Membrane Curvature: The Inseparable Companion of Autophagy
Abstract
1. Introduction
2. Membrane Curvature
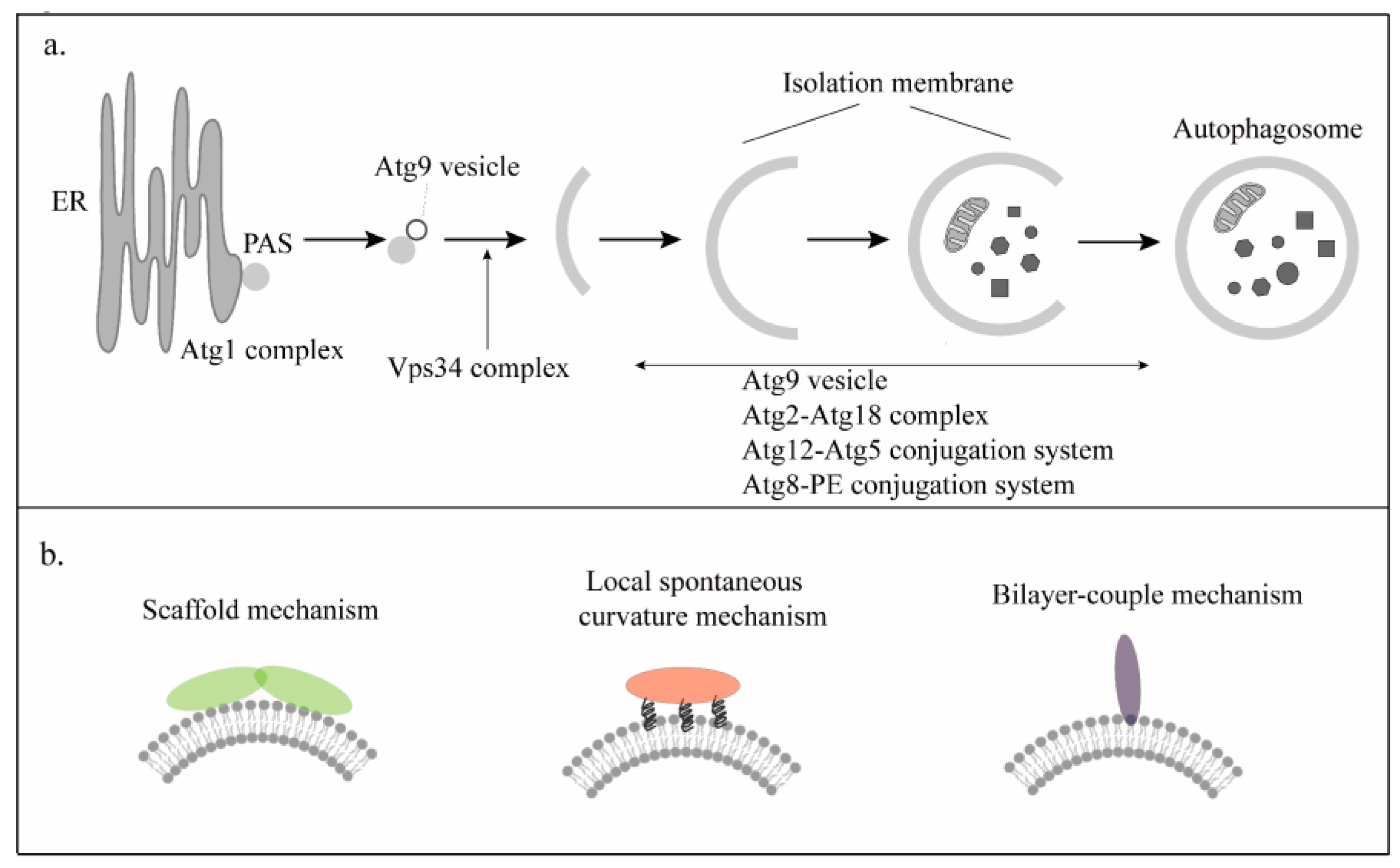
3. Membrane Curvature in the Initiation Phase of Autophagy
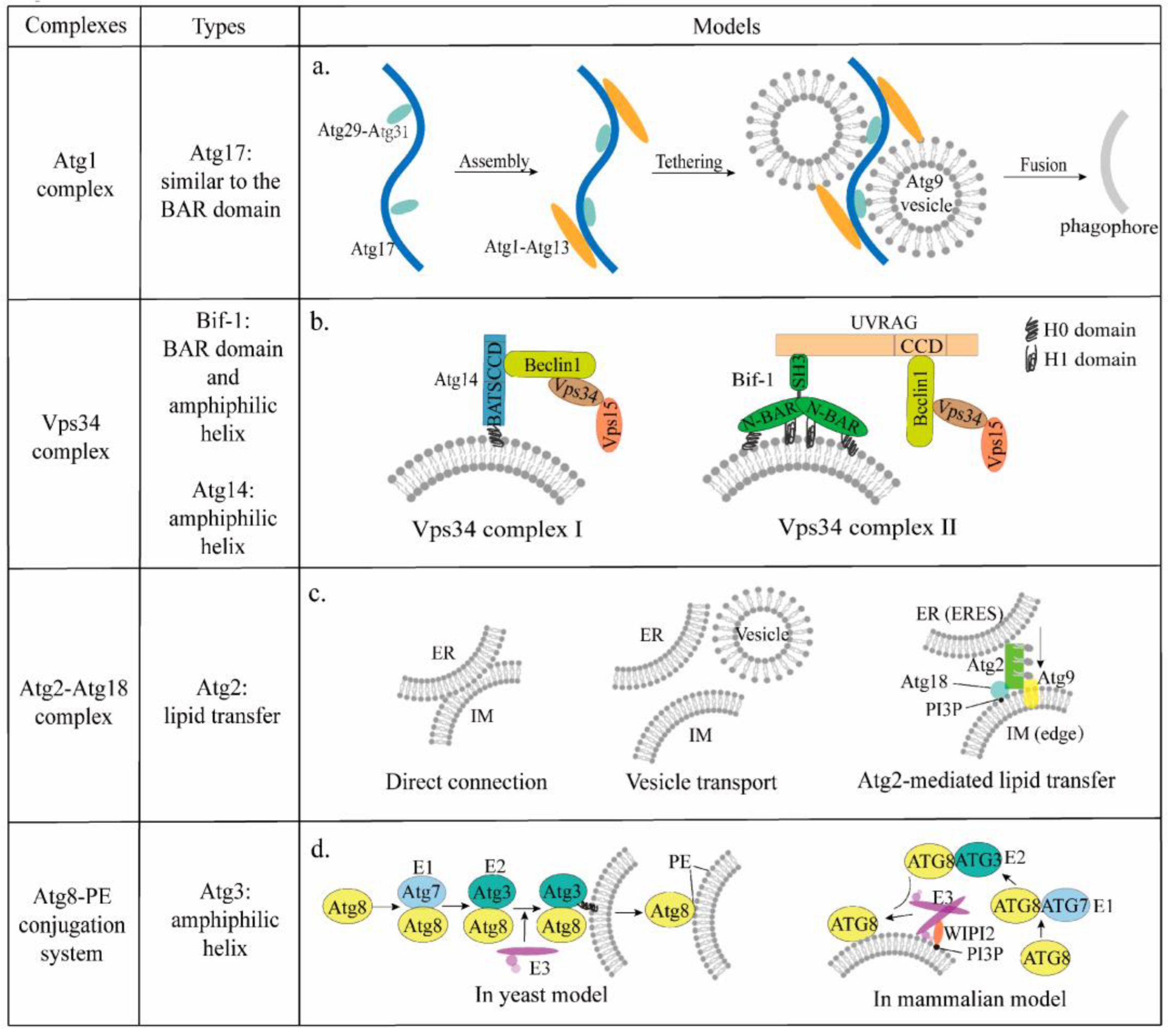
3.1. Atg1 Complex
3.2. Vps34 Complex
3.2.1. Bif-1
3.2.2. Barkor/Atg14
3.3. Atg9
4. Membrane Curvature in the Extension Phase of Autophagy
4.1. Atg2-Atg18 Complex
4.2. Atg8-PE and Atg12-Atg5 Conjugation System
5. Membrane Curvature in the Maturation Phase of Autophagy
6. Membrane Curvature in Selective Autophagy
6.1. ER-Phagy
6.2. Nucleophagy
6.3. Xenophagy
7. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fakhree, M.A.A.; Blum, C.; Claessens, M. Shaping membranes with disordered proteins. Arch Biochem. Biophys. 2019, 677, 108163. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Qiu, Y.; Grace, C.R.R.; Liu, X.; Klionsky, D.J.; Schulman, B.A. A switch element in the autophagy E2 Atg3 mediates allosteric regulation across the lipidation cascade. Nat. Commun. 2019, 10, 3600. [Google Scholar] [CrossRef] [PubMed]
- Has, C.; Sivadas, P.; Das, S.L. Insights into Membrane Curvature Sensing and Membrane Remodeling by Intrinsically Disordered Proteins and Protein Regions. J. Membr. Biol. 2022, 255, 237–259. [Google Scholar] [CrossRef]
- Wu, H.; Voeltz, G.K. Reticulon-3 Promotes Endosome Maturation at ER Membrane Contact Sites. Dev. Cell 2021, 56, 52–66.e57. [Google Scholar] [CrossRef]
- Westrate, L.M.; Lee, J.E.; Prinz, W.A.; Voeltz, G.K. Form follows function: The importance of endoplasmic reticulum shape. Annu. Rev. Biochem. 2015, 84, 791–811. [Google Scholar] [CrossRef]
- Wu, H.; Carvalho, P.; Voeltz, G.K. Here, there, and everywhere: The importance of ER membrane contact sites. Science 2018, 361, eaan5835. [Google Scholar] [CrossRef]
- Berganza, E.; Ebrahimkutty, M.P.; Vasantham, S.K.; Zhong, C.; Wunsch, A.; Navarrete, A.; Galic, M.; Hirtz, M. A multiplexed phospholipid membrane platform for curvature sensitive protein screening. Nanoscale 2021, 13, 12642–12650. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Qian, X.; Hu, M.; Zhang, R.; Liu, N.; Huang, Y.; Yang, J.; Zhang, J.; Bai, H.; Yang, Y.; et al. STYK1 promotes autophagy through enhancing the assembly of autophagy-specific class III phosphatidylinositol 3-kinase complex I. Autophagy 2020, 16, 1786–1806. [Google Scholar] [CrossRef] [PubMed]
- Parzych, K.R.; Klionsky, D.J. An overview of autophagy: Morphology, mechanism, and regulation. Antioxid. Redox Signal. 2014, 20, 460–473. [Google Scholar] [CrossRef]
- Orenstein, S.J.; Cuervo, A.M. Chaperone-mediated autophagy: Molecular mechanisms and physiological relevance. Semin. Cell Dev. Biol. 2010, 21, 719–726. [Google Scholar] [CrossRef]
- Osawa, T.; Alam, J.M.; Noda, N.N. Membrane-binding domains in autophagy. Chem. Phys. Lipids 2019, 218, 1–9. [Google Scholar] [CrossRef]
- Fujioka, Y.; Suzuki, S.W.; Yamamoto, H.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Akada, R.; Inagaki, F.; Ohsumi, Y.; Noda, N.N. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat. Struct. Mol. Biol. 2014, 21, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Zimmerberg, J.; Kozlov, M.M. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 2006, 7, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Roux, A.; Cappello, G.; Cartaud, J.; Prost, J.; Goud, B.; Bassereau, P. A minimal system allowing tubulation with molecular motors pulling on giant liposomes. Proc. Natl. Acad. Sci. USA 2002, 99, 5394–5399. [Google Scholar] [CrossRef] [PubMed]
- Farsad, K.; De Camilli, P. Mechanisms of membrane deformation. Curr. Opin. Cell Biol. 2003, 15, 372–381. [Google Scholar] [CrossRef]
- Mim, C.; Unger, V.M. Membrane curvature and its generation by BAR proteins. Trends Biochem. Sci. 2012, 37, 526–533. [Google Scholar] [CrossRef]
- Qualmann, B.; Koch, D.; Kessels, M.M. Let’s go bananas: Revisiting the endocytic BAR code. EMBO J. 2011, 30, 3501–3515. [Google Scholar] [CrossRef]
- Roberts-Galbraith, R.H.; Gould, K.L. Setting the F-BAR: Functions and regulation of the F-BAR protein family. Cell Cycle 2010, 9, 4091–4097. [Google Scholar] [CrossRef]
- Zhao, H.; Pykalainen, A.; Lappalainen, P. I-BAR domain proteins: Linking actin and plasma membrane dynamics. Curr. Opin. Cell Biol. 2011, 23, 14–21. [Google Scholar] [CrossRef]
- Nepal, B.; Sepehri, A.; Lazaridis, T. Mechanism of negative membrane curvature generation by I-BAR domains. Structure 2021, 29, 1440–1452.e1444. [Google Scholar] [CrossRef]
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010, 22, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Blower, M.D. The endoplasmic reticulum: Structure, function and response to cellular signaling. Cell Mol. Life Sci. 2016, 73, 79–94. [Google Scholar] [CrossRef] [PubMed]
- West, M.; Zurek, N.; Hoenger, A.; Voeltz, G.K. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 2011, 193, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Baumann, O.; Walz, B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 2001, 205, 149–214. [Google Scholar] [CrossRef]
- Wang, N.; Clark, L.D.; Gao, Y.; Kozlov, M.M.; Shemesh, T.; Rapoport, T.A. Mechanism of membrane-curvature generation by ER-tubule shaping proteins. Nat. Commun. 2021, 12, 568. [Google Scholar] [CrossRef]
- Frost, A.; Unger, V.M.; De Camilli, P. The BAR domain superfamily: Membrane-molding macromolecules. Cell 2009, 137, 191–196. [Google Scholar] [CrossRef]
- Peter, B.J.; Kent, H.M.; Mills, I.G.; Vallis, Y.; Butler, P.J.; Evans, P.R.; McMahon, H.T. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 2004, 303, 495–499. [Google Scholar] [CrossRef]
- Kabeya, Y.; Noda, N.N.; Fujioka, Y.; Suzuki, K.; Inagaki, F.; Ohsumi, Y. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2009, 389, 612–615. [Google Scholar] [CrossRef]
- Feng, W.Z.; Wu, T.; Dan, X.Y.; Chen, Y.L.; Li, L.; Chen, S.; Miao, D.; Deng, H.T.; Gong, X.Q.; Yu, L. Phosphorylation of Atg31 is required for autophagy. Protein Cell 2015, 6, 288–296. [Google Scholar] [CrossRef]
- Cao, Y.; Nair, U.; Yasumura-Yorimitsu, K.; Klionsky, D.J. A multiple ATG gene knockout strain for yeast two-hybrid analysis. Autophagy 2009, 5, 699–705. [Google Scholar] [CrossRef]
- Ragusa, M.J.; Stanley, R.E.; Hurley, J.H. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell 2012, 151, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Mao, K.; Yu, A.Y.H.; Omairi-Nasser, A.; Austin, J., 2nd; Glick, B.S.; Yip, C.K.; Klionsky, D.J. The Atg17-Atg31-Atg29 Complex Coordinates with Atg11 to Recruit the Vam7 SNARE and Mediate Autophagosome-Vacuole Fusion. Curr. Biol. 2016, 26, 150–160. [Google Scholar] [CrossRef]
- Kamada, Y.; Funakoshi, T.; Shintani, T.; Nagano, K.; Ohsumi, M.; Ohsumi, Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000, 150, 1507–1513. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, T.; Matsuura, A.; Noda, T.; Ohsumi, Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 1997, 192, 207–213. [Google Scholar] [CrossRef]
- Kabeya, Y.; Kamada, Y.; Baba, M.; Takikawa, H.; Sasaki, M.; Ohsumi, Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 2005, 16, 2544–2553. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Rao, Y.; Perna, M.G.; Hofmann, B.; Beier, V.; Wollert, T. The Atg1-kinase complex tethers Atg9-vesicles to initiate autophagy. Nat. Commun. 2016, 7, 10338. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.H.; Jun, C.B.; Ro, S.H.; Kim, Y.M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 2009, 20, 1992–2003. [Google Scholar] [CrossRef]
- Hosokawa, N.; Sasaki, T.; Iemura, S.; Natsume, T.; Hara, T.; Mizushima, N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 2009, 5, 973–979. [Google Scholar] [CrossRef]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef]
- Puente, C.; Hendrickson, R.C.; Jiang, X. Nutrient-regulated Phosphorylation of ATG13 Inhibits Starvation-induced Autophagy. J. Biol. Chem. 2016, 291, 6026–6035. [Google Scholar] [CrossRef] [PubMed]
- Ganley, I.G.; Lam, D.H.; Wang, J.; Ding, X.; Chen, S.; Jiang, X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009, 284, 12297–12305. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, K.; Saitoh, T.; Tabata, K.; Omori, H.; Satoh, T.; Kurotori, N.; Maejima, I.; Shirahama-Noda, K.; Ichimura, T.; Isobe, T.; et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009, 11, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef]
- Sun, Q.; Fan, W.; Chen, K.; Ding, X.; Chen, S.; Zhong, Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA 2008, 105, 19211–19216. [Google Scholar] [CrossRef]
- Itakura, E.; Kishi, C.; Inoue, K.; Mizushima, N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 2008, 19, 5360–5372. [Google Scholar] [CrossRef] [PubMed]
- Nascimbeni, A.C.; Giordano, F.; Dupont, N.; Grasso, D.; Vaccaro, M.I.; Codogno, P.; Morel, E. ER-plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. EMBO J. 2017, 36, 2018–2033. [Google Scholar] [CrossRef]
- Hamasaki, M.; Furuta, N.; Matsuda, A.; Nezu, A.; Yamamoto, A.; Fujita, N.; Oomori, H.; Noda, T.; Haraguchi, T.; Hiraoka, Y.; et al. Autophagosomes form at ER-mitochondria contact sites. Nature 2013, 495, 389–393. [Google Scholar] [CrossRef]
- Bigay, J.; Antonny, B. Curvature, lipid packing, and electrostatics of membrane organelles: Defining cellular territories in determining specificity. Dev. Cell 2012, 23, 886–895. [Google Scholar] [CrossRef]
- Ohashi, Y.; Tremel, S.; Masson, G.R.; McGinney, L.; Boulanger, J.; Rostislavleva, K.; Johnson, C.M.; Niewczas, I.; Clark, J.; Williams, R.L. Membrane characteristics tune activities of endosomal and autophagic human VPS34 complexes. Elife 2020, 9, e58281. [Google Scholar] [CrossRef]
- Pierrat, B.; Simonen, M.; Cueto, M.; Mestan, J.; Ferrigno, P.; Heim, J. SH3GLB, a new endophilin-related protein family featuring an SH3 domain. Genomics 2001, 71, 222–234. [Google Scholar] [CrossRef]
- Cuddeback, S.M.; Yamaguchi, H.; Komatsu, K.; Miyashita, T.; Yamada, M.; Wu, C.; Singh, S.; Wang, H.G. Molecular cloning and characterization of Bif-1. A novel Src homology 3 domain-containing protein that associates with Bax. J. Biol. Chem. 2001, 276, 20559–20565. [Google Scholar] [CrossRef]
- Yang, J.S.; Zhang, L.; Lee, S.Y.; Gad, H.; Luini, A.; Hsu, V.W. Key components of the fission machinery are interchangeable. Nat. Cell Biol. 2006, 8, 1376–1382. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Meyerkord, C.L.; Wang, H.G. BARgaining membranes for autophagosome formation: Regulation of autophagy and tumorigenesis by Bif-1/Endophilin B1. Autophagy 2008, 4, 121–124. [Google Scholar] [CrossRef]
- Gallop, J.L.; Jao, C.C.; Kent, H.M.; Butler, P.J.; Evans, P.R.; Langen, R.; McMahon, H.T. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006, 25, 2898–2910. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Meyerkord, C.L.; Wang, H.G. Bif-1/endophilin B1: A candidate for crescent driving force in autophagy. Cell Death Differ. 2009, 16, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Ford, M.G.; Mills, I.G.; Peter, B.J.; Vallis, Y.; Praefcke, G.J.; Evans, P.R.; McMahon, H.T. Curvature of clathrin-coated pits driven by epsin. Nature 2002, 419, 361–366. [Google Scholar] [CrossRef]
- Liang, C.; Feng, P.; Ku, B.; Dotan, I.; Canaani, D.; Oh, B.H.; Jung, J.U. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 2006, 8, 688–699. [Google Scholar] [CrossRef]
- Takahashi, Y.; Coppola, D.; Matsushita, N.; Cualing, H.D.; Sun, M.; Sato, Y.; Liang, C.; Jung, J.U.; Cheng, J.Q.; Mule, J.J.; et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 2007, 9, 1142–1151. [Google Scholar] [CrossRef]
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef]
- Fan, W.; Nassiri, A.; Zhong, Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc. Natl. Acad. Sci. USA 2011, 108, 7769–7774. [Google Scholar] [CrossRef]
- Matsunaga, K.; Morita, E.; Saitoh, T.; Akira, S.; Ktistakis, N.T.; Izumi, T.; Noda, T.; Yoshimori, T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell Biol. 2010, 190, 511–521. [Google Scholar] [CrossRef]
- Wilz, L.; Fan, W.; Zhong, Q. Membrane curvature response in autophagy. Autophagy 2011, 7, 1249–1250. [Google Scholar] [CrossRef]
- Guardia, C.M.; Tan, X.F.; Lian, T.; Rana, M.S.; Zhou, W.; Christenson, E.T.; Lowry, A.J.; Faraldo-Gomez, J.D.; Bonifacino, J.S.; Jiang, J.; et al. Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery. Cell Rep. 2020, 31, 107837. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Menon, S.; Yamasaki, A.; Chou, H.T.; Walz, T.; Jiang, Y.; Ferro-Novick, S. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc. Natl. Acad. Sci. USA 2013, 110, 9800–9805. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, Y.; Alam, J.M.; Noshiro, D.; Mouri, K.; Ando, T.; Okada, Y.; May, A.I.; Knorr, R.L.; Suzuki, K.; Ohsumi, Y.; et al. Phase separation organizes the site of autophagosome formation. Nature 2020, 578, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Sanchez, R.; Rose, J.; Guimaraes, R.; Mari, M.; Papinski, D.; Rieter, E.; Geerts, W.J.; Hardenberg, R.; Kraft, C.; Ungermann, C.; et al. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 2018, 217, 2743–2763. [Google Scholar] [CrossRef] [PubMed]
- Osawa, T.; Kotani, T.; Kawaoka, T.; Hirata, E.; Suzuki, K.; Nakatogawa, H.; Ohsumi, Y.; Noda, N.N. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 2019, 26, 281–288. [Google Scholar] [CrossRef]
- Valverde, D.P.; Yu, S.; Boggavarapu, V.; Kumar, N.; Lees, J.A.; Walz, T.; Reinisch, K.M.; Melia, T.J. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 2019, 218, 1787–1798. [Google Scholar] [CrossRef]
- Sharom, F.J. Flipping and flopping--lipids on the move. IUBMB Life 2011, 63, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S.; Sakuragi, T.; Segawa, K. Flippase and scramblase for phosphatidylserine exposure. Curr. Opin. Immunol. 2020, 62, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Matoba, K.; Kotani, T.; Tsutsumi, A.; Tsuji, T.; Mori, T.; Noshiro, D.; Sugita, Y.; Nomura, N.; Iwata, S.; Ohsumi, Y.; et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol. 2020, 27, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Bieber, A.; Capitanio, C.; Erdmann, P.S.; Fiedler, F.; Beck, F.; Lee, C.W.; Li, D.; Hummer, G.; Schulman, B.A.; Baumeister, W.; et al. In situ structural analysis reveals membrane shape transitions during autophagosome formation. Proc. Natl. Acad. Sci. USA 2022, 119, e2209823119. [Google Scholar] [CrossRef] [PubMed]
- Shpilka, T.; Welter, E.; Borovsky, N.; Amar, N.; Mari, M.; Reggiori, F.; Elazar, Z. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J. 2015, 34, 2117–2131. [Google Scholar] [CrossRef]
- Schepers, J.; Behl, C. Lipid droplets and autophagy-links and regulations from yeast to humans. J. Cell Biochem. 2021, 122, 602–611. [Google Scholar] [CrossRef]
- Velazquez, A.P.; Tatsuta, T.; Ghillebert, R.; Drescher, I.; Graef, M. Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J. Cell Biol. 2016, 212, 621–631. [Google Scholar] [CrossRef]
- Ge, L.; Zhang, M.; Kenny, S.J.; Liu, D.; Maeda, M.; Saito, K.; Mathur, A.; Xu, K.; Schekman, R. Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 2017, 18, 1586–1603. [Google Scholar] [CrossRef]
- Shima, T.; Kirisako, H.; Nakatogawa, H. COPII vesicles contribute to autophagosomal membranes. J. Cell Biol. 2019, 218, 1503–1510. [Google Scholar] [CrossRef]
- Osawa, T.; Noda, N.N. Atg2: A novel phospholipid transfer protein that mediates de novo autophagosome biogenesis. Protein Sci. 2019, 28, 1005–1012. [Google Scholar] [CrossRef]
- Kotani, T.; Kirisako, H.; Koizumi, M.; Ohsumi, Y.; Nakatogawa, H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. USA 2018, 115, 10363–10368. [Google Scholar] [CrossRef]
- Leonzino, M.; Reinisch, K.M.; De Camilli, P. Insights into VPS13 properties and function reveal a new mechanism of eukaryotic lipid transport. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 159003. [Google Scholar] [CrossRef] [PubMed]
- Graef, M.; Friedman, J.R.; Graham, C.; Babu, M.; Nunnari, J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell 2013, 24, 2918–2931. [Google Scholar] [CrossRef]
- Prinz, W.A.; Hurley, J.H. A firehose for phospholipids. J. Cell Biol. 2020, 219, e202003132. [Google Scholar] [CrossRef] [PubMed]
- Chumpen Ramirez, S.; Gomez-Sanchez, R.; Verlhac, P.; Hardenberg, R.; Margheritis, E.; Cosentino, K.; Reggiori, F.; Ungermann, C. Atg9 interactions via its transmembrane domains are required for phagophore expansion during autophagy. Autophagy 2022, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ghanbarpour, A.; Valverde, D.P.; Melia, T.J.; Reinisch, K.M. A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2101562118. [Google Scholar] [CrossRef]
- Shpilka, T.; Weidberg, H.; Pietrokovski, S.; Elazar, Z. Atg8: An autophagy-related ubiquitin-like protein family. Genome Biol. 2011, 12, 226. [Google Scholar] [CrossRef]
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A ubiquitin-like system mediates protein lipidation. Nature 2000, 408, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Kirisako, T.; Ichimura, Y.; Okada, H.; Kabeya, Y.; Mizushima, N.; Yoshimori, T.; Ohsumi, M.; Takao, T.; Noda, T.; Ohsumi, Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000, 151, 263–276. [Google Scholar] [CrossRef]
- Kabeya, Y.; Mizushima, N.; Yamamoto, A.; Oshitani-Okamoto, S.; Ohsumi, Y.; Yoshimori, T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004, 117, 2805–2812. [Google Scholar] [CrossRef]
- Qiu, Y.; Zheng, Y.; Grace, C.R.R.; Liu, X.; Klionsky, D.J.; Schulman, B.A. Allosteric regulation through a switch element in the autophagy E2, Atg3. Autophagy 2020, 16, 183–184. [Google Scholar] [CrossRef]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef] [PubMed]
- Lystad, A.H.; Carlsson, S.R.; de la Ballina, L.R.; Kauffman, K.J.; Nag, S.; Yoshimori, T.; Melia, T.J.; Simonsen, A. Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy-related processes. Nat. Cell Biol. 2019, 21, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Dancourt, J.; Shteyn, V.; Puente, G.; Fong, W.M.; Nag, S.; Bewersdorf, J.; Yamamoto, A.; Antonny, B.; Melia, T.J. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat. Cell Biol. 2014, 16, 415–424. [Google Scholar] [CrossRef]
- Antonny, B. Mechanisms of membrane curvature sensing. Annu. Rev. Biochem. 2011, 80, 101–123. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Suzuki, N.N.; Hanada, T.; Ichimura, Y.; Kumeta, H.; Fujioka, Y.; Ohsumi, Y.; Inagaki, F. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J. Biol. Chem. 2007, 282, 8036–8043. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Noda, N.N.; Nakatogawa, H.; Kumeta, H.; Ohsumi, Y.; Inagaki, F. Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J. Biol. Chem. 2010, 285, 29599–29607. [Google Scholar] [CrossRef]
- Taherbhoy, A.M.; Tait, S.W.; Kaiser, S.E.; Williams, A.H.; Deng, A.; Nourse, A.; Hammel, M.; Kurinov, I.; Rock, C.O.; Green, D.R.; et al. Atg8 transfer from Atg7 to Atg3: A distinctive E1-E2 architecture and mechanism in the autophagy pathway. Mol. Cell 2011, 44, 451–461. [Google Scholar] [CrossRef]
- Ye, Y.; Tyndall, E.R.; Bui, V.; Tang, Z.; Shen, Y.; Jiang, X.; Flanagan, J.M.; Wang, H.G.; Tian, F. An N-terminal conserved region in human Atg3 couples membrane curvature sensitivity to conjugase activity during autophagy. Nat. Commun. 2021, 12, 374. [Google Scholar] [CrossRef]
- Hervas, J.H.; Landajuela, A.; Anton, Z.; Shnyrova, A.V.; Goni, F.M.; Alonso, A. Human ATG3 binding to lipid bilayers: Role of lipid geometry, and electric charge. Sci. Rep. 2017, 7, 15614. [Google Scholar] [CrossRef]
- Dooley, H.C.; Razi, M.; Polson, H.E.; Girardin, S.E.; Wilson, M.I.; Tooze, S.A. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell 2014, 55, 238–252. [Google Scholar] [CrossRef]
- Fracchiolla, D.; Chang, C.; Hurley, J.H.; Martens, S. A PI3K-WIPI2 positive feedback loop allosterically activates LC3 lipidation in autophagy. J. Cell Biol. 2020, 219, e201912098. [Google Scholar] [CrossRef]
- Jensen, L.E.; Rao, S.; Schuschnig, M.; Cada, A.K.; Martens, S.; Hummer, G.; Hurley, J.H. Membrane curvature sensing and stabilization by the autophagic LC3 lipidation machinery. Sci. Adv. 2022, 8, eadd1436. [Google Scholar] [CrossRef]
- Liu, R.; Zhi, X.; Zhong, Q. ATG14 controls SNARE-mediated autophagosome fusion with a lysosome. Autophagy 2015, 11, 847–849. [Google Scholar] [CrossRef]
- Diao, J.; Liu, R.; Rong, Y.; Zhao, M.; Zhang, J.; Lai, Y.; Zhou, Q.; Wilz, L.M.; Li, J.; Vivona, S.; et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 2015, 520, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wu, Z.; Du, W.; Que, H.; Wang, Y.; Ouyang, Q.; Jian, F.; Yuan, W.; Zhao, Y.; Tian, R.; et al. Recycling of autophagosomal components from autolysosomes by the recycler complex. Nat. Cell Biol. 2022, 24, 497–512. [Google Scholar] [CrossRef] [PubMed]
- Teasdale, R.D.; Collins, B.M. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: Structures, functions and roles in disease. Biochem. J. 2012, 441, 39–59. [Google Scholar] [CrossRef]
- Khaminets, A.; Heinrich, T.; Mari, M.; Grumati, P.; Huebner, A.K.; Akutsu, M.; Liebmann, L.; Stolz, A.; Nietzsche, S.; Koch, N.; et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 2015, 522, 354–358. [Google Scholar] [CrossRef]
- Fumagalli, F.; Noack, J.; Bergmann, T.J.; Cebollero, E.; Pisoni, G.B.; Fasana, E.; Fregno, I.; Galli, C.; Loi, M.; Solda, T.; et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat. Cell Biol. 2016, 18, 1173–1184. [Google Scholar] [CrossRef]
- Grumati, P.; Morozzi, G.; Holper, S.; Mari, M.; Harwardt, M.I.; Yan, R.; Muller, S.; Reggiori, F.; Heilemann, M.; Dikic, I. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife 2017, 6, e25555. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.D.; Harley, M.E.; Kemp, A.J.; Wills, J.; Lee, M.; Arends, M.; von Kriegsheim, A.; Behrends, C.; Wilkinson, S. CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev. Cell 2018, 44, 217–232.e211. [Google Scholar] [CrossRef]
- Chen, Q.; Xiao, Y.; Chai, P.; Zheng, P.; Teng, J.; Chen, J. ATL3 Is a Tubular ER-Phagy Receptor for GABARAP-Mediated Selective Autophagy. Curr. Biol. 2019, 29, 846–855.e846. [Google Scholar] [CrossRef]
- An, H.; Ordureau, A.; Paulo, J.A.; Shoemaker, C.J.; Denic, V.; Harper, J.W. TEX264 Is an Endoplasmic Reticulum-Resident ATG8-Interacting Protein Critical for ER Remodeling during Nutrient Stress. Mol. Cell 2019, 74, 891–908.e810. [Google Scholar] [CrossRef]
- Mochida, K.; Oikawa, Y.; Kimura, Y.; Kirisako, H.; Hirano, H.; Ohsumi, Y.; Nakatogawa, H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015, 522, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Parashar, S.; Zahoor, M.; Needham, P.G.; Mari, M.; Zhu, M.; Chen, S.; Ho, H.C.; Reggiori, F.; Farhan, H.; et al. A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science 2019, 365, 53–60. [Google Scholar] [CrossRef]
- Bhaskara, R.M.; Grumati, P.; Garcia-Pardo, J.; Kalayil, S.; Covarrubias-Pinto, A.; Chen, W.; Kudryashev, M.; Dikic, I.; Hummer, G. Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER-phagy. Nat. Commun. 2019, 10, 2370. [Google Scholar] [CrossRef]
- Di Lorenzo, G.; Iavarone, F.; Maddaluno, M.; Plata-Gomez, A.B.; Aureli, S.; Quezada Meza, C.P.; Cinque, L.; Palma, A.; Reggio, A.; Cirillo, C.; et al. Phosphorylation of FAM134C by CK2 controls starvation-induced ER-phagy. Sci. Adv. 2022, 8, eabo1215. [Google Scholar] [CrossRef] [PubMed]
- Cinque, L.; De Leonibus, C.; Iavazzo, M.; Krahmer, N.; Intartaglia, D.; Salierno, F.G.; De Cegli, R.; Di Malta, C.; Svelto, M.; Lanzara, C.; et al. MiT/TFE factors control ER-phagy via transcriptional regulation of FAM134B. EMBO J. 2020, 39, e105696. [Google Scholar] [CrossRef]
- Reggio, A.; Buonomo, V.; Berkane, R.; Bhaskara, R.M.; Tellechea, M.; Peluso, I.; Polishchuk, E.; Di Lorenzo, G.; Cirillo, C.; Esposito, M.; et al. Role of FAM134 paralogues in endoplasmic reticulum remodeling, ER-phagy, and Collagen quality control. EMBO Rep. 2021, 22, e52289. [Google Scholar] [CrossRef]
- Kumar, D.; Lak, B.; Suntio, T.; Vihinen, H.; Belevich, I.; Viita, T.; Xiaonan, L.; Vartiainen, A.; Vartiainen, M.; Varjosalo, M.; et al. RTN4B interacting protein FAM134C promotes ER membrane curvature and has a functional role in autophagy. Mol. Biol. Cell 2021, 32, 1158–1170. [Google Scholar] [CrossRef]
- Mochida, K.; Yamasaki, A.; Matoba, K.; Kirisako, H.; Noda, N.N.; Nakatogawa, H. Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes. Nat. Commun. 2020, 11, 3306. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, G.; Tavernarakis, N. Autophagy of the Nucleus in Health and Disease. Front. Cell Dev. Biol. 2021, 9, 814955. [Google Scholar] [CrossRef] [PubMed]
- Mochida, K.; Otani, T.; Katsumata, Y.; Kirisako, H.; Kakuta, C.; Kotani, T.; Nakatogawa, H. Atg39 links and deforms the outer and inner nuclear membranes in selective autophagy of the nucleus. J. Cell Biol. 2022, 221, e202103178. [Google Scholar] [CrossRef]
- Ammanathan, V.; Vats, S.; Abraham, I.M.; Manjithaya, R. Xenophagy in cancer. Semin. Cancer Biol. 2020, 66, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhou, P.; Cheng, S.; Lu, Q.; Nowak, K.; Hopp, A.K.; Li, L.; Shi, X.; Zhou, Z.; Gao, W.; et al. A Bacterial Effector Reveals the V-ATPase-ATG16L1 Axis that Initiates Xenophagy. Cell 2019, 178, 552–566.e20. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Li, M.; Sun, L.; Zhao, H.; Li, Y.; Zhou, L.; Yang, Z.; Zhao, X.; Qu, W.; Xue, H.; et al. VMP1 and TMEM41B are essential for DMV formation during beta-coronavirus infection. J. Cell Biol. 2022, 221, e202112081. [Google Scholar] [CrossRef]
- Oudshoorn, D.; Rijs, K.; Limpens, R.; Groen, K.; Koster, A.J.; Snijder, E.J.; Kikkert, M.; Barcena, M. Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication. mBio 2017, 8, e01658-17. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Sung, B.H.; von Lersner, A.; Guerrero, J.; Krystofiak, E.S.; Inman, D.; Pelletier, R.; Zijlstra, A.; Ponik, S.M.; Weaver, A.M. A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nat. Commun. 2020, 11, 2092. [Google Scholar] [CrossRef]
- Tian, X.; Teng, J.; Chen, J. New insights regarding SNARE proteins in autophagosome-lysosome fusion. Autophagy 2021, 17, 2680–2688. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Warner, B.M.; Michael, D.G.; Nakamura, H.; Odani, T.; Yin, H.; Atsumi, T.; Noguchi, M.; Chiorini, J.A. LAMP3 inhibits autophagy and contributes to cell death by lysosomal membrane permeabilization. Autophagy 2022, 18, 1629–1647. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zou, C.X.; Liu, X.M.; Jiang, Z.D.; Yu, Z.Q.; Suo, F.; Du, T.Y.; Dong, M.Q.; He, W.; Du, L.L. A UPR-Induced Soluble ER-Phagy Receptor Acts with VAPs to Confer ER Stress Resistance. Mol. Cell 2020, 79, 963–977.e963. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Tang, Y.; Zhou, Z.; Huang, Y.; Zhang, R.; Lyu, H.; Xiao, S.; Guo, D.; Ali, D.W.; Michalak, M.; et al. Membrane Curvature: The Inseparable Companion of Autophagy. Cells 2023, 12, 1132. https://doi.org/10.3390/cells12081132
Liu L, Tang Y, Zhou Z, Huang Y, Zhang R, Lyu H, Xiao S, Guo D, Ali DW, Michalak M, et al. Membrane Curvature: The Inseparable Companion of Autophagy. Cells. 2023; 12(8):1132. https://doi.org/10.3390/cells12081132
Chicago/Turabian StyleLiu, Lei, Yu Tang, Zijuan Zhou, Yuan Huang, Rui Zhang, Hao Lyu, Shuai Xiao, Dong Guo, Declan William Ali, Marek Michalak, and et al. 2023. "Membrane Curvature: The Inseparable Companion of Autophagy" Cells 12, no. 8: 1132. https://doi.org/10.3390/cells12081132
APA StyleLiu, L., Tang, Y., Zhou, Z., Huang, Y., Zhang, R., Lyu, H., Xiao, S., Guo, D., Ali, D. W., Michalak, M., Chen, X.-Z., Zhou, C., & Tang, J. (2023). Membrane Curvature: The Inseparable Companion of Autophagy. Cells, 12(8), 1132. https://doi.org/10.3390/cells12081132







