The Regulation and Double-Edged Roles of the Deubiquitinase OTUD5
Abstract
:1. Introduction
2. Structural Characteristics of OTUD5
2.1. Ubiquitination and Deubiquitination Processes
2.2. Structural Characteristics of OTUD5
3. Physiological Processes Regulated by OTUD5
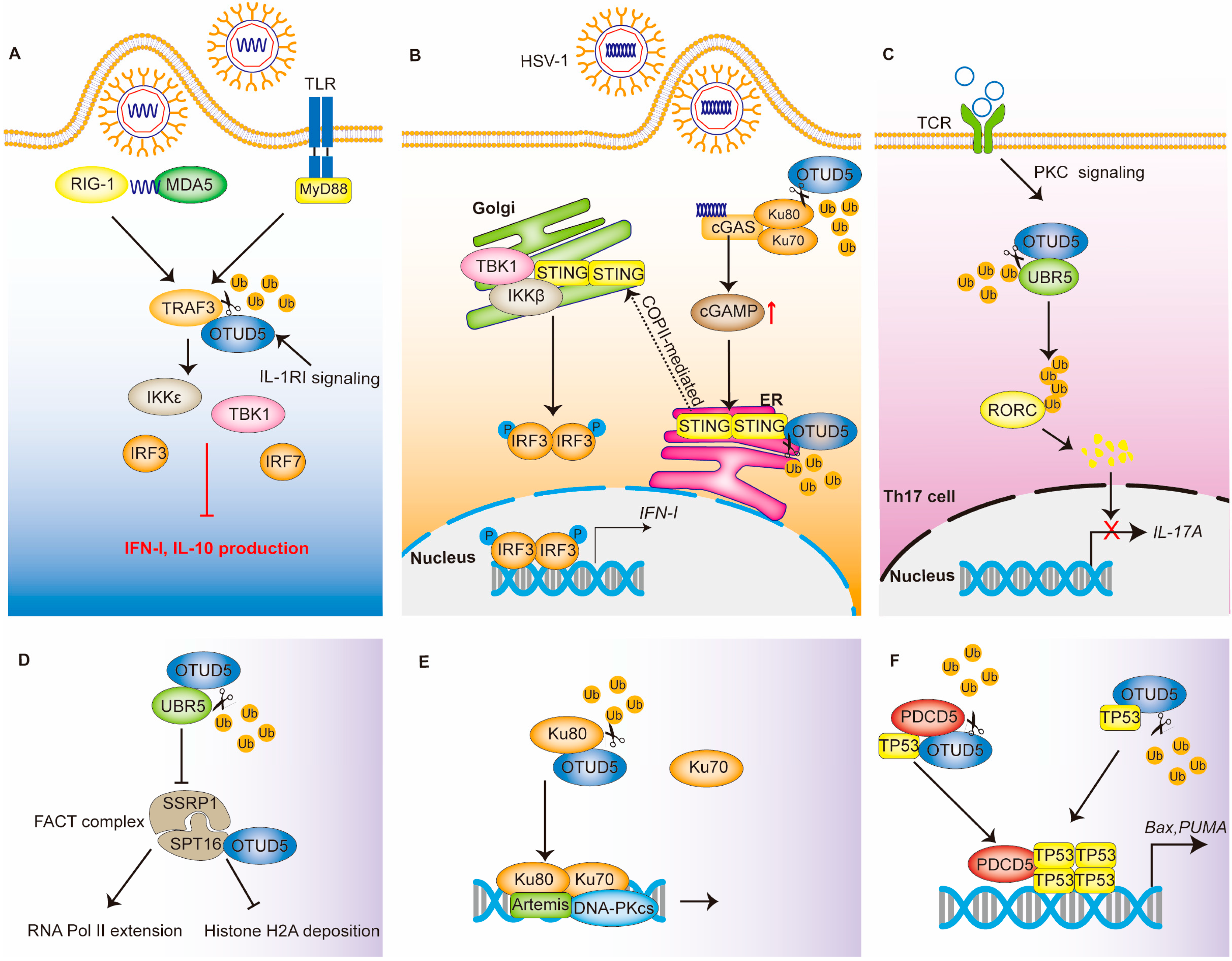
3.1. Immunity
3.1.1. Innate Immunity: The TLR and RLR Signaling Pathway
3.1.2. Innate Immunity: The cGAS-STING Signaling Pathway
3.1.3. Acquired Immunity
3.2. DNA Damage
3.2.1. FACT Complex
3.2.2. Ku Heterodimers
3.2.3. TP53 and PDCD5
4. OTUD5-Related Diseases
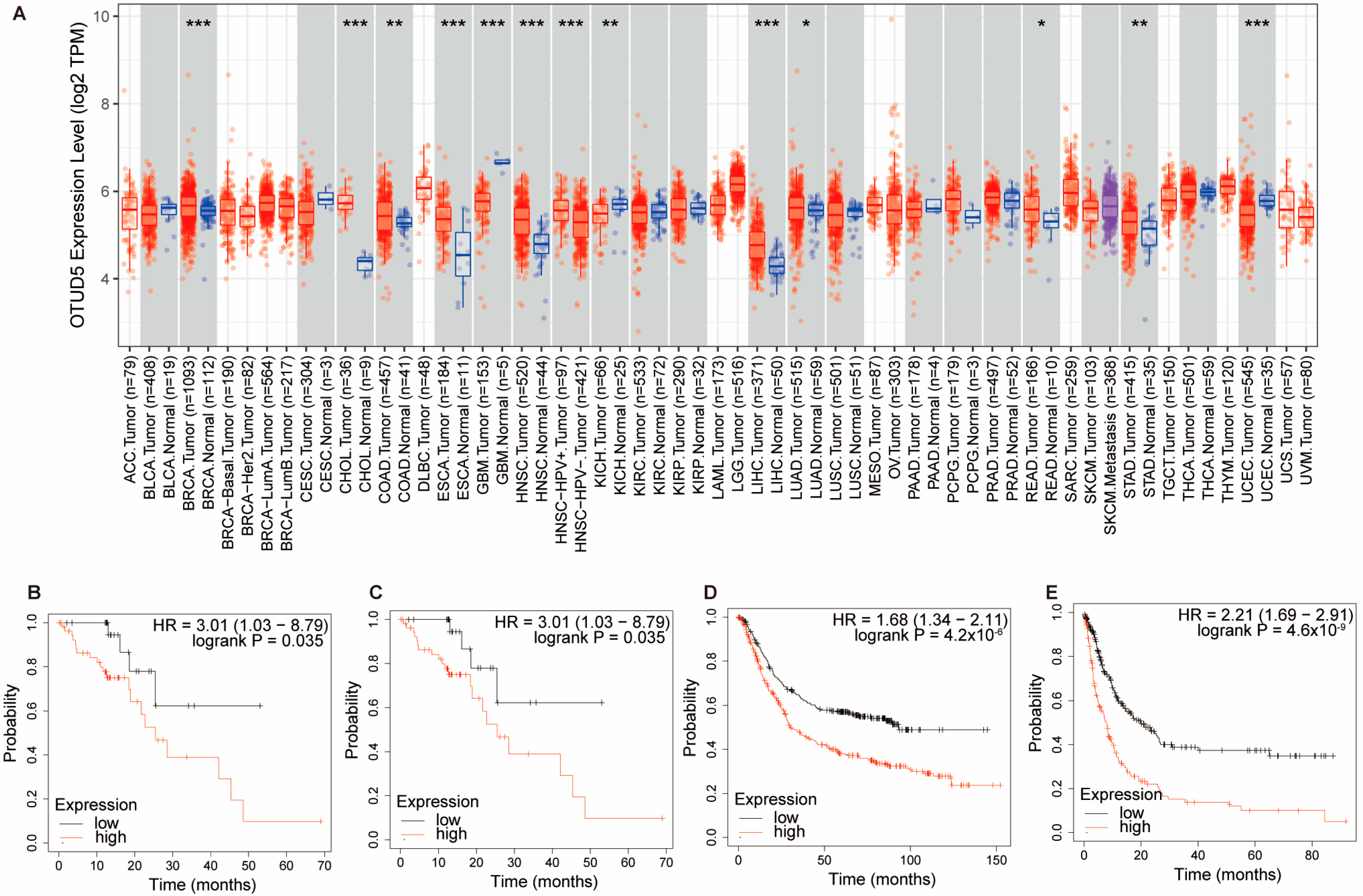
4.1. Tumors
4.1.1. Tumor Promoter
mTOR Pathway
Hippo Pathway
4.1.2. Tumor Inhibitor
TRIM25
TP53 and PDCD5
PTEN and Akt
| Target Substrate | Tumor Type | Result of Deubiquitination | Affected Pathway or Event | Effect | References |
|---|---|---|---|---|---|
| βTrCP1 | Colon cancer Breast cancer | Stable protein | mTOR pathway | Enhanced cancer cell proliferation | [10] |
| RNF186 | Bladder cancer | Stable protein | mTOR pathway | Enhanced cancer cell progression | [11] |
| YAP | Triple-negative breast cancer | Stable protein | Hippo pathway | Enhanced cancer cell metastasis | [12] |
| TRIM25 | Hepatocellular carcinoma and non-small cell lung cancer | Decreased transcriptional activity | - | Reduced tumor growth | [7] |
| P53/PDCD5 | Non-small cell lung cancer | Stable protein | Apoptosis | Reduced cancer cell proliferation and metastasis | [8] |
| PTEN | Non-small cell lung cancer | Stable protein | Akt signaling | Inhibited proliferation, invasion and migration | [9] |
| Akt | Cervical cancer | Stable protein | Akt signaling | Sensitive to radiotherapy | [73] |
4.2. Inflammation in the Digestive System
4.3. Genetic Diseases
5. Regulation of OTUD5 Activity and Expression
5.1. Transcription Level Regulation
5.2. Post-Translational Modifications
6. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OTUD5 | OTU deubiquitinase 5 |
| OTU | Ovarian tumor proteases |
| DUBA | Deubiquitinating enzyme A |
| DUBs | Deubiquitinases |
| IFN | Interferon |
| HCC | Hepatocellular carcinoma |
| NSCLC | Non-small cell lung cancer |
| TRIM25 | Tripartite motif containing 25 |
| TP53 | Tumor protein P53 |
| PDCD5 | Programmed cell death 5 |
| PTEN | Phosphatase and tensin homolog |
| mTOR | Mammalian target of rapamycin |
| USPs | Ubiquitin-specific proteases |
| MJDs | Machado-Joseph domain-containing proteases |
| JAMMs | JAMM/MPN domain-associated Zn-dependent metalloproteases |
| UCHs | Ubiquitin C-terminal hydrolases |
| ZUP1 | Zinc finger-containing ubiquitin peptidase 1 |
| MINDYs | Ubiquitin-containing proteases |
| UIM | Ubiquitin-interacting motif |
| STING | Stimulator of interferon genes |
| SPT16 | Suppressor of Ty 16 Homolog |
| YAP | Yes-associated protein |
| PAMPs | Pathogen-associated molecular patterns |
| PRRs | Pattern recognition receptors |
| TLRs | Toll-like receptors |
| RIG-I | Retinoic acid-inducible gene-I |
| RLRs | RIG-I-like receptors |
| MDA5 | Melanoma differentiation-associated gene 5 |
| LGP2 | Laboratory of genetics and physiology 2 |
| TBK1 | TANK-binding kinase 1 |
| cGAMP | Cyclic GMP-AMP |
| ER | Endoplasmic reticulum |
| HSV-1 | Herpes simplex virus-1 |
| DC | Dendritic cell |
| SAVI | STING-associated vasculopathy with onset in infancy |
| TCR | T cell a ntigen receptor |
| TH17 | T helper type 17 |
| IL-17A | Interleukin-17A |
| RORC | RAR related orphan receptor C |
| FACT | Facilitates Chromatin Transcription |
| DSB | DNA double-strand break |
| SSRP1 | Structure-Specific Recognition Protein 1 |
| NHEJ | Non-homologous end joining |
| HR | Homologous recombination |
| βTrCP1 | β-transducin repeat-containing protein 1 |
| DEPTOR | DEP Domain-Containing MTOR-Interacting Protein |
| MCP-1 | Monocyte chemoattractant protein-1 |
| CCR2 | C-C Motif chemokine receptor 2 |
| PML | Promyelocytic leukemia protein |
| TRIM19 | Tripartite motif containing 19 |
| NBs | Nucleosomes |
| IBD | Inflammatory bowel disease |
| CD | Crohn’s disease |
| UC | Ulcerative colitis |
| LPMC | Lamina propria mononuclear cells |
| GWAS | Genome-wide association studies |
| PBC | Primary biliary cholangitis |
| CP | Chronic pancreatitis |
| Bach2 | BTB and CNC homologous 2 |
| LINKED | Linkage-specific deubiquitylation deficiency-induced embryonic defects |
| HDAC2 | Histone deacetylase 2 |
| XLID | X-linked intellectual disability |
| NMR | Nuclear magnetic resonance |
| PKC | Protein kinase C |
| TNKS1/2 | Tankyrase 1 and 2 |
| SMURF1 | Smad ubiquitination regulatory factor 1 |
References
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popovic, D.; Vucic, D.; Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014, 20, 1242–1253. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Wang, R.; Zhang, Y.; Li, X.; Gan, Y.; Gao, F.; Rong, P.; Wang, W.; Li, W. The role of ubiquitination and deubiquitination in tumor invasion and metastasis. Int. J. Biol. Sci. 2022, 18, 2292–2303. [Google Scholar] [CrossRef]
- Cruz Walma, D.A.; Chen, Z.; Bullock, A.N.; Yamada, K.M. Ubiquitin ligases: Guardians of mammalian development. Nat. Rev. Mol. Cell Biol. 2022, 23, 350–367. [Google Scholar] [CrossRef]
- Kayagaki, N.; Phung, Q.; Chan, S.; Chaudhari, R.; Quan, C.; O’Rourke, K.M.; Eby, M.; Pietras, E.; Cheng, G.; Bazan, J.F.; et al. DUBA: A deubiquitinase that regulates type I interferon production. Science 2007, 318, 1628–1632. [Google Scholar] [CrossRef] [PubMed]
- Rutz, S.; Kayagaki, N.; Phung, Q.T.; Eidenschenk, C.; Noubade, R.; Wang, X.; Lesch, J.; Lu, R.; Newton, K.; Huang, O.W.; et al. Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature 2015, 518, 417–421. [Google Scholar] [CrossRef]
- Li, F.; Sun, Q.; Liu, K.; Zhang, L.; Lin, N.; You, K.; Liu, M.; Kon, N.; Tian, F.; Mao, Z.; et al. OTUD5 cooperates with TRIM25 in transcriptional regulation and tumor progression via deubiquitination activity. Nat. Commun. 2020, 11, 4184. [Google Scholar] [CrossRef]
- Kang, X.Y.; Zhang, J.; Tang, L.; Huang, L.; Tong, J.; Fu, Q. OTU deubiquitinase 5 inhibits the progression of non-small cell lung cancer via regulating p53 and PDCD5. Chem. Biol. Drug Des. 2020, 96, 790–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Lu, B.; Zhang, L.; Yang, J.; Cheng, Y.; Yan, D. Mechanism of OTUD5 in non-small cell lung cancer cell proliferation, invasion, and migration. Bosn. J. Basic Med. Sci. 2022, 22, 901–911. [Google Scholar] [CrossRef]
- Cho, J.H.; Kim, K.; Kim, S.A.; Park, S.; Park, B.O.; Kim, J.H.; Kim, S.Y.; Kwon, M.J.; Han, M.H.; Lee, S.B.; et al. Deubiquitinase OTUD5 is a positive regulator of mTORC1 and mTORC2 signaling pathways. Cell Death Differ. 2021, 28, 900–914. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Dan, W.; Liu, T.; Liu, B.; Wei, Y.; Yue, C.; Que, T.; Ma, B.; Lei, Y.; Wang, Z.; et al. Deubiquitinase OTUD5 modulates mTORC1 signaling to promote bladder cancer progression. Cell Death Dis. 2022, 13, 778. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, Y.; Jing, X.; Zhao, L.; Liu, T.; Wang, L.; Zhang, L.; Gu, S.; Zhao, X.; Teng, Y. OTUD5-mediated deubiquitination of YAP in macrophage promotes M2 phenotype polarization and favors triple-negative breast cancer progression. Cancer Lett. 2021, 504, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I.; Wakatsuki, S.; Walters, K.J. Ubiquitin-binding domains—From structures to functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Cui, C.P.; Zhang, X.; Zhang, L. The functions and regulation of Smurfs in cancers. Semin. Cancer Biol. 2020, 67, 102–116. [Google Scholar] [CrossRef] [PubMed]
- Akutsu, M.; Dikic, I.; Bremm, A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016, 129, 875–880. [Google Scholar] [CrossRef] [Green Version]
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 18, 579–586. [Google Scholar] [CrossRef]
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef] [PubMed]
- Neutzner, M.; Neutzner, A. Enzymes of ubiquitination and deubiquitination. Essays Biochem. 2012, 52, 37–50. [Google Scholar] [CrossRef] [Green Version]
- Clague, M.J.; Urbe, S.; Komander, D. Breaking the chains: Deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 2019, 20, 338–352. [Google Scholar] [CrossRef]
- Du, J.; Fu, L.; Sui, Y.; Zhang, L. The function and regulation of OTU deubiquitinases. Front. Med. 2020, 14, 542–563. [Google Scholar] [CrossRef] [Green Version]
- Schluter, D.; Schulze-Niemand, E.; Stein, M.; Naumann, M. Ovarian tumor domain proteases in pathogen infection. Trends Microbiol. 2022, 30, 22–33. [Google Scholar] [CrossRef]
- Park, S.Y.; Choi, H.K.; Choi, Y.; Kwak, S.; Choi, K.C.; Yoon, H.G. Deubiquitinase OTUD5 mediates the sequential activation of PDCD5 and p53 in response to genotoxic stress. Cancer Lett. 2015, 357, 419–427. [Google Scholar] [CrossRef] [PubMed]
- de Vivo, A.; Sanchez, A.; Yegres, J.; Kim, J.; Emly, S.; Kee, Y. The OTUD5-UBR5 complex regulates FACT-mediated transcription at damaged chromatin. Nucleic Acids Res. 2019, 47, 729–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mevissen, T.E.T.; Komander, D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 2017, 86, 159–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, O.W.; Ma, X.; Yin, J.; Flinders, J.; Maurer, T.; Kayagaki, N.; Phung, Q.; Bosanac, I.; Arnott, D.; Dixit, V.M.; et al. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat. Struct. Mol. Biol. 2012, 19, 171–175. [Google Scholar] [CrossRef]
- Brennan, K.; Bowie, A.G. Activation of host pattern recognition receptors by viruses. Curr. Opin. Microbiol. 2010, 13, 503–507. [Google Scholar] [CrossRef]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef] [Green Version]
- Paludan, S.R.; Bowie, A.G.; Horan, K.A.; Fitzgerald, K.A. Recognition of herpesviruses by the innate immune system. Nat. Rev. Immunol. 2011, 11, 143–154. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal. Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.W.; Yang, Y.K.; Xu, H.; Yang, W.W.; Zhai, Z.H.; Chen, D.Y. Ring finger protein 166 potentiates RNA virus-induced interferon-beta production via enhancing the ubiquitination of TRAF3 and TRAF6. Sci. Rep. 2015, 5, 14770. [Google Scholar] [CrossRef] [Green Version]
- Nakhaei, P.; Mesplede, T.; Solis, M.; Sun, Q.; Zhao, T.; Yang, L.; Chuang, T.H.; Ware, C.F.; Lin, R.; Hiscott, J. The E3 ubiquitin ligase Triad3A negatively regulates the RIG-I/MAVS signaling pathway by targeting TRAF3 for degradation. PLoS Pathog. 2009, 5, e1000650. [Google Scholar] [CrossRef] [Green Version]
- Gong, T.; Liu, L.; Jiang, W.; Zhou, R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 2020, 20, 95–112. [Google Scholar] [CrossRef]
- Decout, A.; Katz, J.D.; Venkatraman, S.; Ablasser, A. The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 2021, 21, 548–569. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.; Bakhoum, S.F. The Cytosolic DNA-Sensing cGAS-STING Pathway in Cancer. Cancer Discov. 2020, 10, 26–39. [Google Scholar] [CrossRef]
- Guo, Y.; Jiang, F.; Kong, L.; Wu, H.; Zhang, H.; Chen, X.; Zhao, J.; Cai, B.; Li, Y.; Ma, C.; et al. OTUD5 promotes innate antiviral and antitumor immunity through deubiquitinating and stabilizing STING. Cell. Mol. Immunol. 2021, 18, 1945–1955. [Google Scholar] [CrossRef] [PubMed]
- Jeremiah, N.; Neven, B.; Gentili, M.; Callebaut, I.; Maschalidi, S.; Stolzenberg, M.C.; Goudin, N.; Fremond, M.L.; Nitschke, P.; Molina, T.J.; et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Investig. 2014, 124, 5516–5520. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jesus, A.A.; Marrero, B.; Yang, D.; Ramsey, S.E.; Sanchez, G.A.M.; Tenbrock, K.; Wittkowski, H.; Jones, O.Y.; Kuehn, H.S.; et al. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 2014, 371, 507–518. [Google Scholar] [CrossRef] [Green Version]
- Yasuda, K.; Takeuchi, Y.; Hirota, K. The pathogenicity of Th17 cells in autoimmune diseases. Semin. Immunopathol. 2019, 41, 283–297. [Google Scholar] [CrossRef]
- Kumar, R.; Theiss, A.L.; Venuprasad, K. RORgammat protein modifications and IL-17-mediated inflammation. Trends Immunol. 2021, 42, 1037–1050. [Google Scholar] [CrossRef]
- Carusillo, A.; Mussolino, C. DNA Damage: From Threat to Treatment. Cells 2020, 9, 1665. [Google Scholar] [CrossRef]
- Ui, A.; Chiba, N.; Yasui, A. Relationship among DNA double-strand break (DSB), DSB repair, and transcription prevents genome instability and cancer. Cancer Sci. 2020, 111, 1443–1451. [Google Scholar] [CrossRef] [PubMed]
- Kwak, H.; Lis, J.T. Control of transcriptional elongation. Annu. Rev. Genet. 2013, 47, 483–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Chen, M.; Lee, D.; Law, C.T.; Wei, L.; Tsang, F.H.; Chin, D.W.; Cheng, C.L.; Lee, J.M.; Ng, I.O.; et al. Histone chaperone FACT complex mediates oxidative stress response to promote liver cancer progression. Gut 2020, 69, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.G.; Shin, H.C.; Seo, T.; Nawale, L.; Han, G.; Kim, B.Y.; Kim, S.J.; Cha-Molstad, H. Signaling Pathways Regulated by UBR Box-Containing E3 Ligases. Int. J. Mol. Sci. 2021, 22, 8323. [Google Scholar] [CrossRef]
- Fu, L.; Zhang, L. Physiological functions of CKIP-1: From molecular mechanisms to therapy implications. Ageing Res. Rev. 2019, 53, 100908. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, D. Repair pathway choice for double-strand breaks. Essays Biochem. 2020, 64, 765–777. [Google Scholar] [CrossRef]
- Frit, P.; Ropars, V.; Modesti, M.; Charbonnier, J.B.; Calsou, P. Plugged into the Ku-DNA hub: The NHEJ network. Prog. Biophys. Mol. Biol. 2019, 147, 62–76. [Google Scholar] [CrossRef]
- Postow, L. Destroying the ring: Freeing DNA from Ku with ubiquitin. FEBS Lett. 2011, 585, 2876–2882. [Google Scholar] [CrossRef] [Green Version]
- Postow, L.; Ghenoiu, C.; Woo, E.M.; Krutchinsky, A.N.; Chait, B.T.; Funabiki, H. Ku80 removal from DNA through double strand break-induced ubiquitylation. J. Cell Biol. 2008, 182, 467–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Sun, Q.; Liu, K.; Han, H.; Lin, N.; Cheng, Z.; Cai, Y.; Tian, F.; Mao, Z.; Tong, T.; et al. The deubiquitinase OTUD5 regulates Ku80 stability and non-homologous end joining. Cell. Mol. Life Sci. 2019, 76, 3861–3873. [Google Scholar] [CrossRef]
- Chapman, J.R.; Taylor, M.R.; Boulton, S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 2012, 47, 497–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, X.; Song, J.; Song, Y.; Zhang, Y.; Yang, J.; Zhang, P.; Zhang, D.; Chen, D.; Sun, Q. Ku proteins promote DNA binding and condensation of cyclic GMP-AMP synthase. Cell Rep. 2022, 40, 111310. [Google Scholar] [CrossRef]
- Williams, A.B.; Schumacher, B. p53 in the DNA-Damage-Repair Process. Cold Spring Harb. Perspect. Med. 2016, 6, a026070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.; Lu, Z.; Lu, X.; Chen, L.; Cao, J.; Zhang, S.; Ling, Y.; Zhou, X. OTUD5 regulates p53 stability by deubiquitinating p53. PLoS ONE 2013, 8, e77682. [Google Scholar] [CrossRef]
- Xu, L.; Hu, J.; Zhao, Y.; Hu, J.; Xiao, J.; Wang, Y.; Ma, D.; Chen, Y. PDCD5 interacts with p53 and functions as a positive regulator in the p53 pathway. Apoptosis 2012, 17, 1235–1245. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Che, Y.; Lu, K.; Fu, L. Analysis of deubiquitinase OTUD5 as a biomarker and therapeutic target for cervical cancer by bioinformatic analysis. PeerJ 2020, 8, e9146. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [Green Version]
- Peterson, T.R.; Laplante, M.; Thoreen, C.C.; Sancak, Y.; Kang, S.A.; Kuehl, W.M.; Gray, N.S.; Sabatini, D.M. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009, 137, 873–886. [Google Scholar] [CrossRef] [Green Version]
- Lear, T.B.; Lockwood, K.C.; Ouyang, Y.; Evankovich, J.W.; Larsen, M.B.; Lin, B.; Liu, Y.; Chen, B.B. The RING-type E3 ligase RNF186 ubiquitinates Sestrin-2 and thereby controls nutrient sensing. J. Biol. Chem. 2019, 294, 16527–16534. [Google Scholar] [CrossRef]
- Wolfson, R.L.; Chantranupong, L.; Saxton, R.A.; Shen, K.; Scaria, S.M.; Cantor, J.R.; Sabatini, D.M. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016, 351, 43–48. [Google Scholar] [CrossRef] [Green Version]
- Nie, L.; Wang, C.; Li, N.; Feng, X.; Lee, N.; Su, D.; Tang, M.; Yao, F.; Chen, J. Proteome-wide Analysis Reveals Substrates of E3 Ligase RNF146 Targeted for Degradation. Mol. Cell. Proteom. 2020, 19, 2015–2030. [Google Scholar] [CrossRef] [PubMed]
- Janse van Rensburg, H.J.; Azad, T.; Ling, M.; Hao, Y.; Snetsinger, B.; Khanal, P.; Minassian, L.M.; Graham, C.H.; Rauh, M.J.; Yang, X. The Hippo Pathway Component TAZ Promotes Immune Evasion in Human Cancer through PD-L1. Cancer Res. 2018, 78, 1457–1470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locati, M.; Curtale, G.; Mantovani, A. Diversity, Mechanisms, and Significance of Macrophage Plasticity. Annu. Rev. Pathol. 2020, 15, 123–147. [Google Scholar] [CrossRef] [Green Version]
- Heikel, G.; Choudhury, N.R.; Michlewski, G. The role of Trim25 in development, disease and RNA metabolism. Biochem. Soc. Trans. 2016, 44, 1045–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, L.; Zhou, X.; Jiao, Q.; Chen, X. The Functions of TRIM56 in Antiviral Innate Immunity and Tumorigenesis. Int. J. Mol. Sci. 2023, 24, 5046. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Sui, Y.; Fu, L. Identification of TRIM56 as a Potential Biomarker for Lung Adenocarcinoma. Cancer Manag. Res. 2021, 13, 2201–2213. [Google Scholar] [CrossRef]
- Walsh, L.A.; Alvarez, M.J.; Sabio, E.Y.; Reyngold, M.; Makarov, V.; Mukherjee, S.; Lee, K.W.; Desrichard, A.; Turcan, S.; Dalin, M.G.; et al. An Integrated Systems Biology Approach Identifies TRIM25 as a Key Determinant of Breast Cancer Metastasis. Cell Rep. 2017, 20, 1623–1640. [Google Scholar] [CrossRef] [Green Version]
- Lallemand-Breitenbach, V. PML nuclear bodies: From architecture to function. Curr. Opin. Cell Biol. 2018, 52, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Hernandez Borrero, L.J.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef] [PubMed]
- Perez-Ramirez, C.; Canadas-Garre, M.; Molina, M.A.; Faus-Dader, M.J.; Calleja-Hernandez, M.A. PTEN and PI3K/AKT in non-small-cell lung cancer. Pharmacogenomics 2015, 16, 1843–1862. [Google Scholar] [CrossRef]
- Yin, F.; He, H.; Zhang, B.; Zheng, J.; Wang, M.; Zhang, M.; Cui, H. Effect of Deubiquitinase Ovarian Tumor Domain-Containing Protein 5 (OTUD5) on Radiosensitivity of Cervical Cancer by Regulating the Ubiquitination of Akt and its Mechanism. Med. Sci. Monit. 2019, 25, 3469–3475. [Google Scholar] [CrossRef] [PubMed]
- Yehia, L.; Keel, E.; Eng, C. The Clinical Spectrum of PTEN Mutations. Annu. Rev. Med. 2020, 71, 103–116. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, G.G. The global burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Geremia, A.; Biancheri, P.; Allan, P.; Corazza, G.R.; Di Sabatino, A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun. Rev. 2014, 13, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Dinallo, V.; Di Fusco, D.; Di Grazia, A.; Laudisi, F.; Troncone, E.; Di Maggio, G.; Franze, E.; Marafini, I.; Colantoni, A.; Ortenzi, A.; et al. The Deubiquitinating Enzyme OTUD5 Sustains Inflammatory Cytokine Response in Inflammatory Bowel Disease. J. Crohns Colitis 2022, 16, 122–132. [Google Scholar] [CrossRef]
- Asselta, R.; Paraboschi, E.M.; Gerussi, A.; Cordell, H.J.; Mells, G.F.; Sandford, R.N.; Jones, D.E.; Nakamura, M.; Ueno, K.; Hitomi, Y.; et al. X Chromosome Contribution to the Genetic Architecture of Primary Biliary Cholangitis. Gastroenterology 2021, 160, 2483–2495. [Google Scholar] [CrossRef]
- Sakaguchi, S.; Miyara, M.; Costantino, C.M.; Hafler, D.A. FOXP3+ regulatory T cells in the human immune system. Nat. Rev. Immunol. 2010, 10, 490–500. [Google Scholar] [CrossRef]
- Beyer, G.; Habtezion, A.; Werner, J.; Lerch, M.M.; Mayerle, J. Chronic pancreatitis. Lancet 2020, 396, 499–512. [Google Scholar] [CrossRef]
- Sasikala, M.; Ravikanth, V.V.; Murali Manohar, K.; Deshpande, N.; Singh, S.; Pavan Kumar, P.; Talukdar, R.; Ghosh, S.; Aslam, M.; Rao, G.V.; et al. Bach2 repression mediates Th17 cell induced inflammation and associates with clinical features of advanced disease in chronic pancreatitis. United Eur. Gastroenterol. J. 2018, 6, 272–282. [Google Scholar] [CrossRef] [Green Version]
- Roth, T.L.; Marson, A. Genetic Disease and Therapy. Annu. Rev. Pathol. 2021, 16, 145–166. [Google Scholar] [CrossRef]
- Pogue, R.E.; Cavalcanti, D.P.; Shanker, S.; Andrade, R.V.; Aguiar, L.R.; de Carvalho, J.L.; Costa, F.F. Rare genetic diseases: Update on diagnosis, treatment and online resources. Drug. Discov. Today 2018, 23, 187–195. [Google Scholar] [CrossRef]
- Sui, Y.; Lu, K.; Fu, L. Prediction and analysis of novel key genes ITGAX, LAPTM5, SERPINE1 in clear cell renal cell carcinoma through bioinformatics analysis. PeerJ 2021, 9, e11272. [Google Scholar] [CrossRef]
- Tripolszki, K.; Sasaki, E.; Hotakainen, R.; Kassim, A.H.; Pereira, C.; Rolfs, A.; Bauer, P.; Reardon, W.; Bertoli-Avella, A.M. An X-linked syndrome with severe neurodevelopmental delay, hydrocephalus, and early lethality caused by a missense variation in the OTUD5 gene. Clin. Genet. 2021, 99, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Beck, D.B.; Basar, M.A.; Asmar, A.J.; Thompson, J.J.; Oda, H.; Uehara, D.T.; Saida, K.; Pajusalu, S.; Talvik, I.; D’Souza, P.; et al. Linkage-specific deubiquitylation by OTUD5 defines an embryonic pathway intolerant to genomic variation. Sci. Adv. 2021, 7, eabe2116. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Cavalli, V. Neuroproteomics approaches to decipher neuronal regeneration and degeneration. Mol. Cell Proteom. 2010, 9, 963–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, F.; Li, H.; Jiao, Q.; Li, C.; Fu, L.; Cui, C.; Jiang, H.; Zhang, L. Deubiquitylase OTUD3 prevents Parkinson’s disease through stabilizing iron regulatory protein 2. Cell Death Dis. 2022, 13, 418. [Google Scholar] [CrossRef]
- Huang, L.; Jolly, L.A.; Willis-Owen, S.; Gardner, A.; Kumar, R.; Douglas, E.; Shoubridge, C.; Wieczorek, D.; Tzschach, A.; Cohen, M.; et al. A noncoding, regulatory mutation implicates HCFC1 in nonsyndromic intellectual disability. Am. J. Hum. Genet. 2012, 91, 694–702. [Google Scholar] [CrossRef] [Green Version]
- Wagner, V.F.; Hillman, P.R.; Britt, A.D.; Ray, J.W.; Farach, L.S. A De novo HDAC2 variant in a patient with features consistent with Cornelia de Lange syndrome phenotype. Am. J. Med. Genet. A 2019, 179, 852–856. [Google Scholar] [CrossRef]
- Odnokoz, O.; Wavelet-Vermuse, C.; Hophan, S.L.; Bulun, S.; Wan, Y. ARID1 proteins: From transcriptional and post-translational regulation to carcinogenesis and potential therapeutics. Epigenomics 2021, 13, 809–823. [Google Scholar] [CrossRef]
- Gurley, J.M.; Gmyrek, G.B.; Hargis, E.A.; Bishop, G.A.; Carr, D.J.J.; Elliott, M.H. The Chx10-Traf3 Knockout Mouse as a Viable Model to Study Neuronal Immune Regulation. Cells 2021, 10, 2068. [Google Scholar] [CrossRef] [PubMed]
- Saida, K.; Fukuda, T.; Scott, D.A.; Sengoku, T.; Ogata, K.; Nicosia, A.; Hernandez-Garcia, A.; Lalani, S.R.; Azamian, M.S.; Streff, H.; et al. OTUD5 Variants Associated With X-Linked Intellectual Disability and Congenital Malformation. Front. Cell Dev. Biol. 2021, 9, 631428. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Lanczky, A.; Gyorffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Gonzalez-Navajas, J.M.; Law, J.; Nguyen, K.P.; Bhargava, M.; Corr, M.P.; Varki, N.; Eckmann, L.; Hoffman, H.M.; Lee, J.; Raz, E. Interleukin 1 receptor signaling regulates DUBA expression and facilitates Toll-like receptor 9-driven antiinflammatory cytokine production. J. Exp. Med. 2010, 207, 2799–2807. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Zhang, L. Serotonylation: A novel histone H3 marker. Signal Transduct. Target. Ther. 2019, 4, 15. [Google Scholar] [CrossRef] [Green Version]
- Kabra, A.; Li, Y. Conformational Dynamics of Deubiquitinase A and Functional Implications. Biochemistry 2021, 60, 201–209. [Google Scholar] [CrossRef]
- Kim, Y.C.; Guan, K.L. mTOR: A pharmacologic target for autophagy regulation. J. Clin. Investig. 2015, 125, 25–32. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, S.; Mickanin, C.; Feng, Y.; Charlat, O.; Michaud, G.A.; Schirle, M.; Shi, X.; Hild, M.; Bauer, A.; et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 2011, 13, 623–629. [Google Scholar] [CrossRef]
- Gao, Y.; Song, C.; Hui, L.; Li, C.Y.; Wang, J.; Tian, Y.; Han, X.; Chen, Y.; Tian, D.L.; Qiu, X.; et al. Overexpression of RNF146 in non-small cell lung cancer enhances proliferation and invasion of tumors through the Wnt/beta-catenin signaling pathway. PLoS ONE 2014, 9, e85377. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhu, Y.; Gu, Y.; Ma, M.; Wang, Y.; Qi, S.; Zeng, Y.; Zhu, R.; Wang, X.; Yu, P.; et al. Stabilization of Motin family proteins in NF2-deficient cells prevents full activation of YAP/TAZ and rapid tumorigenesis. Cell Rep. 2021, 36, 109596. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; He, M.; Kong, X.; Jiang, P.; Liu, X.; Diao, L.; Zhang, X.; Li, H.; Ling, X.; et al. UbiBrowser 2.0: A comprehensive resource for proteome-wide known and predicted ubiquitin ligase/deubiquitinase-substrate interactions in eukaryotic species. Nucleic Acids Res. 2022, 50, D719–D728. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, L.; Li, X.F. Hypoxia and the Tumor Microenvironment. Technol. Cancer Res. Treat. 2021, 20, 15330338211036304. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Bae, J.S. Tumor-Associated Macrophages and Neutrophils in Tumor Microenvironment. Mediat. Inflamm. 2016, 2016, 6058147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitt, J.M.; Marabelle, A.; Eggermont, A.; Soria, J.C.; Kroemer, G.; Zitvogel, L. Targeting the tumor microenvironment: Removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol. 2016, 27, 1482–1492. [Google Scholar] [CrossRef]
- Chen, Q.; Sun, L.; Chen, Z.J. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016, 17, 1142–1149. [Google Scholar] [CrossRef]
- Deng, L.; Liang, H.; Xu, M.; Yang, X.; Burnette, B.; Arina, A.; Li, X.D.; Mauceri, H.; Beckett, M.; Darga, T.; et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity 2014, 41, 843–852. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Bai, X.C.; Chen, Z.J. Structures and Mechanisms in the cGAS-STING Innate Immunity Pathway. Immunity 2020, 53, 43–53. [Google Scholar] [CrossRef]
- Mao, X.; Xu, J.; Wang, W.; Liang, C.; Hua, J.; Liu, J.; Zhang, B.; Meng, Q.; Yu, X.; Shi, S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: New findings and future perspectives. Mol. Cancer 2021, 20, 131. [Google Scholar] [CrossRef] [PubMed]
- Bastounis, E.E.; Serrano-Alcalde, F.; Radhakrishnan, P.; Engstrom, P.; Gomez-Benito, M.J.; Oswald, M.S.; Yeh, Y.T.; Smith, J.G.; Welch, M.D.; Garcia-Aznar, J.M.; et al. Mechanical competition triggered by innate immune signaling drives the collective extrusion of bacterially infected epithelial cells. Dev. Cell 2021, 56, 443–460.e11. [Google Scholar] [CrossRef] [PubMed]


| Numbers | Clinical Manifestation | OTUD5 Variant | Protein Change | Age | Status | References |
|---|---|---|---|---|---|---|
| n = 13 | Neurodevelopmental delay, hydrocephalus and early lethality. | 598G > A | Glu200Lys | 4 days–37 y | 12 deceased (from infancy (4 days–2 y), 6 y, 37 y) and 1 alive | [85] |
| n = 10 | Global developmental delay with brain malformations, hirsutism, genitourinary defects and early lethality. | 482_490del, 766G > A, 820C > T, 1055 T > C, 1210C > T, 1480 G > A | 161_164del, Asp256Asn, Arg274Trp, Leu352Pro, Arg404Trp, Gly494Ser | 2–14 y | 4 deceased (from infancy (1–13 m), 1 deceased in utero) and 6 alive | [86] |
| n = 3 | Severe short stature refractory epilepsy and congenital anomalies. | 878A > T 1210C > T | Asn293Ile Arg404Trp | 2–49 y | Alive | [93] |
| Gene Symbol (E3) | Domain_ Likelihood Ratio | Go_ Likelihood Ratio | Network_ Likelihood Ratio | Motif_ Likelihood Ratio | Confidence Score |
|---|---|---|---|---|---|
| SMURF2 | 1 | 3.63 | 3.83 | 4.09 | 0.853 |
| NEDD4 | 1 | 2.78 | 3.83 | 4.09 | 0.837 |
| MARCHF7 | 1 | 1 | 3.83 | 9.23 | 0.825 |
| SMURF1 | 1 | 8.56 | 1 | 4.09 | 0.824 |
| UBE4B | 1 | 3.63 | 3.83 | 2.28 | 0.818 |
| ITCH | 1 | 1.99 | 3.83 | 4.09 | 0.817 |
| SYTL4 | 1 | 8.56 | 1 | 3.35 | 0.811 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, L.; Lu, K.; Jiao, Q.; Chen, X.; Jia, F. The Regulation and Double-Edged Roles of the Deubiquitinase OTUD5. Cells 2023, 12, 1161. https://doi.org/10.3390/cells12081161
Fu L, Lu K, Jiao Q, Chen X, Jia F. The Regulation and Double-Edged Roles of the Deubiquitinase OTUD5. Cells. 2023; 12(8):1161. https://doi.org/10.3390/cells12081161
Chicago/Turabian StyleFu, Lin, Kun Lu, Qian Jiao, Xi Chen, and Fengju Jia. 2023. "The Regulation and Double-Edged Roles of the Deubiquitinase OTUD5" Cells 12, no. 8: 1161. https://doi.org/10.3390/cells12081161
APA StyleFu, L., Lu, K., Jiao, Q., Chen, X., & Jia, F. (2023). The Regulation and Double-Edged Roles of the Deubiquitinase OTUD5. Cells, 12(8), 1161. https://doi.org/10.3390/cells12081161





