Untargeted Metabolomics Reveals a Multi-Faceted Resistance Response to Fusarium Head Blight Mediated by the Thinopyrum elongatum Fhb7E Locus Transferred via Chromosome Engineering into Wheat
Abstract
1. Introduction
2. Materials and Methods
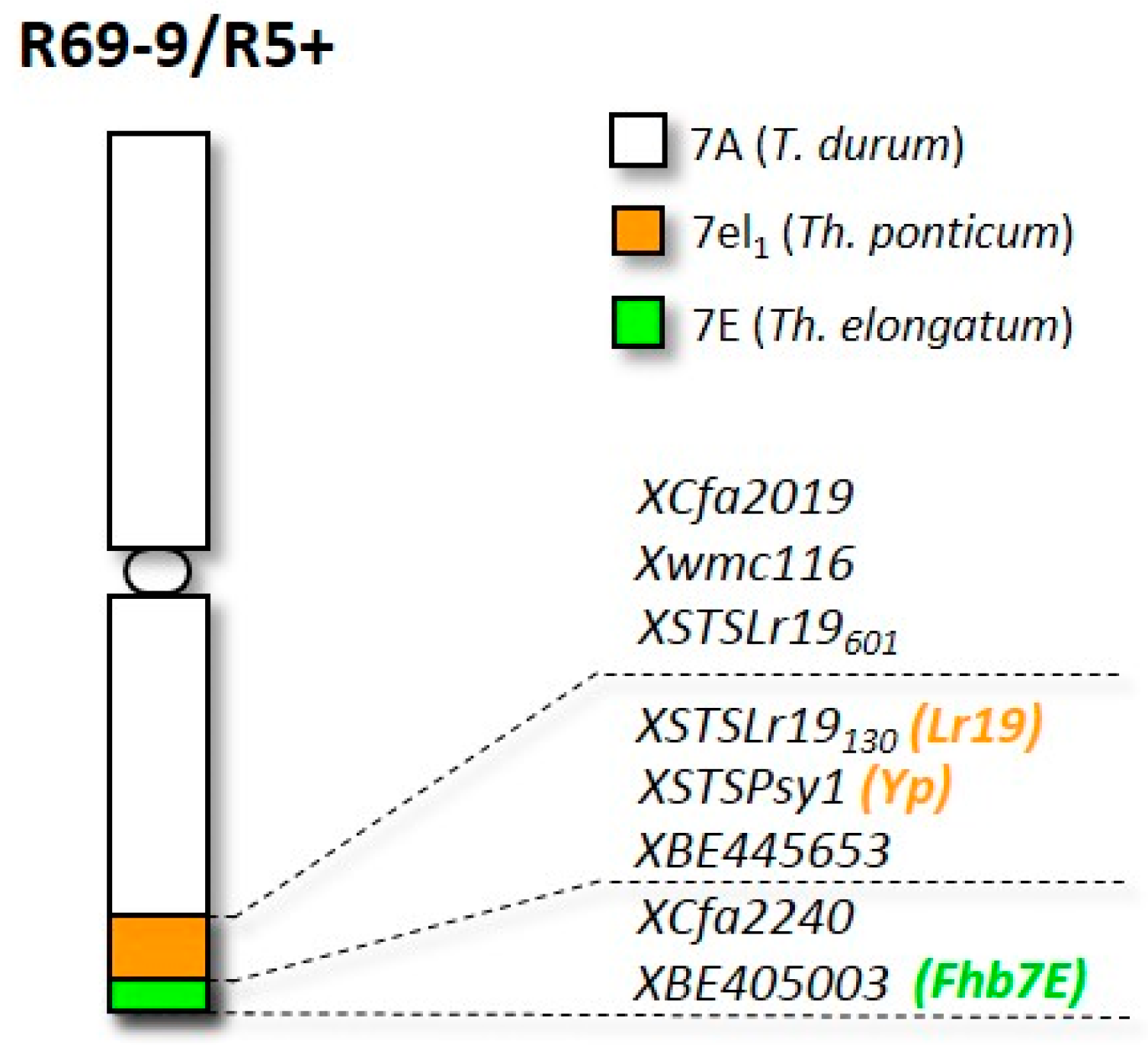
2.1. Plant Materials
2.2. Fusarium graminearum (Fg) Inoculation
2.3. Tissue Sampling, Metabolite Extraction and UHPLC-MS Analysis
2.4. Metabolomic Data Processing and Statistical Analysis
3. Results
3.1. Visual Observation of Symptoms on F. graminearum-Inoculated Spikes
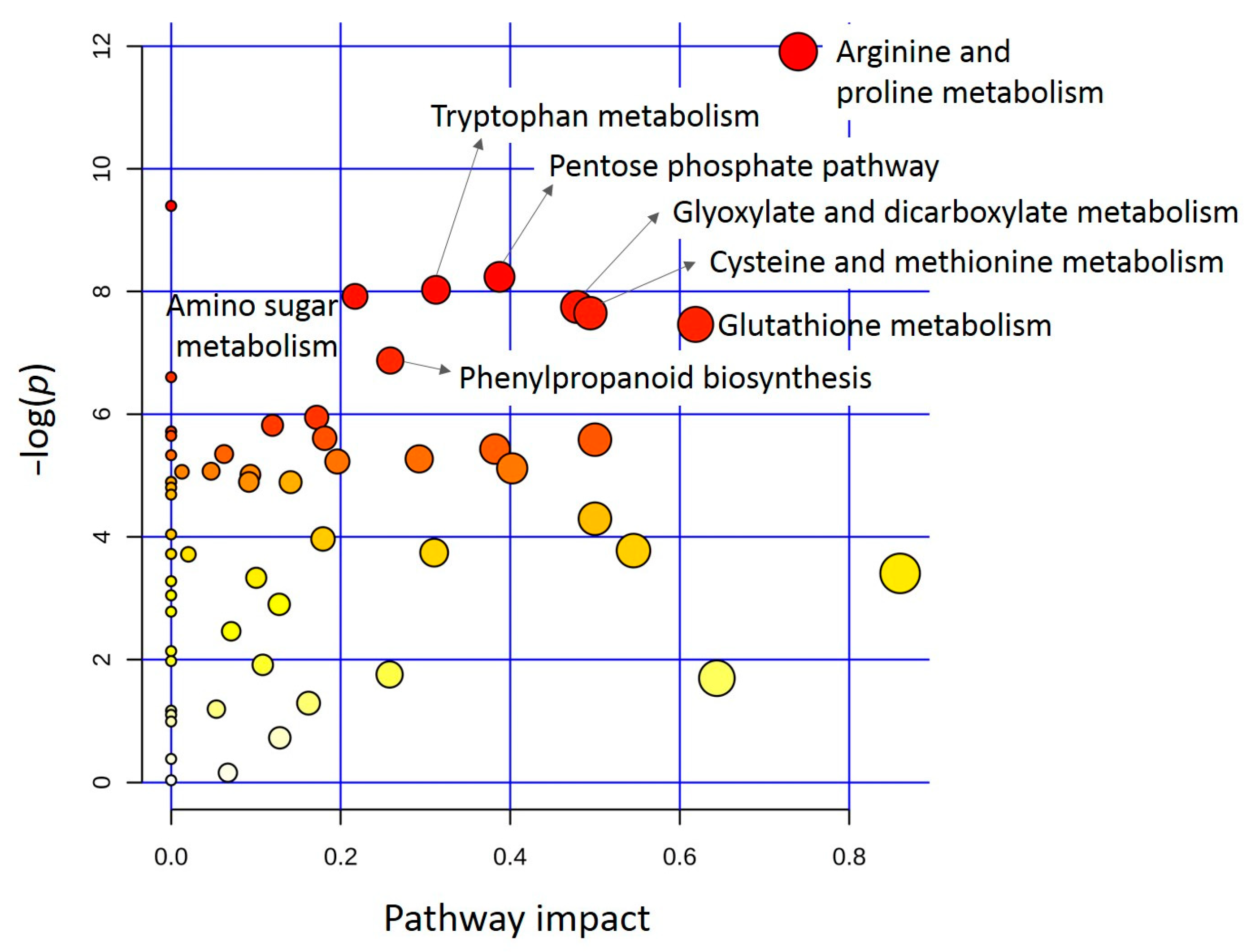
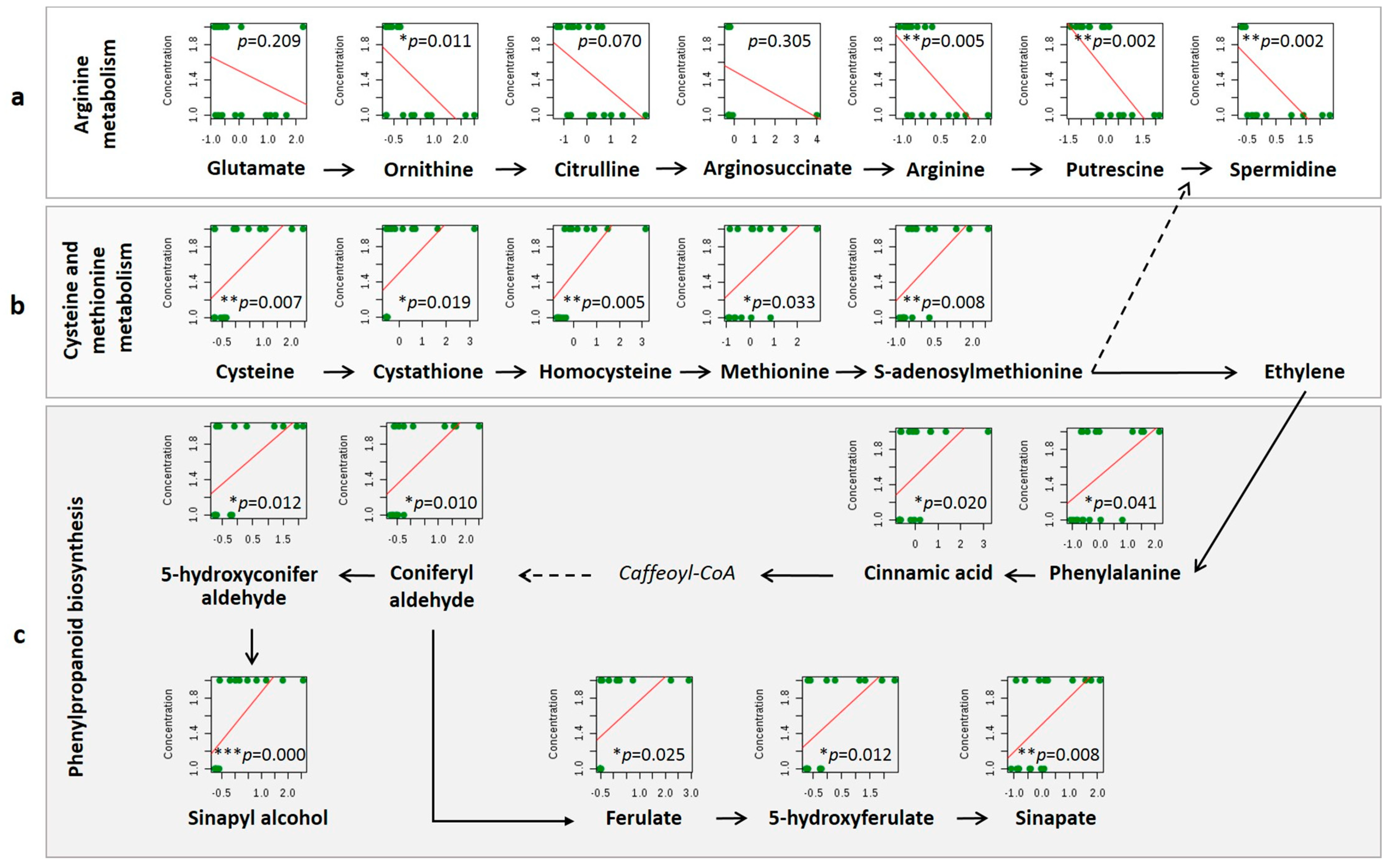
3.2. Differentially Accumulated Metabolites in Rachis
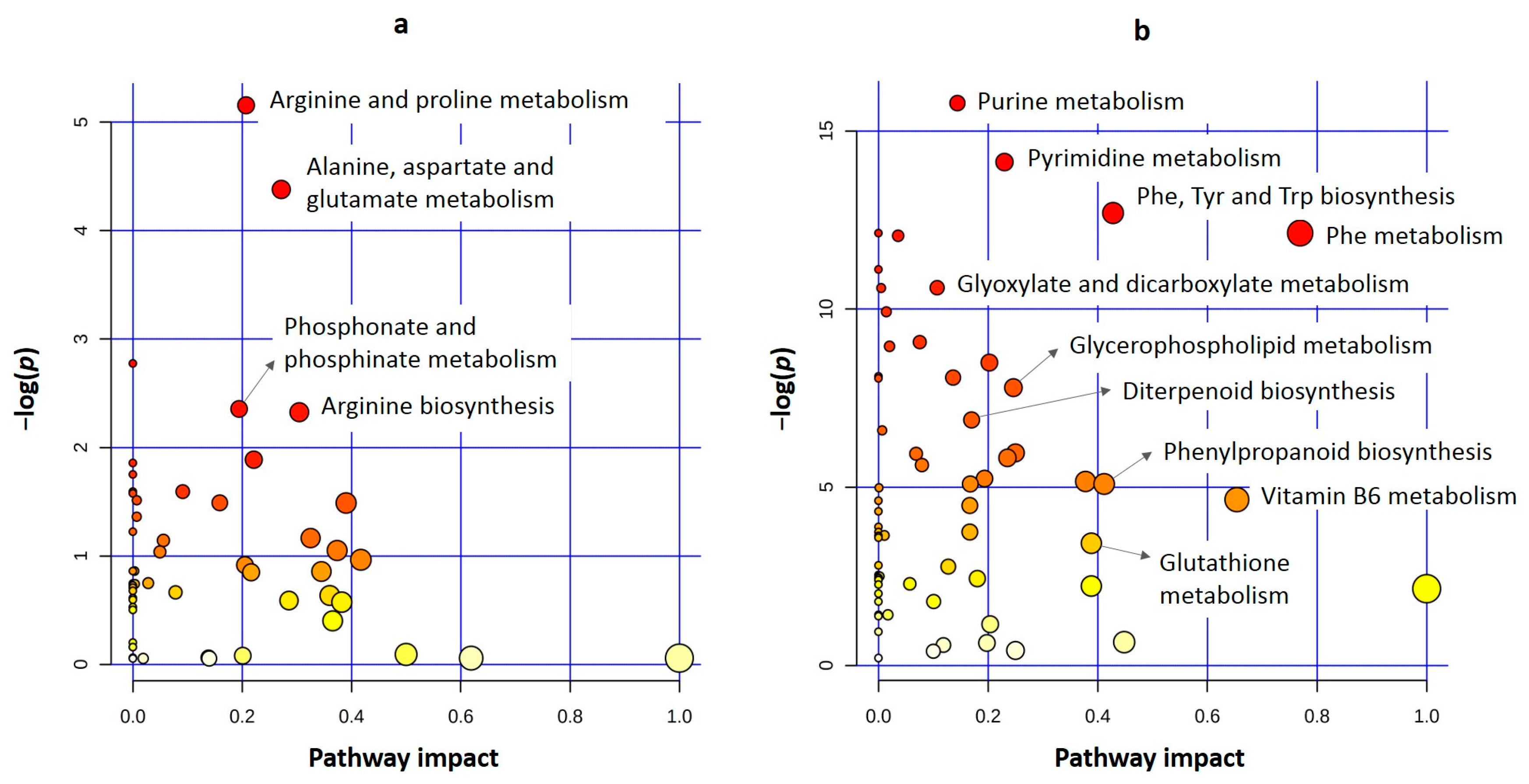
Pathway Analysis
3.3. Grain Tissue Metabolite Profiling
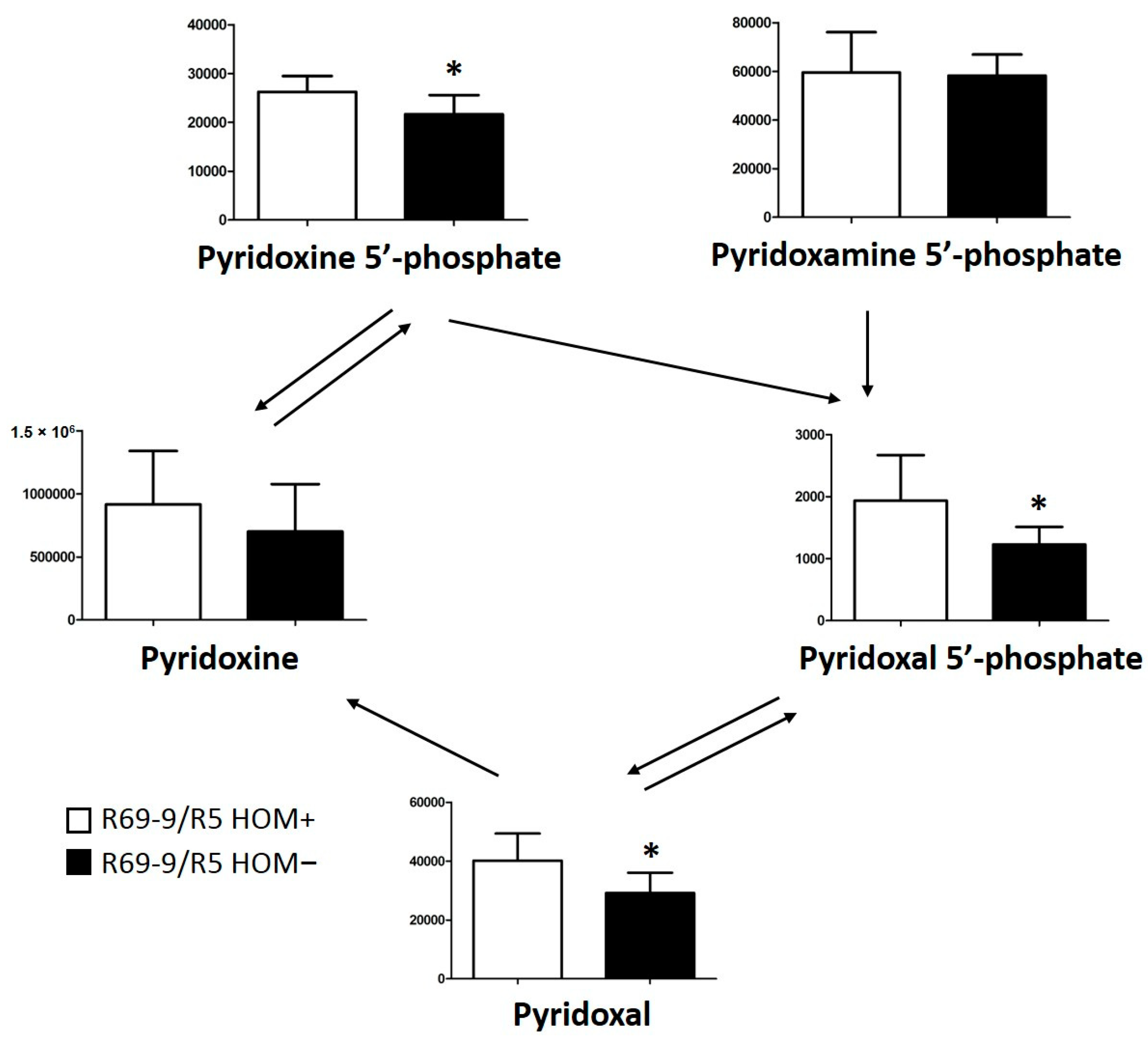
3.4. DON Detoxification Mechanisms in Rachis and Grain Tissue
4. Discussion
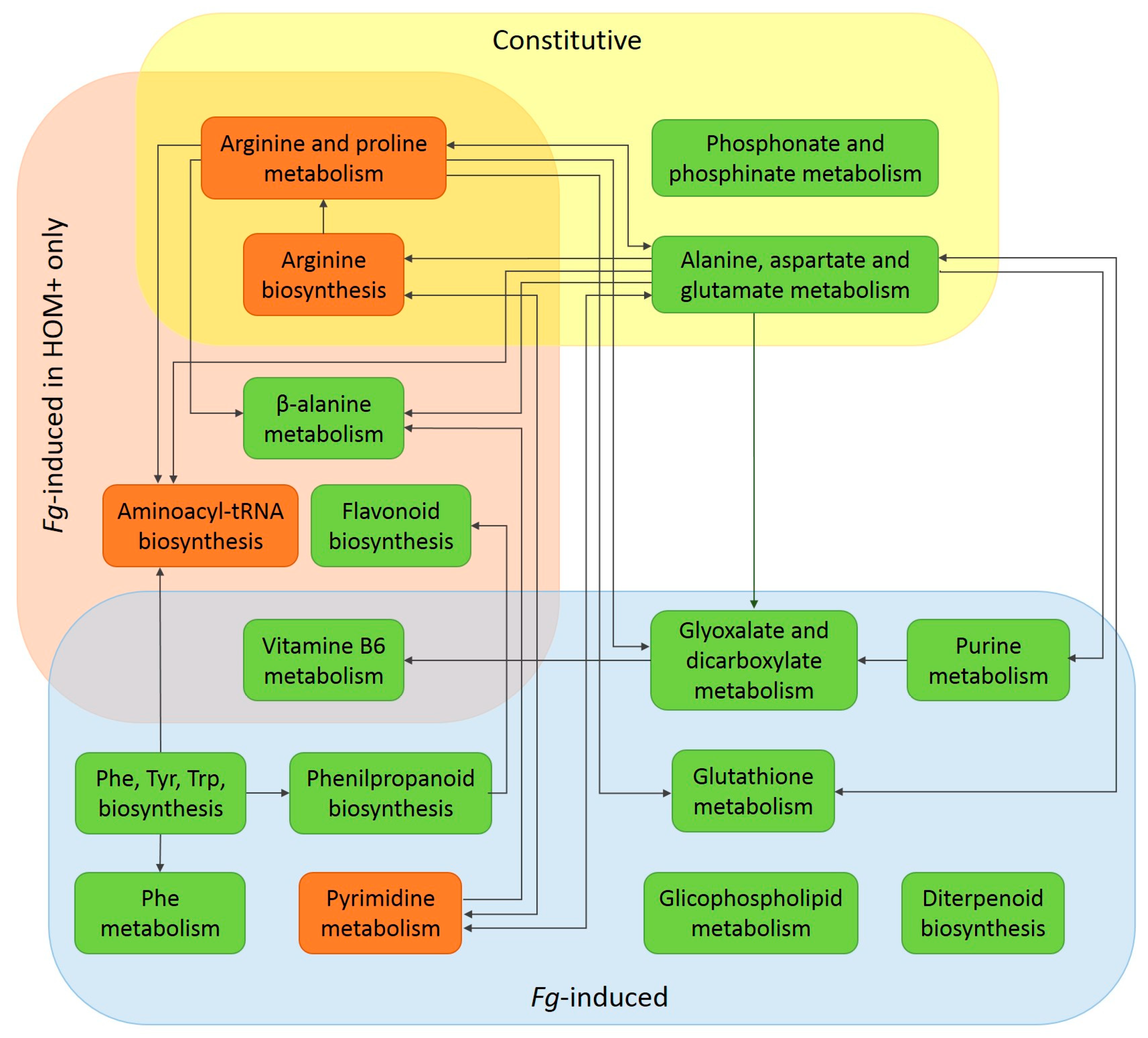
4.1. Constitutive Defence Potential of Fhb7E-Carrier Lines
4.2. Induced Response of Fhb7E-Carrier Lines to Fg Infection
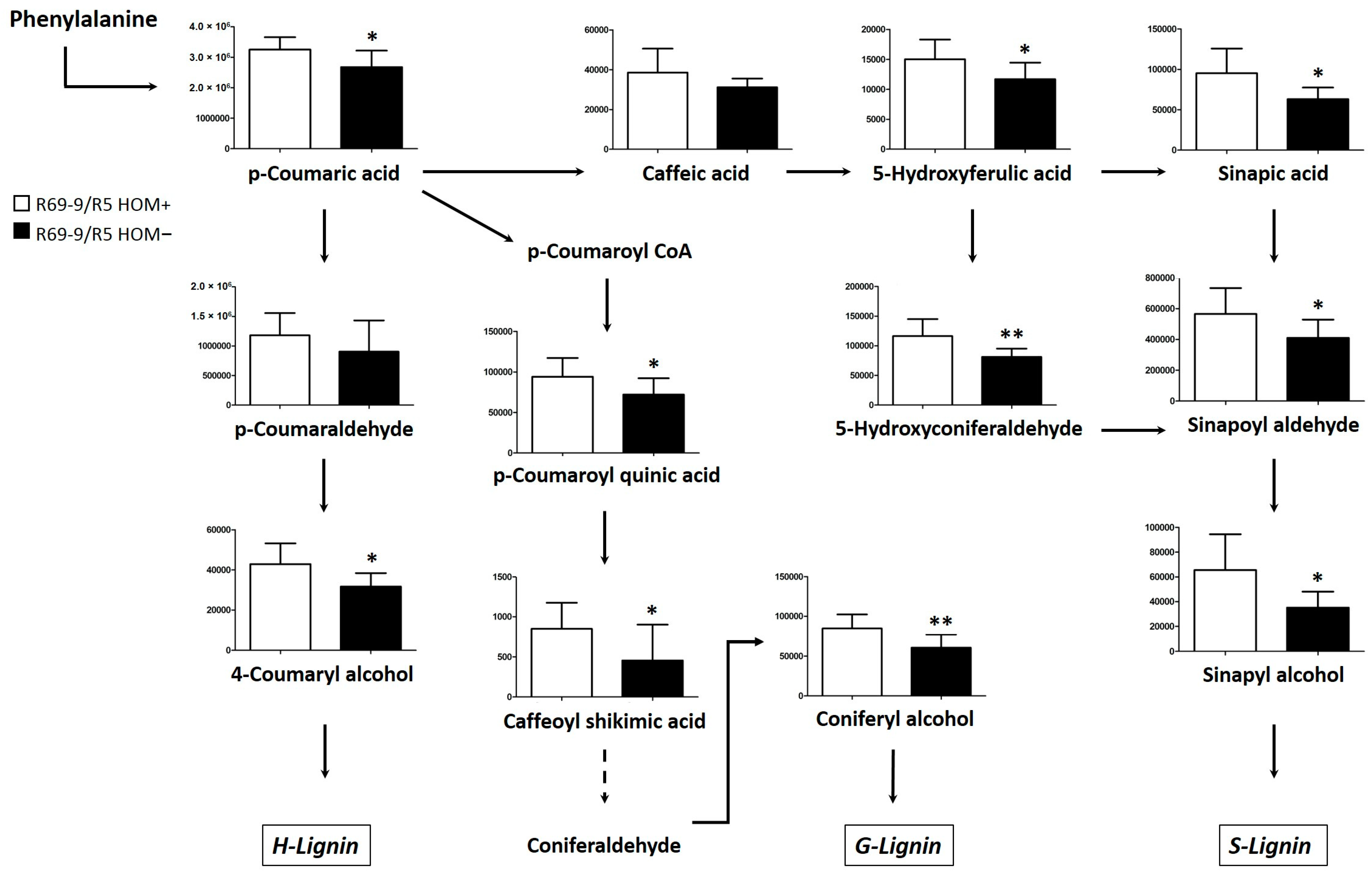
4.2.1. Fg-Induced Metabolic Changes in Rachis
4.2.2. Fg-Induced Metabolic Changes in Grains
4.3. Novel Insights into the Metabolomics of FHB Resistance Determined by the Fhb7E Locus
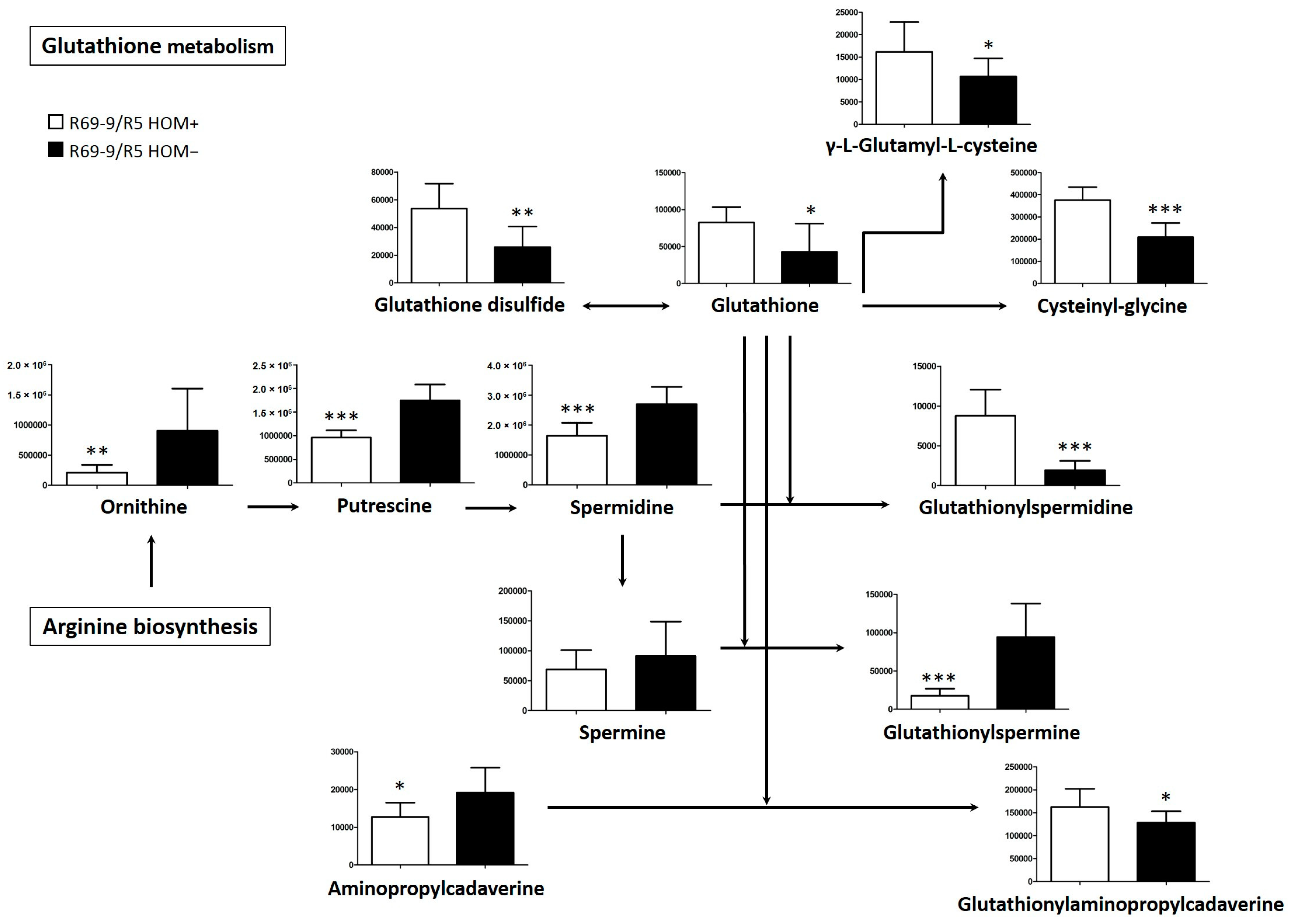
4.3.1. Polyamine Biosynthesis and Conjugation with Glutathione
4.3.2. VB6 Metabolism
4.3.3. Multiple DON Detoxification Mechanisms
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO, Food and Agriculture Organisation of the United Nations. FAOSTAT—Crops and Livestock Data. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 28 January 2022).
- OECD-FAO. OECD-FAO Agricultural Outlook 2021–2030; Chapter 3 Cereals; OECD Publishing: Paris, France, 2021; pp. 124–137. [Google Scholar] [CrossRef]
- Wing, I.S.; De Cian, E.; Mistry, M.N. Global vulnerability of crop yields to climate change. J. Environ. Econ. Manag. 2021, 109, 102462. [Google Scholar] [CrossRef]
- Ceoloni, C.; Kuzmanović, L.; Forte, P.; Gennaro, A.; Bitti, A. Targeted exploitation of gene pools of alien Triticeae species for sustainable and multi-faceted improvement of the durum wheat crop. Crop Pasture Sci. 2014, 65, 96–111. [Google Scholar] [CrossRef]
- Sloat, L.L.; Davis, S.J.; Gerber, J.S.; Moore, F.C.; Ray, D.K.; West, P.C.; Mueller, N.D. Climate adaptation by crop migration. Nat. Commun. 2020, 11, 1243. [Google Scholar] [CrossRef] [PubMed]
- Bebber, D.P.; Ramotowski, M.A.T.; Gurr, S.J. Crop pests and pathogens move polewards in a warming world. Nat. Clim. Chang. 2013, 3, 985–988. [Google Scholar] [CrossRef]
- Fones, H.N.; Gurr, S.J. NOXious gases and the unpredictability of emerging plant pathogens under climate change. BMC Biol. 2017, 15, 36. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Petronaitis, T.; Simpfendorfer, S.; Hüberli, D. Importance of Fusarium spp. in wheat to food security: A global perspective. In Plant Diseases and Food Security in the 21st Century; Scott, P., Strange, R., Korsten, L., Gullino, M.L., Eds.; Plant Pathology in the 21st Century; Springer: Cham, Switzerland, 2021; Volume 10, pp. 127–159. [Google Scholar] [CrossRef]
- Alahmad, S.; Kang, Y.; Dinglasan, E.; Mazzucotelli, E.; Voss-Fels, K.P.; Able, J.A.; Christopher, J.; Bassi, F.M.; Hickey, L.T. Adaptive traits to improve durum wheat yield in drought and crown rot environments. Int. J. Mol. Sci. 2020, 21, 5260. [Google Scholar] [CrossRef]
- Fakhfakh, M.; Yahyaoui, A.; Rezgui, S.; Elias, E.M.; Daaloul, A. Identification and pathogenicity assessment of Fusarium spp. sampled from durum wheat fields in Tunisia. Afr. J. Biotechnol. 2011, 10, 6529–6539. [Google Scholar] [CrossRef]
- Scala, V.; Aureli, G.; Cesarano, G.; Incerti, G.; Fanelli, C.; Scala, F.; Reverberi, M.; Bonanomi, G. Climate, soil management, and cultivar affect Fusarium head blight incidence and deoxynivalenol accumulation in durum wheat of Southern Italy. Front. Microbiol. 2016, 7, 1014. [Google Scholar] [CrossRef]
- Bouanaka, H.; Bellil, I.; Khelifi, D. Multiple methods for varietal resistance assessment of durum wheat cultivars against Fusarium culmorum the causal agent of Fusarium head blight and crown rot in Algeria. Physiol. Mol. Plant Pathol. 2021, 115, 101683. [Google Scholar] [CrossRef]
- Haile, J.K.; N’Diaye, A.; Walkowiak, S.; Nilsen, K.T.; Clarke, J.M.; Kutcher, H.R.; Steiner, B.; Buerstmayr, H.; Pozniak, C.J. Fusarium head blight in durum wheat: Recent status, breeding directions, and future research prospects. Phytopathology 2019, 109, 1664–1675. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Xie, Q.; Li, G.; Jia, H.; Zhou, J.; Kong, Z.; Li, N.; Yuan, Y. Germplasms, genetics and genomics for better control of disastrous wheat Fusarium head blight. Theor. Appl. Genet. 2020, 133, 1541–1568. [Google Scholar] [CrossRef]
- Buerstmayr, M.; Steiner, B.; Buerstmayr, H. Breeding for Fusarium head blight resistance in wheat—Progress and challenges. Plant Breed. 2020, 139, 429–454. [Google Scholar] [CrossRef]
- Khan, M.K.; Pandey, A.; Athar, T.; Choudhary, S.; Deval, R.; Gezgin, S.; Hamurcu, M.; Topal, A.; Atmaca, E.; Santos, P.A.; et al. Fusarium head blight in wheat: Contemporary status and molecular approaches. 3 Biotech 2020, 10, 172. [Google Scholar] [CrossRef] [PubMed]
- Beres, B.L.; Brûlé-Babel, A.L.; Ye, Z.; Graf, R.J.; Turkington, T.K.; Harding, M.W.; Kutcher, H.R.; Hooker, D.C. Exploring genotype × environment × management synergies to manage Fusarium head blight in wheat. Can. J. Plant Pathol. 2018, 40, 179–188. [Google Scholar] [CrossRef]
- Fernando, W.G.D.; Oghenekaro, A.O.; Tucker, J.R.; Badea, A. Building on a foundation: Advances in epidemiology, resistance breeding, and forecasting research for reducing the impact of Fusarium head blight in wheat and barley. Can. J. Plant Pathol. 2021, 43, 495–526. [Google Scholar] [CrossRef]
- van der Lee, T.; Zhang, H.; van Diepeningen, A.; Waalwijk, C. Biogeography of Fusarium graminearum species complex and chemotypes: A review. Food Addit. Contam. Part A 2015, 32, 453–460. [Google Scholar] [CrossRef]
- Fabre, F.; Bormann, J.; Urbach, S.; Roche, S.; Langin, T.; Bonhomme, L. Unbalanced roles of fungal aggressiveness and host cultivars in the establishment of the Fusarium head blight in bread wheat. Front. Microbiol. 2019, 10, 2857. [Google Scholar] [CrossRef]
- Moreno-Amores, J.; Michel, S.; Löschenberger, F.; Buerstmayr, H. Dissecting the contribution of environmental influences, plant phenology, and disease resistance to improving genomic predictions for Fusarium head blight resistance in wheat. Agronomy 2020, 10, 2008. [Google Scholar] [CrossRef]
- Bönnighausen, J.; Schauer, N.; Schäfer, W.; Bormann, J. Metabolic profiling of wheat rachis node infection by Fusarium graminearum—Decoding deoxynivalenol-dependent susceptibility. New Phytol. 2019, 221, 459–469. [Google Scholar] [CrossRef]
- Nilsen, K.T.; Walkowiak, S.; Kumar, S.; Molina, O.I.; Randhawa, H.S.; Dhariwal, R.; Byrns, B.; Pozniak, C.J.; Henriquez, M.A. Histology and RNA sequencing provide insights into Fusarium head blight resistance in AAC tenacious. Front. Plant Sci. 2021, 11, 570418. [Google Scholar] [CrossRef] [PubMed]
- Brar, G.S.; Karunakaran, C.; Bond, T.; Stobbs, J.; Liu, N.; Hucl, P.J.; Kutcher, H.R. Showcasing the application of synchrotron-based X-ray computed tomography in host–pathogen interactions: The role of wheat rachilla and rachis nodes in Type-II resistance to Fusarium graminearum. Plant Cell Environ. 2019, 42, 509–526. [Google Scholar] [CrossRef] [PubMed]
- Gunnaiah, R.; Kushalappa, A.C.; Duggavathi, R.; Fox, S.; Somers, D.J. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS ONE 2012, 7, e40695. [Google Scholar] [CrossRef] [PubMed]
- Dhokane, D.; Karre, S.; Kushalappa, A.C.; McCartney, C. Integrated metabolo-transcriptomics reveals Fusarium head blight candidate resistance genes in wheat QTL-Fhb2. PLoS ONE 2016, 11, e0155851. [Google Scholar] [CrossRef] [PubMed]
- Lahlali, R.; Kumar, S.; Wang, L.; Forseille, L.; Sylvain, N.; Korbas, M.; Muir, D.; Swerhone, G.; Lawrence, J.R.; Fobert, P.R.; et al. Cell wall biomolecular composition plays a potential role in the host type II resistance to Fusarium head blight in wheat. Front. Microbiol. 2016, 7, 910. [Google Scholar] [CrossRef] [PubMed]
- Kage, U.; Karre, S.; Kushalappa, A.C.; McCartney, C. Identification and characterization of a fusarium head blight resistance gene TaACT in wheat QTL-2DL. Plant Biotechnol. J. 2017, 15, 447–457. [Google Scholar] [CrossRef]
- Soni, N.; Hegde, N.; Dhariwal, A.; Kushalappa, A.C. Role of laccase gene in wheat NILs differing at QTL-Fhb1 for resistance against Fusarium head blight. Plant Sci. 2020, 298, 110574. [Google Scholar] [CrossRef] [PubMed]
- Buerstmayr, M.; Wagner, C.; Nosenko, T.; Omony, J.; Steiner, B.; Nussbaumer, T.; Mayer, K.F.X.; Buerstmayr, H. Fusarium head blight resistance in European winter wheat: Insights from genome-wide transcriptome analysis. BMC Genom. 2021, 22, 470. [Google Scholar] [CrossRef] [PubMed]
- Mesterhazy, A. Updating the breeding philosophy of wheat to Fusarium head blight (FHB): Resistance components, QTL identification, and phenotyping—A review. Plants 2020, 9, 1702. [Google Scholar] [CrossRef]
- Kluger, B.; Bueschl, C.; Lemmens, M.; Berthiller, F.; Häubl, G.; Jaunecker, G.; Adam, G.; Krska, R.; Schuhmacher, R. Stable isotopic labelling-assisted untargeted metabolic profiling reveals novel conjugates of the mycotoxin deoxynivalenol in wheat. Anal. Bioanal. Chem. 2013, 405, 5031–5036. [Google Scholar] [CrossRef]
- Kluger, B.; Bueschl, C.; Lemmens, M.; Michlmayr, H.; Malachova, A.; Koutnik, A.; Maloku, I.; Berthiller, F.; Adam, G.; Krska, R.; et al. Biotransformation of the mycotoxin deoxynivalenol in Fusarium resistant and susceptible near isogenic wheat lines. PLoS ONE 2015, 10, e0119656. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Glutathione in plants: Biosynthesis and physiological role in environmental stress tolerance. Physiol. Mol. Biol. Plants 2017, 23, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Ma, J.; Zhang, Q.; Wu, C.; Zhao, H.; Wu, Y.; Yang, G.; He, G. Genome-wide identification and expression profiling of glutathione transferase gene family under multiple stresses and hormone treatments in wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 986. [Google Scholar] [CrossRef]
- Pan, Y.; Liu, Z.; Rocheleau, H.; Fauteux, F.; Wang, Y.; McCartney, C.; Ouellet, T. Transcriptome dynamics associated with resistance and susceptibility against fusarium head blight in four wheat genotypes. BMC Genom. 2018, 19, 642. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, S.; Ge, W.; Zhao, L.; Hou, B.; Wang, K.; Lyu, Z.; Chen, L.; Xu, S.; Guo, J.; et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, 6493. [Google Scholar] [CrossRef]
- Gunupuru, L.R.; Perochon, A.; Doohan, F.M. Deoxynivalenol resistance as a component of FHB resistance. Trop. Plant Pathol. 2017, 42, 175–183. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Venske, E.; dos Santos, R.S.; Farias, D.d.R.; Rother, V.; Maia, L.C.d.; Pegoraro, C.; Costa de Oliveira, A. Meta-Analysis of the QTLome of Fusarium head blight resistance in bread wheat: Refining the current puzzle. Front. Plant Sci. 2019, 10, 727. [Google Scholar] [CrossRef]
- Zheng, T.; Hua, C.; Li, L.; Sun, Z.; Yuan, M.; Bai, G.; Humphreys, G.; Li, T. Integration of meta-QTL discovery with omics: Towards a molecular breeding platform for improving wheat resistance to Fusarium head blight. Crop J. 2021, 9, 739–749. [Google Scholar] [CrossRef]
- Cuthbert, P.A.; Somers, D.J.; Thomas, J.; Cloutier, S.; Brule-Babel, A. Fine mapping Fhb1, a major gene controlling fusarium head blight resistance in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2006, 112, 1465–1472. [Google Scholar] [CrossRef]
- Schweiger, W.; Steiner, B.; Vautrin, S.; Nussbaumer, T.; Siegwart, G.; Zamini, M.; Jungreithmeier, F.; Gratl, V.; Lemmens, M.; Mayer, K.F.X.; et al. Suppressed recombination and unique candidate genes in the divergent haplotype encoding Fhb1, a major Fusarium head blight resistance locus in wheat. Theor. Appl. Genet. 2016, 129, 1607–1623. [Google Scholar] [CrossRef] [PubMed]
- Rawat, N.; Pumphrey, M.O.; Liu, S.; Zhang, X.; Tiwari, V.K.; Ando, K.; Trick, H.N.; Bockus, W.W.; Akhunov, E.; Anderson, J.A.; et al. Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat. Genet. 2016, 48, 1576–1580. [Google Scholar] [CrossRef]
- Li, G.; Zhou, J.; Jia, H.; Gao, Z.; Fan, M.; Luo, Y.; Zhao, P.; Xue, S.; Li, N.; Yuan, Y.; et al. Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat. Genet. 2019, 51, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Bernardo, A.; Tian, B.; Chen, H.; Wang, S.; Ma, H.; Cai, S.; Liu, D.; Zhang, D.; Li, T.; et al. A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat. Genet. 2019, 51, 1099–1105. [Google Scholar] [CrossRef]
- Paudel, B.; Zhuang, Y.; Galla, A.; Dahal, S.; Qiu, Y.; Ma, A.; Raihan, T.; Yien, Y. WFhb1-1 plays an important role in resistance against Fusarium head blight in wheat. Sci. Rep. 2020, 10, 7794. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Zhou, J.; Xue, S.; Li, G.; Yan, H.; Ran, C.; Zhang, Y.; Shi, J.; Jia, L.; Wang, X.; et al. A journey to understand wheat Fusarium head blight resistance in the Chinese wheat landrace Wangshuibai. Crop J. 2018, 6, 48–59. [Google Scholar] [CrossRef]
- Lagudah, E.S.; Krattinger, S.G. A new player contributing to durable Fusarium resistance. Nat. Genet. 2019, 51, 1070–1071. [Google Scholar] [CrossRef]
- Gunnaiah, R.; Kushalappa, A.C. Metabolomics deciphers the host resistance mechanisms in wheat cultivar Sumai-3, against trichothecene producing and non-producing isolates of Fusarium graminearum. Plant Physiol. Bioch. 2014, 83, 40–50. [Google Scholar] [CrossRef]
- Warth, B.; Parich, A.; Bueschl, C.; Schoefbeck, D.; Neumann, N.K.N.; Kluger, B.; Schuster, K.; Krska, R.; Adam, G.; Lemmens, M.; et al. GC–MS based targeted metabolic profiling identifies changes in the wheat metabolome following deoxynivalenol treatment. Metabolomics 2015, 11, 722–738. [Google Scholar] [CrossRef]
- Cuperlovic-Culf, M.; Wang, L.; Forseille, L.; Boyle, K.; Merkley, N.; Burton, I.; Fobert, P.R. Metabolic biomarker panels of response to Fusarium head blight infection in different wheat varieties. PLoS ONE 2016, 11, e0153642. [Google Scholar] [CrossRef]
- Lemmens, M.; Scholz, U.; Berthiller, F.; Dall’Asta, C.; Koutnik, A.; Schuhmacher, R.; Adam, G.; Buerstmayr, H.; Mesterházy, Á.; Krska, R.; et al. The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol. Plant-Microbe Interact. 2005, 18, 1318–1324. [Google Scholar] [CrossRef] [PubMed]
- Cuthbert, P.A.; Somers, D.J.; Brulé-Babel, A. Mapping of Fhb2 on chromosome 6BS: A gene controlling Fusarium head blight field resistance in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2007, 114, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Ceoloni, C.; Kuzmanović, L.; Gennaro, A.; Forte, P.; Giorgi, D.; Grossi, M.R.; Bitti, A. Genomes, chromosomes and genes of perennial Triticeae of the genus Thinopyrum: The value of their transfer into wheat for gains in cytogenomic knowledge and ‘precision’ breeding. In Genomics of Plant Genetic Resources; Tuberosa, R., Graner, A., Frison, E., Eds.; Springer Science: Dordrecht, The Netherlands, 2014; pp. 333–358. [Google Scholar] [CrossRef]
- Ceoloni, C.; Kuzmanović, L.; Forte, P.; Virili, M.E.; Bitti, A. Wheat-perennial Triticeae introgressions: Major achievements and prospects. In Alien Introgression in Wheat—Cytogenetics, Molecular Biology, and Genomics; Molnár-Láng, M., Ceoloni, C., Doležel, J., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 273–313. [Google Scholar] [CrossRef]
- Ceoloni, C.; Kuzmanović, L.; Ruggeri, R.; Rossini, F.; Forte, P.; Cuccurullo, A.; Bitti, A. Harnessing genetic diversity of wild gene pools to enhance wheat crop production and sustainability: Challenges and opportunities. Diversity 2017, 9, 55. [Google Scholar] [CrossRef]
- Zhu, Z.; Hao, Y.; Mergoum, M.; Bai, G.; Humphreys, G.; Cloutier, S.; Xia, X.; He, Z. Breeding wheat for resistance to Fusarium head blight in the Global North: China, USA, and Canada. Crop J. 2019, 7, 730–738. [Google Scholar] [CrossRef]
- Forte, P.; Virili, M.E.; Kuzmanović, L.; Moscetti, I.; Gennaro, A.; D’Ovidio, R.; Ceoloni, C. A novel assembly of Thinopyrum ponticum genes into the durum wheat genome: Pyramiding Fusarium head blight resistance onto recombinant lines previously engineered for other beneficial traits from the same alien species. Mol. Breed. 2014, 34, 1701–1716. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, X.; Hou, Y.; Cai, J.; Shen, X.; Zhou, T.; Xu, H.; Ohm, H.W.; Wang, H.; Li, A.; et al. High-density mapping of the major FHB resistance gene Fhb7 derived from Thinopyrum ponticum and its pyramiding with Fhb1 by marker-assisted selection. Theor. Appl. Genet. 2015, 128, 2301–2316. [Google Scholar] [CrossRef]
- Ceoloni, C.; Forte, P.; Kuzmanović, L.; Tundo, S.; Moscetti, I.; De Vita, P.; Virili, M.E.; D’Ovidio, R. Cytogenetic mapping of a major locus for resistance to Fusarium head blight and crown rot of wheat on Thinopyrum elongatum 7EL and its pyramiding with valuable genes from a Th. ponticum homoeologous arm onto bread wheat 7DL. Theor. Appl. Genet. 2017, 130, 2005–2024. [Google Scholar] [CrossRef]
- Guo, X.; Shi, Q.; Yuan, J.; Zhang, J.; Wang, M.; Wang, J.; Wang, C.; Fu, S.; Su, H.; Liu, Y.; et al. Alien chromatin other than the GST-encoding Fhb7 candidate confers Fusarium head blight resistance in wheat breeding. bioRxiv 2021, 02.03.429547. [Google Scholar] [CrossRef]
- Guo, X.; Wang, M.; Kang, H.; Zhou, Y.; Han, F. Distribution, polymorphism and function characteristics of the GST-encoding Fhb7 in Triticeae. Plants 2022, 11, 2074. [Google Scholar] [CrossRef]
- Konkin, D.; Hsueh, Y.-C.; Kirzinger, M.; Kubaláková, M.; Haldar, A.; Balcerzak, M.; Han, F.; Fedak, G.; Doležel, J.; Sharpe, A.; et al. Genomic sequencing of Thinopyrum elongatum chromosome arm 7EL, carrying fusarium head blight resistance, and characterization of its impact on the transcriptome of the introgressed line CS-7EL. BMC Genom. 2022, 23, 228. [Google Scholar] [CrossRef]
- Kuzmanović, L.; Mandalà, G.; Tundo, S.; Ciorba, R.; Frangella, M.; Ruggeri, R.; Rossini, F.; Gevi, F.; Rinalducci, S.; Ceoloni, C. Equipping durum wheat—Thinopyrum ponticum recombinant lines with a Thinopyrum elongatum major QTL for resistance to Fusarium diseases through a cytogenetic strategy. Front. Plant Sci. 2019, 10, 1324. [Google Scholar] [CrossRef]
- Uhlig, S.; Stanic, A.; Hofgaard, I.S.; Kluger, B.; Schuhmacher, R.; Miles, C.O. Glutathione-conjugates of deoxynivalenol in naturally contaminated grain are primarily linked via the epoxide group. Toxins 2016, 8, 329. [Google Scholar] [CrossRef]
- Wang, J.R.; Wang, L.; Gulden, S.; Rocheleau, H.; Balcerzak, M.; Hattori, J.; Cao, W.; Han, F.; Zheng, Y.-L.; Fedak, G.; et al. RNA profiling of fusarium head blight-resistant wheat addition lines containing the Thinopyrum elongatum chromosome 7E. Can. J. Plant Pathol. 2010, 32, 188–214. [Google Scholar] [CrossRef]
- Miller, S.S.; Watson, E.M.; Lazebnik, J.; Gulden, S.; Balcerzak, M.; Fedak, G.; Ouellet, T. Characterization of an alien source of resistance to Fusarium head blight transferred to Chinese spring wheat. Botany 2011, 89, 301–311. [Google Scholar] [CrossRef]
- Hadinezhad, M.; Miller, S.S. Response to Fusarium graminearum infection in the rachis of a resistant and a susceptible wheat genotype. Can. J. Plant Pathol. 2019, 42, 265–278. [Google Scholar] [CrossRef]
- Kuzmanović, L.; Giovenali, G.; Ruggeri, R.; Rossini, F.; Ceoloni, C. Small “nested” introgressions from wild Thinopyrum species, conferring effective resistance to Fusarium diseases, positively impact durum wheat yield potential. Plants 2021, 10, 579. [Google Scholar] [CrossRef]
- Tundo, S.; Janni, M.; Moscetti, I.; Mandalà, G.; Savatin, D.; Blechl, A.; Favaron, F.; D’Ovidio, R. PvPGIP2 accumulation in specific floral tissues but not in the endosperm limits Fusarium graminearum infection in wheat. Mol. Plant Microbe. Interact. 2016, 29, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Gevi, F.; Fanelli, G.; Zolla, L.; Rinalducci, S. Untargeted metabolomics of plant leaf tissues. Methods Mol. Biol. 2019, 1978, 187–195. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. MetPA: A web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 2010, 26, 2342–2344. [Google Scholar] [CrossRef]
- Zhang, Y.; Jin, X.; Ouyang, Z.; Li, X.; Liu, B.; Huang, L.; Hong, Y.; Zhang, H.; Song, F.; Li, D. Vitamin B6 contributes to disease resistance against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea in Arabidopsis thaliana. J. Plant Physiol. 2015, 175, 21–25. [Google Scholar] [CrossRef]
- Kim, D.R.; Jeon, C.W.; Cho, G.; Thomashow, L.S.; Weller, D.M.; Paik, M.J.; Lee, Y.B.; Kwak, Y.S. Glutamic acid reshapes the plant microbiota to protect plants against pathogens. Microbiome 2021, 9, 244. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.S.; Chung, Y.H.; Hsieh, M.H. Glutamate: A multifunctional amino acid in plants. Plant Sci. 2022, 318, 111238. [Google Scholar] [CrossRef] [PubMed]
- Nussbaumer, T.; Warth, B.; Sharma, S.; Ametz, C.; Bueschl, C.; Parich, A.; Pfeifer, M.; Siegwart, G.; Steiner, B.; Lemmens, M.; et al. Joint transcriptomic and metabolomic analyses reveal changes in the primary metabolism and imbalances in the subgenome orchestration in the bread wheat molecular response to Fusarium graminearum. G3-Genes Genom. Genet. 2015, 5, 2579–2592. [Google Scholar] [CrossRef] [PubMed]
- De Bruyne, L.; Höfte, M.; De Vleesschauwer, D. Connecting growth and defense: The emerging roles of brassinosteroids and gibberellins in plant innate immunity. Mol. Plant 2014, 7, 943–959. [Google Scholar] [CrossRef]
- Berkels, R.; Taubert, D.; Gründemann, D.; Schömig, E. Agmatine signaling: Odds and threads. Cardiovasc. Drug Rev. 2004, 22, 7–16. [Google Scholar] [CrossRef]
- Gardiner, D.M.; Kazan, K.; Praud, S.; Torney, F.J.; Rusu, A.; Manners, J.M. Early activation of wheat polyamine biosynthesis during Fusarium head blight implicates putrescine as an inducer of trichothecene mycotoxin production. BMC Plant Biol. 2010, 10, 289. [Google Scholar] [CrossRef]
- Pasquali, M.; Serchi, T.; Cocco, E.; Leclercq, C.C.; Planchon, S.; Guignard, C.; Renaut, J.; Hoffmann, L. A Fusarium graminearum strain-comparative proteomic approach identifies regulatory changes triggered by agmatine. J. Proteom. 2016, 137, 107–116. [Google Scholar] [CrossRef]
- Nakajima, Y.; Akasaka, M.; Shiobara, T.; Kitou, Y.; Maeda, K.; Kanamaru, K.; Ohsato, S.; Kobayashi, T.; Nishiuchi, T.; Kimura, M. Impact of nitrogen metabolism-associated culture pH changes on regulation of Fusarium trichothecene biosynthesis: Revision of roles of polyamine agmatine and transcription factor AreA. Curr. Genet. 2020, 66, 1179–1190. [Google Scholar] [CrossRef]
- Jiménez-Bremont, J.F.; Marina, M.; Guerrero-González, M.L.; Rossi, F.R.; Sánchez-Rangel, D.; Rodríguez-Kessler, M.; Ruiz, O.A.; Gárriz, A. Physiological and molecular implications of plant polyamine metabolism during biotic interactions. Front. Plant Sci. 2014, 5, 95. [Google Scholar] [CrossRef]
- Su, P.; Zhao, L.; Li, W.; Zhao, J.; Yan, J.; Ma, X.; Li, A.; Wang, H.; Kong, L. Integrated metabolo-transcriptomics and functional characterization reveals that the wheat auxin receptor TIR1 negatively regulates defense against Fusarium graminearum. J. Integr. Plant Biol. 2021, 63, 340–352. [Google Scholar] [CrossRef]
- Gauthier, L.; Atanasova-Penichon, V.; Chéreau, S.; Richard-Forget, F. Metabolomics to decipher the chemical defense of cereals against Fusarium graminearum and deoxynivalenol accumulation. Int. J. Mol. Sci. 2015, 16, 24839–24872. [Google Scholar] [CrossRef]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef] [PubMed]
- Treutter, D. Significance of flavonoids in plant resistance: A review. Environ. Chem. Lett. 2006, 4, 147–157. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic acid biosynthesis in plants. Front. Plant Sci. 2020, 11, 338. [Google Scholar] [CrossRef]
- Hao, Q.; Wang, W.; Han, X.; Wu, J.; Lyu, B.; Chen, F.; Caplan, A.; Li, C.; Wu, J.; Wang, W.; et al. Isochorismate-based salicylic acid biosynthesis confers basal resistance to Fusarium graminearum in barley. Mol. Plant Pathol. 2018, 19, 1995–2010. [Google Scholar] [CrossRef]
- Whitney, K.; Gracia-Gomez, G.; Anderson, J.A.; Simsek, S. Time course metabolite profiling of Fusarium Head Blight-infected hard red spring wheat using Ultra-High-Performance Liquid Chromatography coupled with quadrupole time of flight/MS. J. Agric. Food Chem. 2022, 70, 4152–4163. [Google Scholar] [CrossRef]
- Wang, L.; Li, Q.; Liu, Z.; Surendra, A.; Pan, Y.; Li, Y.; Zaharia, L.I.; Ouellet, T.; Fobert, P.R. Integrated transcriptome and hormone profiling highlight the role of multiple phytohormone pathways in wheat resistance against fusarium head blight. PLoS ONE 2018, 13, e0207036. [Google Scholar] [CrossRef]
- del Río, J.C.; Rencoret, J.; Gutiérrez, A.; Elder, T.; Kim, H.; Ralph, J. Lignin monomers from beyond the canonical monolignol biosynthetic pathway: Another brick in the wall. ACS Sustain. Chem. Eng. 2020, 8, 4997–5012. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]
- Mnich, E.; Bjarnholt, N.; Eudes, A.; Harholt, J.; Holland, C.; Jørgensen, B.; Larsen, F.H.; Liu, M.; Manat, R.; Meyer, A.S.; et al. Phenolic cross-links: Building and de-constructing the plant cell wall. Nat. Prod. Rep. 2020, 37, 919–961. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Wang, Z.; Wei, C.; Amo, A.; Ahmed, B.; Yang, X.; Zhang, X. Phenylpropanoid pathway engineering: An emerging approach towards plant defense. Pathogens 2020, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Fu, J.; Hiromasa, Y.; Pan, H.; Bai, G. Differentially expressed proteins associated with Fusarium head blight resistance in wheat. PLoS ONE 2013, 8, e82079. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Eudes, F.; Laroche, A. Identification of differentially regulated proteins in response to a compatible interaction between the pathogen Fusarium graminearum and its host, Triticum aestivum. Proteomics 2006, 6, 4599–4609. [Google Scholar] [CrossRef] [PubMed]
- Awika, J.M.; Rooney, L.W.; Wu, X.; Prior, R.L.; Cisneros-Zevallos, L. Screening methods to measure antioxidant activity of sorghum (sorghum bicolor) and sorghum products. J. Agr. Food Chem. 2003, 51, 6657–6662. [Google Scholar] [CrossRef]
- Gorinstein, S.; Lojek, A.; Číž, M.; Pawelzik, E.; Delgado-Licon, E.; Medina, O.J.; Moreno, M.; Salas, I.A.; Goshev, I. Comparison of composition and antioxidant capacity of some cereals and pseudocereals. Int. J. Food Sci. Tech. 2008, 43, 629–637. [Google Scholar] [CrossRef]
- Fernandez-Orozco, R.; Li, L.; Harflett, C.; Shewry, P.R.; Ward, J.L. Effects of environment and genotype on phenolic acids in wheat in the HEALTHGRAIN diversity screen. J. Agric. Food Chem. 2010, 58, 9341–9352. [Google Scholar] [CrossRef]
- McKeehen, J.D.; Busch, R.H.; Fulcher, R.G. Evaluation of wheat (Triticum aestivum L.) phenolic acids during grain development and their contribution to Fusarium resistance. J. Agric. Food Chem. 1999, 47, 1476–1482. [Google Scholar] [CrossRef]
- Siranidou, E.; Kang, Z.; Buchenauer, H. Studies on symptom development, phenolic compounds and morphological defence responses in wheat cultivars differing in resistance to Fusarium head blight. J. Phytopathol. 2003, 150, 200–208. [Google Scholar] [CrossRef]
- Husain, T.; Fatima, A.; Suhel, M.; Singh, S.; Sharma, A.; Prasad, S.M.; Singh, V.P. A brief appraisal of ethylene signaling under abiotic stress in plants. Plant Signal. Behav. 2020, 15, 1782051. [Google Scholar] [CrossRef]
- Keunen, E.; Schellingen, K.; Vangronsveld, J.; Cuypers, A. Ethylene and metal stress: Small molecule, big impact. Front. Plant Sci. 2016, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Hyodo, H.; Yang, S.F. Ethylene-enhanced synthesis of phenylalanine ammonia-lyase in pea seedlings. Plant Physiol. 1971, 47, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Nambeesan, S.; Abuqamar, S.; Laluk, K.; Mattoo, A.K.; Mickelbart, M.V.; Ferruzzi, M.G.; Mengiste, T.; Handa, A.K. Polyamines attenuate ethylene-mediated defense responses to abrogate resistance to Botrytis cinerea in tomato. Plant Physiol. 2012, 158, 1034–1045. [Google Scholar] [CrossRef] [PubMed]
- Baró-Montel, N.; Vall-Llaura, N.; Giné-Bordonaba, J.; Usall, J.; Serrano-Prieto, S.; Teixidó, N.; Torres, R. Double-sided battle: The role of ethylene during Monilinia spp. infection in peach at different phenological stages. Plant Physiol. Bioch. 2019, 144, 324–333. [Google Scholar] [CrossRef]
- Pal, M.; Janda, T. Role of polyamine metabolism in plant pathogen interactions. .J Plant Sci. Phytopathol. 2017, 1, 95–100. [Google Scholar] [CrossRef]
- Gupta, K.; Sengupta, A.; Chakraborty, M.; Gupta, B. Hydrogen Peroxide and Polyamines Act as Double Edged Swords in Plant Abiotic Stress Responses. Front. Plant Sci. 2016, 7, 1343. [Google Scholar] [CrossRef]
- Gerlin, L.; Baroukh, C.; Genin, S. Polyamines: Double agents in disease and plant immunity. Trends Plant Sci. 2021, 26, 1061–1071. [Google Scholar] [CrossRef]
- Cuperlovic-Culf, M.; Loewen, M.; Rajagopalan, N.; Surendra, A. Perspectives on the specific targeting of Fusarium graminearum for the development of alternative head blight treatment approaches. Plant Pathol. 2017, 66, 1391–1403. [Google Scholar] [CrossRef]
- Crespo-Sempere, A.; Estiarte, N.; Marin, S.; Sanchis, V.; Ramos, A.J. Targeting Fusarium graminearum control via polyamine enzyme inhibitors and polyamine analogs. Food Microbiol. 2015, 49, 95–103. [Google Scholar] [CrossRef]
- Rocha, R.O.; Wilson, R.A. Essential, deadly, enigmatic: Polyamine metabolism and roles in fungal cells. Fungal Biol. Rev. 2019, 33, 47–57. [Google Scholar] [CrossRef]
- Tang, G.; Xia, H.; Liang, J.; Ma, Z.; Liu, W. Spermidine is critical for growth, development, environmental adaptation and virulence in Fusarium graminearum. Front. Microbiol. 2021, 19, 3484. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.C.; Chiang, B.Y.; Chou, C.C.; Chen, T.C.; Chen, Y.J.; Chen, Y.J.; Lin, C.H. Glutathionylspermidine in the modification of protein SH groups: The enzymology and its application to study protein glutathionylation. Molecules 2015, 20, 1452–1474. [Google Scholar] [CrossRef] [PubMed]
- Comini, M.A. Polyamine-based thiols in pathogens. In Redox Chemistry and Biology of Thiols; Academic Press: New York, NY, USA, 2022; pp. 555–584. [Google Scholar]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef]
- Jancewicz, A.L.; Gibbs, N.M.; Masson, P.H. Cadaverine’s functional role in plant development and environmental response. Front. Plant Sci. 2016, 7, 870. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, C.F.; Ugena, L.; Humplík, J.F.; Polák, M.; Ćavar Zeljković’, S.; Podlešáková, K.; Fürst, T.; De Diego, N.; Spíchal, L. A novel image-based screening method to study water-deficit response and recovery of barley populations using canopy dynamics phenotyping and simple metabolite profiling. Front. Plant Sci. 2019, 10, 1252. [Google Scholar] [CrossRef]
- Czégény, G.; Kőrösi, L.; Strid, Å.; Hideg, É. Multiple roles for Vitamin B in plant acclimation to UV-B. Sci. Rep. 2019, 9, 1259. [Google Scholar] [CrossRef]
- Parra, M.; Stahl, S.; Hellmann, H. Vitamin B6 and its role in cell metabolism and physiology. Cells 2018, 7, 84. [Google Scholar] [CrossRef]
- Mangel, N.; Fudge, J.B.; Li, K.T.; Wu, T.Y.; Tohge, T.; Fernie, A.R.; Szurek, B.; Fitzpatrick, T.B.; Gruissem, W.; Vanderschuren, H. Enhancement of vitamin B6 levels in rice expressing Arabidopsis vitamin B6 biosynthesis de novo genes. Plant J. 2019, 99, 1047–1065. [Google Scholar] [CrossRef]
- Vanderschuren, H.; Boycheva, S.; Li, K.-T.; Szydlowski, N.; Gruissem, W.; Fitzpatrick, T.B. Strategies for vitamin B6 biofortification of plants: A dual role as a micronutrient and a stress protectant. Front. Plant Sci. 2013, 4, 143. [Google Scholar] [CrossRef]
- Fudge, J.; Mangel, N.; Gruissem, W.; Vanderschuren, H.; Fitzpatrick, T.B. Rationalising vitamin B6 biofortification in crop plants. Curr. Opin. Biotech. 2017, 44, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Samsatly, J.; Copley, T.R.; Jabaji, S.H. Antioxidant genes of plants and fungal pathogens are distinctly regulated during disease development in different Rhizoctonia solani pathosystems. PLoS ONE 2018, 13, e0192682. [Google Scholar] [CrossRef] [PubMed]
- Dell’Aglio, E.; Boycheva, S.; Fitzpatrick, T.B. The pseudoenzyme PDX1.2 sustains vitamin B6 biosynthesis as a function of heat stress. Plant Physiol. 2017, 174, 2098–2112. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Li, W.; Zhang, W.; Jin, X.; Shenkute, A.G.; Aynalem, T.; Xu, S.; Wang, W. Transcriptome sequencing analysis provides insights into the response to Fusarium oxysporum in Lilium pumilum. Evol. Bioinform. 2019, 15, 1176934319838818. [Google Scholar] [CrossRef]
- De la Cruz, M.T.; Hernández, E.E.; González, J.A.; Zozaya, R.D.; García, J.A.; Rodríguez, M.L. Bioinformatic analysis deciphers the molecular toolbox in the endophytic/pathogenic behaviour in F. oxysporum f. sp. vanillae—V. planifolia Jacks interaction. bioRxiv 2021, 03.23.436347. [Google Scholar] [CrossRef]
- Kosová, K.; Chrpová, J.; Šantrůček, J.; Hynek, R.; Štěrbová, L.; Vítámvás, P.; Bradová, J.; Prášil, I.T. The effect of Fusarium culmorum infection and deoxynivalenol (DON) application on proteome response in barley cultivars Chevron and Pedant. J. Proteom. 2017, 169, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Jansen, C.; von Wettstein, D.; Schäfer, W.; Kogel, K.-H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef]
- Ilgen, P.; Hadeler, B.; Maier, F.J.; Schäfer, W. Developing kernel and rachis node induce the trichothecene pathway of Fusarium graminearum during wheat head infection. Mol. Plant-Microbe Interact. 2009, 22, 899–908. [Google Scholar] [CrossRef]
- Yu, G.; Chen, Q.; Chen, F.; Liu, H.; Lin, J.; Chen, R.; Ren, C.; Wei, J.; Zhang, Y.; Yang, F.; et al. Glutathione promotes degradation and metabolism of residual fungicides by inducing UDP-glycosyltransferase genes in tomato. Front. Plant Sci. 2022, 13, 893508. [Google Scholar] [CrossRef]
- Foroud, N.A.; Baines, D.; Gagkaeva, T.Y.; Thakor, N.; Badea, A.; Steiner, B.; Bürstmayr, M.; Bürstmayr, H. Trichothecenes in cereal grains—An update. Toxins 2019, 11, 634. [Google Scholar] [CrossRef]
- Shinozuka, H.; Hettiarachchige, I.K.; Shinozuka, M.; Cogan, N.O.I.; Spangenberg, G.C.; Cocks, B.G.; Forster, J.W.; Sawbridge, T.I. Horizontal transfer of a ß-1,6-glucanase gene from an ancestral species of fungal endophyte to a cool-season grass host. Sci. Rep. 2017, 7, 9024. [Google Scholar] [CrossRef] [PubMed]
- Shinozuka, H.; Shinozuka, M.; de Vries, E.M.; Sawbridge, T.I.; Spangenberg, G.C.; Cocks, B.G. Fungus-originated genes in the genomes of cereal and pasture grasses acquired through ancient lateral transfer. Sci. Rep. 2020, 10, 19883. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Bae, H. Horizontal gene transfer and endophytes: An implication for the acquisition of novel traits. Plants 2020, 9, 305. [Google Scholar] [CrossRef]
- Ma, J.; Wang, S.; Zhu, X.; Sun, G.; Chang, G.; Li, L.; Hu, X.; Zhang, S.; Zhou, Y.; Song, C.-P.; et al. Major episodes of horizontal gene transfer drove the evolution of land plants. Mol. Plant 2022, 15, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lathe, R.S.; Li, J.; He, H.; Bhalerao, R.P. Towards understanding the biological foundations of perenniality. Trends. Plant. Sci. 2022, 27, 56–68. [Google Scholar] [CrossRef]
- Guo, X.; Shi, Q.; Wang, M.; Yuan, J.; Zhang, J.; Wang, J.; Liu, Y.; Su, H.; Wang, Z.; Li, J.; et al. Functional analysis of the glutathione S-transferases from Thinopyrum and its derivatives on wheat Fusarium head blight resistance. Plant Biotechnol. J. 2023. [Google Scholar] [CrossRef]
- Perochon, A.; Benbow, H.R.; Ślęczka-Brady, K.; Malla, K.B.; Doohan, F.M. Analysis of the chromosomal clustering of Fusarium-responsive wheat genes uncovers new players in the defence against head blight disease. Sci. Rep. 2021, 11, 7446. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fanelli, G.; Kuzmanović, L.; Giovenali, G.; Tundo, S.; Mandalà, G.; Rinalducci, S.; Ceoloni, C. Untargeted Metabolomics Reveals a Multi-Faceted Resistance Response to Fusarium Head Blight Mediated by the Thinopyrum elongatum Fhb7E Locus Transferred via Chromosome Engineering into Wheat. Cells 2023, 12, 1113. https://doi.org/10.3390/cells12081113
Fanelli G, Kuzmanović L, Giovenali G, Tundo S, Mandalà G, Rinalducci S, Ceoloni C. Untargeted Metabolomics Reveals a Multi-Faceted Resistance Response to Fusarium Head Blight Mediated by the Thinopyrum elongatum Fhb7E Locus Transferred via Chromosome Engineering into Wheat. Cells. 2023; 12(8):1113. https://doi.org/10.3390/cells12081113
Chicago/Turabian StyleFanelli, Giuseppina, Ljiljana Kuzmanović, Gloria Giovenali, Silvio Tundo, Giulia Mandalà, Sara Rinalducci, and Carla Ceoloni. 2023. "Untargeted Metabolomics Reveals a Multi-Faceted Resistance Response to Fusarium Head Blight Mediated by the Thinopyrum elongatum Fhb7E Locus Transferred via Chromosome Engineering into Wheat" Cells 12, no. 8: 1113. https://doi.org/10.3390/cells12081113
APA StyleFanelli, G., Kuzmanović, L., Giovenali, G., Tundo, S., Mandalà, G., Rinalducci, S., & Ceoloni, C. (2023). Untargeted Metabolomics Reveals a Multi-Faceted Resistance Response to Fusarium Head Blight Mediated by the Thinopyrum elongatum Fhb7E Locus Transferred via Chromosome Engineering into Wheat. Cells, 12(8), 1113. https://doi.org/10.3390/cells12081113








