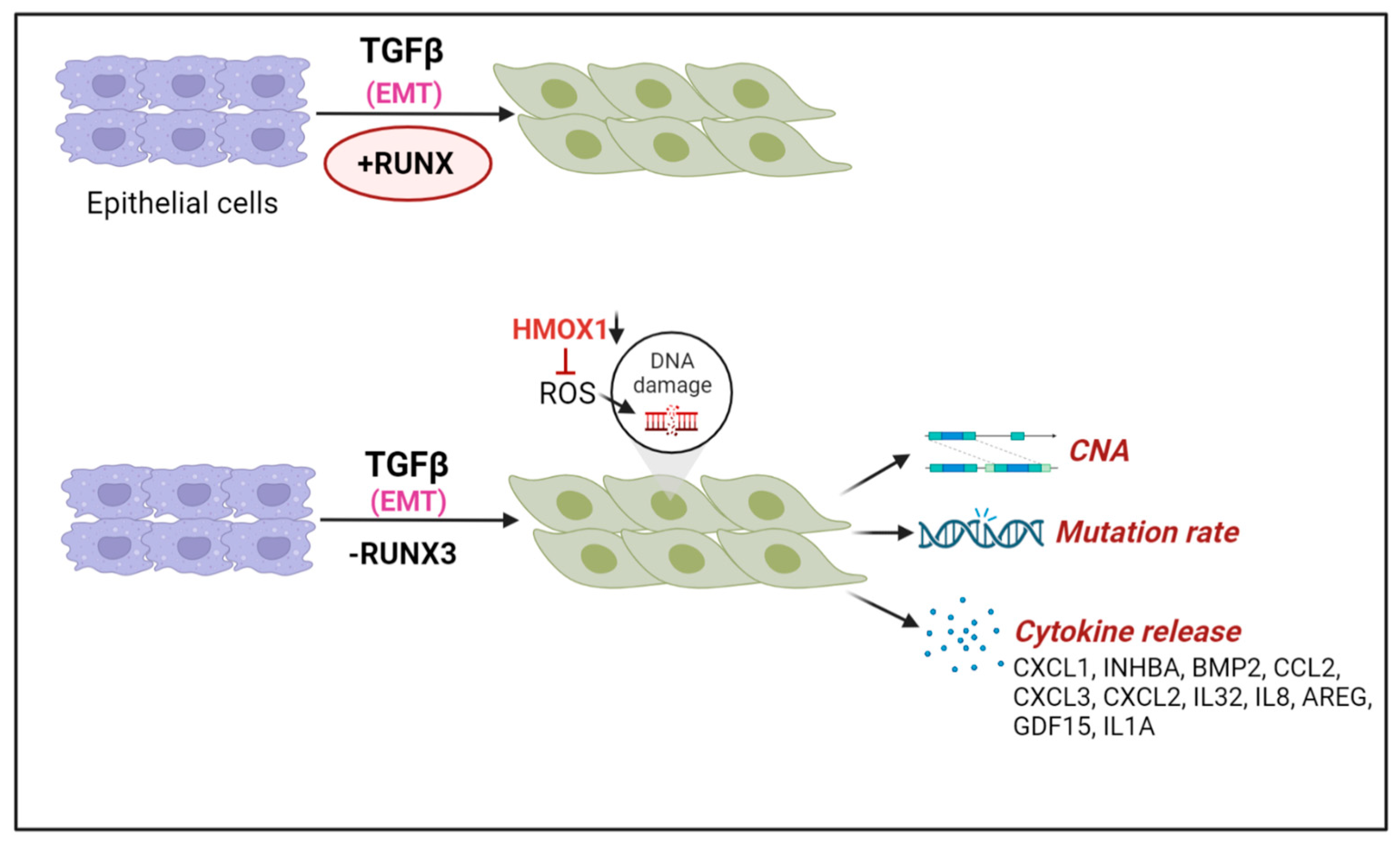
6.4. TGFβ Driven Epithelial-Mesenchymal Transition (EMT), RUNX1/RUNX3 Loss, and DNA Damage
6.5. Oxidative Stress and RUNX Factors
6.6. RUNX Deficiency, Inflammation, and DNA Damage
7. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ito, Y.; Bae, S.-C.; Chuang, L.S.H. The RUNX family: Developmental regulators in cancer. Nat. Rev. Cancer 2015, 15, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Mevel, R.; Draper, J.E.; Lie-A-Ling, M.; Kouskoff, V.; Lacaud, G. RUNX transcription factors: Orchestrators of development. Development 2019, 146, dev148296. [Google Scholar] [CrossRef] [Green Version]
- Tahirov, T.H.; Inoue-Bungo, T.; Morii, H.; Fujikawa, A.; Sasaki, M.; Kimura, K.; Shiina, M.; Sato, K.; Kumasaka, T.; Yamamoto, M.; et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell 2001, 104, 755–767. [Google Scholar] [CrossRef]
- Ito, Y. RUNX Genes in Development and Cancer: Regulation of Viral Gene Expression and the Discovery of RUNX Family Genes. Adv. Cancer Res. 2008, 99, 33–76. [Google Scholar] [CrossRef]
- Chuang, L.S.H.; Ito, K.; Ito, Y. RUNX family: Regulation and diversification of roles through interacting proteins. Int. J. Cancer 2013, 132, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic instability—An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef] [Green Version]
- Peterson, L.F.; Zhang, D.-E. The 8;21 translocation in leukemogenesis. Oncogene 2004, 23, 4255–4262. [Google Scholar] [CrossRef] [Green Version]
- Zelent, A.; Greaves, M.; Enver, T. Role of the TEL-AML1 fusion gene in the molecular pathogenesis of childhood acute lymphoblastic leukaemia. Oncogene 2004, 23, 4275–4283. [Google Scholar] [CrossRef] [Green Version]
- Rubin, C.M.; Larson, R.; Anastasi, J.; Winter, J.N.; Thangavelu, M.; Vardiman, J.W.; Rowley, J.D.; Le Beau, M.M. t(3;21)(q26;q22): A recurring chromosomal abnormality in therapy-related myelodysplastic syndrome and acute myeloid leukemia. Blood 1990, 76, 2594–2598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Tarle, S.A.; Hajra, A.; Claxton, D.F.; Marlton, P.; Freedman, M.; Siciliano, M.J.; Collins, F.S. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science 1993, 261, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Sood, R.; Kamikubo, Y.; Liu, P. Role of RUNX1 in hematological malignancies. Blood 2017, 129, 2070–2082. [Google Scholar] [CrossRef] [Green Version]
- Lam, K.; Zhang, D.E. RUNX1 and RUNX1-ETO: Roles in hematopoiesis and leukemogenesis. Front. Biosci. 2012, 17, 1120–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.-J.; Wang, Z.; Wang, L.; Jiang, Y.; Kost, N.; Soong, T.D.; Chen, W.-Y.; Tang, Z.; Nakadai, T.; Elemento, O.; et al. A stable transcription factor complex nucleated by oligomeric AML1–ETO controls leukaemogenesis. Nature 2013, 500, 93–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ptasinska, A.; Assi, S.A.; Martinez-Soria, N.; Imperato, M.R.; Piper, J.; Cauchy, P.; Pickin, A.; James, S.R.; Hoogenkamp, M.; Williamson, D.; et al. Identification of a Dynamic Core Transcriptional Network in t(8;21) AML that Regulates Differentiation Block and Self-Renewal. Cell Rep. 2014, 8, 1974–1988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stengel, K.R.; Ellis, J.D.; Spielman, C.L.; Bomber, M.L.; Hiebert, S.W. Definition of a small core transcriptional circuit regulated by AML1-ETO. Mol. Cell 2020, 81, 530–545.e5. [Google Scholar] [CrossRef]
- Thiel, V.N.; Giaimo, B.D.; Schwarz, P.; Soller, K.; Vas, V.; Bartkuhn, M.; Blätte, T.J.; Döhner, K.; Bullinger, L.; Borggrefe, L.; et al. Heterodimerization of AML1/ETO with CBFbeta is required for leukemogenesis but not for myeloproliferation. Leukemia 2017, 31, 2491–2502. [Google Scholar] [CrossRef] [Green Version]
- Rejeski, K.; Duque-Afonso, J.; Lubbert, M. AML1/ETO and its function as a regulator of gene transcription via epigenetic mechanisms. Oncogene 2021, 40, 5665–5676. [Google Scholar] [CrossRef]
- Miyoshi, H.; Shimizu, K.; Kozu, T.; Maseki, N.; Kaneko, Y.; Ohki, M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. USA 1991, 88, 10431–10434. [Google Scholar] [CrossRef] [Green Version]
- Mrózek, K.; Bloomfield, C.D. Clinical Significance of the Most Common Chromosome Translocations in Adult Acute Myeloid Leukemia. JNCI Monogr. 2008, 2008, 52–57. [Google Scholar] [CrossRef] [Green Version]
- Alcalay, M.; Meani, N.; Gelmetti, V.; Fantozzi, A.; Fagioli, M.; Orleth, A.; Riganelli, D.; Sebastiani, C.; Cappelli, E.; Casciari, C.; et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J. Clin. Investig. 2003, 112, 1751–1761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krejci, O.; Wunderlich, M.; Geiger, H.; Chou, F.-S.; Schleimer, D.; Jansen, M.; Andreassen, P.R.; Mulloy, J.C. p53 signaling in response to increased DNA damage sensitizes AML1-ETO cells to stress-induced death. Blood 2008, 111, 2190–2199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.K.; Chan, N.P.H.; Wan, T.S.K.; Lam, L.Y.; Cheung, C.H.Y.; Wong, T.H.Y.; Ip, R.K.L.; Wong, R.S.M.; Ng, M.H.L. Helicase-like transcription factor is a RUNX1 target whose downregulation promotes genomic instability and correlates with complex cytogenetic features in acute myeloid leukemia. Haematologica 2016, 101, 448–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, G.; Kermi, C.; Stoy, H.; Schiltz, C.J.; Bacal, J.; Zaino, A.M.; Hadden, M.K.; Eichman, B.F.; Lopes, M.; Cimprich, K.A. HLTF Promotes Fork Reversal, Limiting Replication Stress Resistance and Preventing Multiple Mechanisms of Unrestrained DNA Synthesis. Mol. Cell 2020, 78, 1237–1251.e7. [Google Scholar] [CrossRef] [PubMed]
- Esposito, M.T.; Zhao, L.; Fung, T.K.; Rane, J.K.; Wilson, A.; Martin, N.; Gil, J.; Leung, A.Y.; Ashworth, A.; So, C.W.E. Synthetic lethal targeting of oncogenic transcription factors in acute leukemia by PARP inhibitors. Nat. Med. 2015, 21, 1481–1490. [Google Scholar] [CrossRef] [PubMed]
- Forster, V.J.; Nahari, M.H.; Martinez-Soria, N.; Bradburn, A.K.; Ptasinska, A.; Assi, S.A.; Fordham, S.E.; McNeil, H.; Bonifer, C.; Heidenreich, O.; et al. The leukemia-associated RUNX1/ETO oncoprotein confers a mutator phenotype. Leukemia 2015, 30, 251–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandell, J.D.; Fisk, J.N.; Cyrenne, E.; Xu, M.L.; Cannataro, V.L.; Townsend, J.P. Not only mutations but also tumorigenesis can be substantially attributed to DNA damage from reactive oxygen species in RUNX1::RUNX1T1-fusion-positive acute myeloid leukemia. Leukemia 2022, 36, 2931–2933. [Google Scholar] [CrossRef]
- Gunnarsson, R.; Yang, M.; Olsson-Arvidsson, L.; Biloglav, A.; Behrendtz, M.; Castor, A.; Paulsson, K.; Johansson, B. Single base substitution mutational signatures in pediatric acute myeloid leukemia based on whole genome sequencing. Leukemia 2021, 35, 1485–1489. [Google Scholar] [CrossRef] [PubMed]
- Romana, S.P.; Mauchauffé, M.; Le Coniat, M.; Chumakov, I.; Le Paslier, D.; Berger, R.; Bernard, O. The t(12;21) of acute lymphoblastic leukemia results in a tel-AML1 gene fusion. Blood 1995, 85, 3662–3670. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Hernández, G.; Casado-García, A.; Isidro-Hernández, M.; Picard, D.; Raboso-Gallego, J.; Alemán-Arteaga, S.; Orfao, A.; Blanco, O.; Riesco, S.; Prieto-Matos, P.; et al. The Second Oncogenic Hit Determines the Cell Fate of ETV6-RUNX1 Positive Leukemia. Front. Cell Dev. Biol. 2021, 9, 704591. [Google Scholar] [CrossRef] [PubMed]
- Kantner, H.P.; Warsch, W.; Delogu, A.; Bauer, E.; Esterbauer, H.; Casanova, E.; Sexl, V.; Stoiber, D. ETV6/RUNX1 induces reactive oxygen species and drives the accumulation of DNA damage in B cells. Neoplasia 2013, 15, 1292–1300. [Google Scholar] [CrossRef] [Green Version]
- Fuka, G.; Kauer, M.; Kofler, R.; Haas, O.A.; Panzer-Grumayer, R. The leukemia-specific fusion gene ETV6/RUNX1 perturbs distinct key biological functions primarily by gene repression. PLoS ONE 2011, 6, e26348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaindl, U.; Morak, M.; Portsmouth, C.; Mecklenbräuker, A.; Kauer, M.; Zeginigg, M.; Attarbaschi, A.; Haas, O.A.; R Panzer-Grümayer, R. Blocking ETV6/RUNX1-induced MDM2 overexpression by Nutlin-3 reactivates p53 signaling in childhood leukemia. Leukemia 2014, 28, 600–608. [Google Scholar] [CrossRef] [Green Version]
- Nucifora, G.; Rowley, J. The AML1 Gene in the 8;21 and 3;21 Translocations in Chronic and Acute Myeloid Leukemia. Cold Spring Harb. Symp. Quant. Biol. 1994, 59, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.C.; Cortes, J.; Barkoh, B.; Hayes, K.; Kantarjian, H.; Jones, D. t(3;21)(q26;q22) in myeloid leukemia: An aggressive syndrome of blast transformation associated with hydroxyurea or antimetabolite therapy. Cancer 2006, 106, 1730–1738. [Google Scholar]
- Kellaway, S.G.; Keane, P.; Kennett, E.; Bonifer, C. RUNX1-EVI1 disrupts lineage determination and the cell cycle by interfering with RUNX1 and EVI1 driven gene regulatory networks. Haematologica 2020, 106, 1569–1580. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Zhu, J.; Chen, F.; Lin, W.; Cai, J.; Zhong, J. RUNX1-Evi-1 fusion gene inhibited differentiation and apoptosis in myelopoiesis: An in vivo study. BMC Cancer 2015, 15, 970. [Google Scholar] [CrossRef] [Green Version]
- Bard-Chapeau, E.A.; Gunaratne, J.; Kumar, P.; Chua, B.Q.; Muller, J.; Bard, F.A.; Blackstock, W.; Copeland, N.G.; Jenkins, N.A. EVI1 oncoprotein interacts with a large and complex network of proteins and integrates signals through protein phosphorylation. Proc. Natl. Acad. Sci. USA 2013, 110, E2885–E2894. [Google Scholar] [CrossRef] [Green Version]
- Bellissimo, D.; Speck, N.A. RUNX1 Mutations in Inherited and Sporadic Leukemia. Front. Cell Dev. Biol. 2017, 5, 111. [Google Scholar] [CrossRef] [PubMed]
- Jacob, B.; Osato, M.; Yamashita, N.; Wang, C.Q.; Taniuchi, I.; Littman, D.R.; Asou, N.; Ito, Y. Stem cell exhaustion due to Runx1 deficiency is prevented by Evi5 activation in leukemogenesis. Blood 2010, 115, 1610–1620. [Google Scholar] [CrossRef] [Green Version]
- Motoda, L.; Osato, M.; Yamashita, N.; Jacob, B.; Chen, L.Q.; Yanagida, M.; Ida, H.; Wee, H.-J.; Sun, A.X.; Taniuchi, I.; et al. Runx1 Protects Hematopoietic Stem/Progenitor Cells from Oncogenic Insult. Stem Cells 2007, 25, 2976–2986. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, N.; Arai, S.; Ichikawa, M.; Nakagawa, M.; Goyama, S.; Kumano, K.; Takahashi, T.; Kamikubo, Y.; Imai, Y.; Kurokawa, M. Loss of AML1/Runx1 accelerates the development of MLL-ENL leukemia through down-regulation of p19ARF. Blood 2011, 118, 2541–25450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, C.P.; Wang, C.Q.; Ng, C.E.L.; Ito, Y.; Araki, M.; Tergaonkar, V.; Huang, G.; Osato, M. RUNX1 meets MLL: Epigenetic regulation of hematopoiesis by two leukemia genes. Leukemia 2013, 27, 1793–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satoh, Y.; Matsumura, I.; Tanaka, H.; Harada, H.; Harada, Y.; Matsui, K.; Shibata, M.; Mizuki, M.; Kanakura, Y. C-terminal mutation of RUNX1 attenuates the DNA-damage repair response in hematopoietic stem cells. Leukemia 2011, 26, 303–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antony-Debré, I.; Manchev, V.T.; Balayn, N.; Bluteau, D.; Tomowiak, C.; Legrand, C.; Langlois, T.; Bawa, O.; Tosca, L.; Tachdjian, G.; et al. Level of RUNX1 activity is critical for leukemic predisposition but not for thrombocytopenia. Blood 2015, 125, 930–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adnan, S.A.; Dufva, O.; Ianevski, A.; Ghimire, B.; Koski, J.; Maliniemi, P.; Thomson, D.; Schreiber, A.; Heckman, C.A.; Koskenvesa, P.; et al. RUNX1 mutations in blast-phase chronic myeloid leukemia associate with distinct phenotypes, transcriptional profiles, and drug responses. Leukemia 2021, 35, 1087–1099. [Google Scholar] [CrossRef]
- Kaisrlikova, M.; Vesela, J.; Kundrat, D.; Votavova, H.; Merkerova, M.D.; Krejcik, Z.; Divoky, V.; Jedlicka, M.; Fric, J.; Klema, J.; et al. RUNX1 mutations contribute to the progression of MDS due to disruption of antitumor cellular defense: A study on patients with lower-risk MDS. Leukemia 2022, 36, 1898–1906. [Google Scholar] [CrossRef]
- Fujii, S.; Ito, K.; Ito, Y.; Ochiai, A. Enhancer of Zeste Homologue 2 (EZH2) Down-regulates RUNX3 by Increasing Histone H3 Methylation. J. Biol. Chem. 2008, 283, 17324–17332. [Google Scholar] [CrossRef] [Green Version]
- Goh, Y.-M.; Cinghu, S.; Hong, E.T.H.; Lee, Y.-S.; Kim, J.-H.; Jang, J.-W.; Li, Y.-H.; Chi, X.-Z.; Lee, K.-S.; Wee, H.; et al. Src Kinase Phosphorylates RUNX3 at Tyrosine Residues and Localizes the Protein in the Cytoplasm. J. Biol. Chem. 2010, 285, 10122–10129. [Google Scholar] [CrossRef] [Green Version]
- Douchi, D.; Yamamura, A.; Matsuo, J.; Lee, J.-W.; Nuttonmanit, N.; Lim, Y.H.M.; Suda, K.; Shimura, M.; Chen, S.; Pang, S.; et al. A Point Mutation R122C in RUNX3 Promotes the Expansion of Isthmus Stem Cells and Inhibits Their Differentiation in the Stomach. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 1317–1345. [Google Scholar] [CrossRef]
- Chen, L.-M.; Nergard, J.C.; Ni, L.; Rosser, C.J.; Chai, K.X. Long-Term Exposure to Cigarette Smoke Extract Induces Hypomethylation at the RUNX3 and IGF2-H19 Loci in Immortalized Human Urothelial Cells. PLoS ONE 2013, 8, e65513. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.A.; Zhang, R.; Kim, G.Y.; Bae, S.C.; Hyun, J.W. Epigenetic changes induced by oxidative stress in colorectal cancer cells: Methylation of tumor suppressor RUNX3. Tumor Biol. 2012, 33, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kim, J.; Kim, W.-H.; Lee, Y.M. Hypoxic silencing of tumor suppressor RUNX3 by histone modification in gastric cancer cells. Oncogene 2008, 28, 184–194. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Hyeon, D.Y.; Yoon, S.-H.; Jeong, J.-H.; Han, S.-M.; Jang, J.-W.; Nguyen, M.P.; Chi, X.-Z.; An, S.; Hyun, K.-G.; et al. RUNX3 methylation drives hypoxia-induced cell proliferation and antiapoptosis in early tumorigenesis. Cell Death Differ. 2020, 28, 1251–1269. [Google Scholar] [CrossRef]
- Kolinjivadi, A.M.; Sankar, H.; Choudhary, R.; Tay, L.S.; Tan, T.Z.; Murata-Kamiya, N.; Voon, D.C.-C.; Kappei, D.; Hatakeyama, M.; Krishnan, V.; et al. The H. pylori CagA Oncoprotein Induces DNA Double Strand Breaks through Fanconi Anemia Pathway Downregulation and Replication Fork Collapse. Int. J. Mol. Sci. 2022, 23, 1661. [Google Scholar] [CrossRef] [PubMed]
- Koeppel, M.; Garcia-Alcalde, F.; Glowinski, F.; Schlaermann, P.; Meyer, T.F. Helicobacter pylori Infection Causes Characteristic DNA Damage Patterns in Human Cells. Cell Rep. 2015, 11, 1703–1713. [Google Scholar] [CrossRef] [Green Version]
- Wolff, E.M.; Liang, G.; Cortez, C.C.; Tsai, Y.C.; Castelao, J.E.; Cortessis, V.K.; Tsao-Wei, D.D.; Groshen, S.; Jones, P.A. RUNX3 Methylation Reveals that Bladder Tumors Are Older in Patients with a History of Smoking. Cancer Res 2008, 68, 6208–6214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curti, L.; Campaner, S. MYC-Induced Replicative Stress: A Double-Edged Sword for Cancer Development and Treatment. Int. J. Mol. Sci. 2021, 22, 6168. [Google Scholar] [CrossRef]
- Tay, L.S.; Krishnan, V.; Sankar, H.; Chong, Y.L.; Chuang, L.S.H.; Tan, T.Z.; Kolinjivadi, A.M.; Kappei, D.; Ito, Y. RUNX Poly(ADP-Ribosyl)ation and BLM Interaction Facilitate the Fanconi Anemia Pathway of DNA Repair. Cell Rep. 2018, 24, 1747–1755. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Ma, Q.; Long, B.; Sun, Z.; Liu, L.; Lin, D.; Zhao, M. Runt-Related Transcription Factor 3 Promotes Acute Myeloid Leukemia Progression. Front. Oncol. 2021, 11, 4065. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Solal, K.A.; Boregowda, R.K.; Lasfar, A. RUNX2 and the PI3K/AKT axis reciprocal activation as a driving force for tumor progression. Mol. Cancer 2015, 14, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaidi, S.K.; Pande, S.; Pratap, J.; Gaur, T.; Grigoriu, S.; Ali, S.A.; Stein, J.L.; Lian, J.B.; van Wijnen, A.J.; Stein, G.S. Runx2 deficiency and defective subnuclear targeting bypass senescence to promote immortalization and tumorigenic potential. Proc. Natl. Acad. Sci. USA 2007, 104, 19861–19866. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Quaresma, A.J.; Nickerson, J.A.; Green, K.M.; Shaffer, S.A.; Imbalzano, A.N.; Martin-Buley, L.A.; Lian, J.B.; Stein, J.L.; van Wijnen, A.J.; et al. Subnuclear domain proteins in cancer cells support the functions of RUNX2 in the DNA damage response. J. Cell Sci. 2015, 128, 728–740. [Google Scholar] [CrossRef] [Green Version]
- Cobb, A.M.; Yusoff, S.; Hayward, R.; Ahmad, S.; Sun, M.; Verhulst, A.; D’Haese, P.C.; Shanahan, C.M. Runx2 (Runt-Related Transcription Factor 2) Links the DNA Damage Response to Osteogenic Reprogramming and Apoptosis of Vascular Smooth Muscle Cells. Arter. Thromb. Vasc. Biol. 2021, 41, 1339–1357. [Google Scholar] [CrossRef]
- Ozaki, T.; Sugimoto, H.; Nakamura, M.; Hiraoka, K.; Yoda, H.; Sang, M.; Fujiwara, K.; Nagase, H. Runt-related transcription factor 2 attenuates the transcriptional activity as well as DNA damage-mediated induction of pro-apoptotic TAp73 to regulate chemosensitivity. FEBS J. 2014, 282, 114–128. [Google Scholar] [CrossRef] [Green Version]
- Ozaki, T.; Nakagawara, A.; Nagase, H. RUNX Family Participates in the Regulation of p53-Dependent DNA Damage Response. Int. J. Genom. 2013, 2013, 271347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, D.; Ozaki, T.; Yoshihara, Y.; Kubo, N.; Nakagawara, A. Runt-related Transcription Factor 1 (RUNX1) Stimulates Tumor Suppressor p53 Protein in Response to DNA Damage through Complex Formation and Acetylation. J. Biol. Chem. 2013, 288, 1353–1364. [Google Scholar] [CrossRef] [Green Version]
- Morita, K.; Noura, M.; Tokushige, C.; Maeda, S.; Kiyose, H.; Kashiwazaki, G.; Taniguchi, J.; Bando, T.; Yoshida, K.; Ozaki, T.; et al. Autonomous feedback loop of RUNX1-p53-CBFB in acute myeloid leukemia cells. Sci. Rep. 2017, 7, 16604. [Google Scholar] [CrossRef] [Green Version]
- Yamada, C.; Ozaki, T.; Ando, K.; Suenaga, Y.; Inoue, K.-I.; Ito, Y.; Okoshi, R.; Kageyama, H.; Kimura, H.; Miyazaki, M.; et al. RUNX3 Modulates DNA Damage-mediated Phosphorylation of Tumor Suppressor p53 at Ser-15 and Acts as a Co-activator for p53. J. Biol. Chem. 2010, 285, 16693–16703. [Google Scholar] [CrossRef] [Green Version]
- Bae, S.-C.; Kolinjivadi, A.M.; Ito, Y. Functional relationship between p53 and RUNX proteins. J. Mol. Cell Biol. 2018, 11, 224–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozaki, T.; Wu, D.; Sugimoto, H.; Nagase, H.; Nakagawara, A. Runt-related transcription factor 2 (RUNX2) inhibits p53-dependent apoptosis through the collaboration with HDAC6 in response to DNA damage. Cell Death Dis. 2013, 4, e610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrd, R.S.; Zwerdling, T.; Moghaddam, B.; Pinter, J.D.; Steinfeld, M.B. Monosomy 21q22.11-q22.13 presenting as a Fanconi anemia phenotype. Am. J. Med Genet. Part A 2010, 155, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Click, E.S.; Cox, B.; Olson, S.B.; Grompe, M.; Akkari, Y.; Moreau, L.A.; Shimamura, A.; Sternen, D.L.; Liu, Y.J.; Leppig, K.A.; et al. Fanconi anemia-like presentation in an infant with constitutional deletion of 21q including the RUNX1 gene. Am. J. Med Genet. Part A 2011, 155, 1673–1679. [Google Scholar] [CrossRef]
- Nalepa, G.; Clapp, D.W. Fanconi anaemia and cancer: An intricate relationship. Nat. Rev. Cancer 2018, 18, 168–185. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Q.; Krishnan, V.; Tay, L.S.; Chin, D.W.L.; Koh, C.P.; Chooi, J.Y.; Nah, G.S.S.; Du, L.; Jacob, B.; Yamashita, N.; et al. Disruption of Runx1 and Runx3 Leads to Bone Marrow Failure and Leukemia Predisposition due to Transcriptional and DNA Repair Defects. Cell Rep. 2014, 8, 767–782. [Google Scholar] [CrossRef] [Green Version]
- Chao, M.M.; Thomay, K.; Goehring, G.; Wlodarski, M.; Pastor, V.; Schlegelberger, B.; Schindler, D.; Kratz, C.P.; Niemeyer, C. Mutational Spectrum of Fanconi Anemia Associated Myeloid Neoplasms. Klin Padiatr. 2017, 229, 329–334. [Google Scholar] [CrossRef]
- Quentin, S.; Cuccuini, W.; Ceccaldi, R.; Nibourel, O.; Pondarre, C.; Pagès, M.P.; Vasquez, N.; d’Enghien, C.D.; Larghero, J.; de Latour, R.P.; et al. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood 2011, 117, e161–e170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlacher, K.; Wu, H.; Jasin, M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 2012, 22, 106–116. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Imamura, J.; Kanai, F.; Ichimura, T.; Isobe, T.; Koike, M.; Kudo, Y.; Tateishi, K.; Ikenoue, T.; Ijichi, H.; et al. Runx3 interacts with DNA repair protein Ku70. Exp. Cell Res. 2007, 313, 3251–3260. [Google Scholar] [CrossRef]
- Puustinen, P.; Keldsbo, A.; Corcelle-Termeau, E.; Ngoei, K.; Sønder, S.L.; Farkas, T.; Andersen, K.K.; Oakhill, J.; Jäättelä, M. DNA-dependent protein kinase regulates lysosomal AMP-dependent protein kinase activation and autophagy. Autophagy 2020, 16, 1871–1888. [Google Scholar] [CrossRef]
- Lee, K.-J.; Lin, Y.-F.; Chou, H.-Y.; Yajima, H.; Fattah, K.R.; Lee, S.-C.; Chen, B.P. Involvement of DNA-dependent Protein Kinase in Normal Cell Cycle Progression through Mitosis. J. Biol. Chem. 2011, 286, 12796–12802. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Mersaoui, S.Y.; Guitton-Sert, L.; Coulombe, Y.; Song, J.; Masson, J.-Y.; Richard, S. DDX5 resolves R-loops at DNA double-strand breaks to promote DNA repair and avoid chromosomal deletions. NAR Cancer 2020, 2, zcaa028. [Google Scholar] [CrossRef]
- Lara-Gonzalez, P.; Pines, J.; Desai, A. Spindle assembly checkpoint activation and silencing at kinetochores. Semin. Cell Dev. Biol. 2021, 117, 86–98. [Google Scholar] [CrossRef]
- Carvajal, L.A.; Hamard, P.-J.; Tonnessen, C.; Manfredi, J.J. E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev. 2012, 26, 1533–1545. [Google Scholar] [CrossRef] [Green Version]
- Panneerselvam, J.; Wang, H.; Zhang, J.; Che, R.; Yu, H.; Fei, P. BLM promotes the activation of Fanconi Anemia signaling pathway. Oncotarget 2016, 7, 32351–32361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Acosta, D.; Blanco-Romero, E.; Ubieto-Capella, P.; Mutreja, K.; Míguez, S.; Llanos, S.; García, F.; Muñoz, J.; Blanco, L.; Lopes, M.; et al. PrimPol-mediated repriming facilitates replication traverse of DNA interstrand crosslinks. EMBO J. 2021, 40, e106355. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V.; Chong, Y.L.; Tan, T.Z.; Kulkarni, M.; Bin Rahmat, M.B.; Tay, L.S.; Sankar, H.; Jokhun, D.S.; Ganesan, A.; Chuang, L.S.H.; et al. TGFbeta Promotes Genomic Instability after Loss of RUNX3. Cancer Res. 2018, 78, 88–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnan, V.; Ito, Y. RUNX3 loss turns on the dark side of TGF-beta signaling. Oncoscience 2017, 4, 156–157. [Google Scholar] [CrossRef]
- Liddiard, K.; Hills, R.; Burnett, A.K.; Darley, R.L.; Tonks, A. OGG1 is a novel prognostic indicator in acute myeloid leukaemia. Oncogene 2009, 29, 2005–2012. [Google Scholar] [CrossRef] [Green Version]
- Wolyniec, K.; Wotton, S.; Kilbey, A.; Jenkins, A.; Terry, A.; Peters, G.; Stocking, C.; Cameron, E.; Neil, J.C. RUNX1 and its fusion oncoprotein derivative, RUNX1-ETO, induce senescence-like growth arrest independently of replicative stress. Oncogene 2009, 28, 2502–2512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Brugge, J.S.; Janes, K.A. Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc. Natl. Acad. Sci. USA 2011, 108, E803–E812. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Shim, J.; Bae, J.; Kim, Y.-J.; Lee, J. Stabilization of RNT-1 Protein, Runt-related Transcription Factor (RUNX) Protein Homolog of Caenorhabditis elegans, by Oxidative Stress through Mitogen-activated Protein Kinase Pathway*. J. Biol. Chem. 2012, 287, 10444–10452. [Google Scholar] [CrossRef] [Green Version]
- Beneforti, L.; Dander, E.; Bresolin, S.; Bueno, C.; Acunzo, D.; Bertagna, M.; Ford, A.; Gentner, B.; Kronnie, G.T.L.; Vergani, P.; et al. Pro-inflammatory cytokines favor the emergence of ETV6-RUNX1-positive pre-leukemic cells in a model of mesenchymal niche. Br. J. Haematol. 2020, 190, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Fitch, B.A.; Zhou, M.; Situ, J.; Surianarayanan, S.; Reeves, M.Q.; Hermiston, M.L.; Wiemels, J.L.; Kogan, S.C. Decreased IL-10 accelerates B-cell leukemia/lymphoma in a mouse model of pediatric lymphoid leukemia. Blood Adv. 2022, 6, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Seo, W.; Shimizu, K.; Kojo, S.; Okeke, A.; Kohwi-Shigematsu, T.; Fujii, S.-I.; Taniuchi, I. Runx-mediated regulation of CCL5 via antagonizing two enhancers influences immune cell function and anti-tumor immunity. Nat. Commun. 2020, 11, 1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, X.; Velasco-Velázquez, M.A.; Wang, M.; Li, Z.; Rui, H.; Peck, A.R.; Korkola, J.E.; Chen, X.; Xu, S.; DuHadaway, J.B.; et al. CCR5 Governs DNA Damage Repair and Breast Cancer Stem Cell Expansion. Cancer Res 2018, 78, 1657–1671. [Google Scholar] [CrossRef] [Green Version]

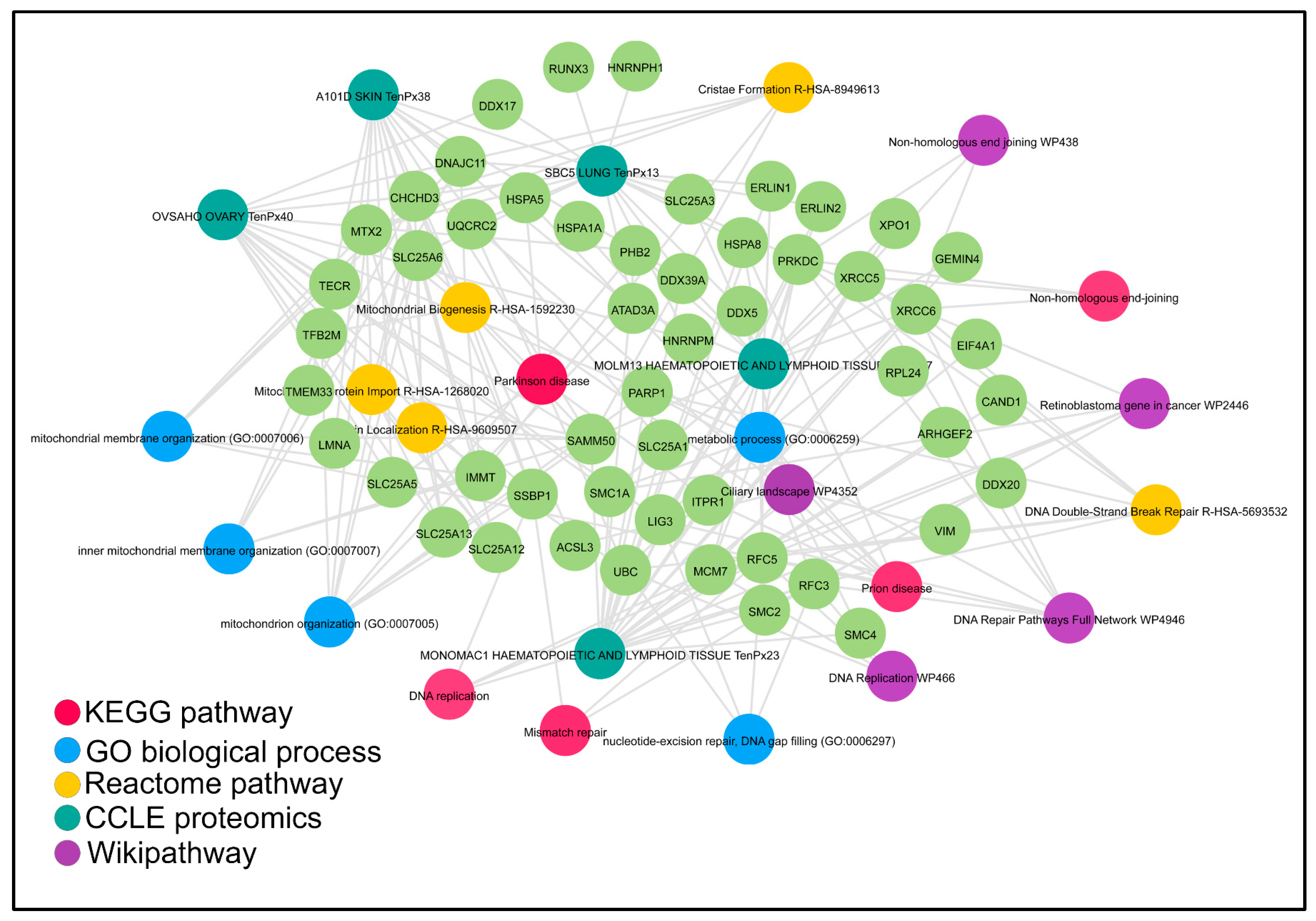
| RUNX Models (Human Cancer/In Vivo/In Vitro) | Effects on DNA Repair Gene Expression/Genomic Integrity | References | |
|---|---|---|---|
| RUNX1 fusion genes | RUNX1-ETO t(8;21) | DNA damage, DNA repair gene transcriptional impairment, mutator phenotype; ROS-associated SBS18 mutational signature; Impaired FANCD2 recruitment; PARPi sensitivity | [22,23,24,25,26,27,28,29,76] |
| ETV6-RUNX1 t(12;21) | DNA repair gene transcriptional impairment and p53 pathway deregulation | [33,34] | |
| RUNX1-EVI1 t(3;21) | DNA repair gene transcriptional impairment (zebrafish model) | [38] | |
| RUNX1 mutations | RUNX1-R174Q | DNA repair gene transcriptional impairment (iPSC model) | [46] |
| RUNX1-C terminal deletions | DNA damage accumulation (murine stem cell model) | [45] | |
| RUNX1-R162K, R204Q, R107C | Mutational signature 9 in CML blast crisis patient samples | [47] | |
| RUNX1-G141, R142 | Impaired FANCD2 recruitment expression of breast cancer walker domain mutations | [60] | |
| RUNX3 mutation | RUNX3-R122C | Upregulation of MYC, a known activator of DNA replication stress (mouse model) | [51] |
| RUNX3 altered expression | RUNX3-transcriptional silencing | Copy number alterations and mutational accumulation (bladder, lung, liver, esophageal cancers) | [60] |
| RUNX3 overexpression | Higher resistance to DNA damage-induced apoptosis in AML | [61] | |
| RUNX2 upregulation | RUNX2 overexpression | Chemoresistance to adriamycin | [66] |
| Mouse model | Runx1/Runx3 double knockout mice | Phenotypic resemblance to human Fanconi anemia | [76] |
| Runx2-null osteoblasts | DNA damage accumulation and loss of telomeric integrity | [63] | |
| Biochemical studies | RUNX interactome | Interaction of distinct DNA repair complexes in the presence and absence of DNA damage; RUNX1 interaction with Primpol | [60,88] |
| RUNX interaction with p53 | Regulates the transactivation of p53 target genes | [67,68,69,70,71,72] | |
| Cytokine exposed cell model and human cancer | RUNX3 downregulation and cytokine exposure | Higher oxidative stress and DNA damage accumulation in RUNX3-deficient lung cancers exposed to TGF-beta | [89] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krishnan, V. The RUNX Family of Proteins, DNA Repair, and Cancer. Cells 2023, 12, 1106. https://doi.org/10.3390/cells12081106
Krishnan V. The RUNX Family of Proteins, DNA Repair, and Cancer. Cells. 2023; 12(8):1106. https://doi.org/10.3390/cells12081106
Chicago/Turabian StyleKrishnan, Vaidehi. 2023. "The RUNX Family of Proteins, DNA Repair, and Cancer" Cells 12, no. 8: 1106. https://doi.org/10.3390/cells12081106





