Essential Protein PHB2 and Its Regulatory Mechanisms in Cancer
Abstract
1. Introduction
1.1. History of Prohibitin
1.2. Prohibitin Family and Structure
1.3. Prohibitin Expression
1.4. PHB1
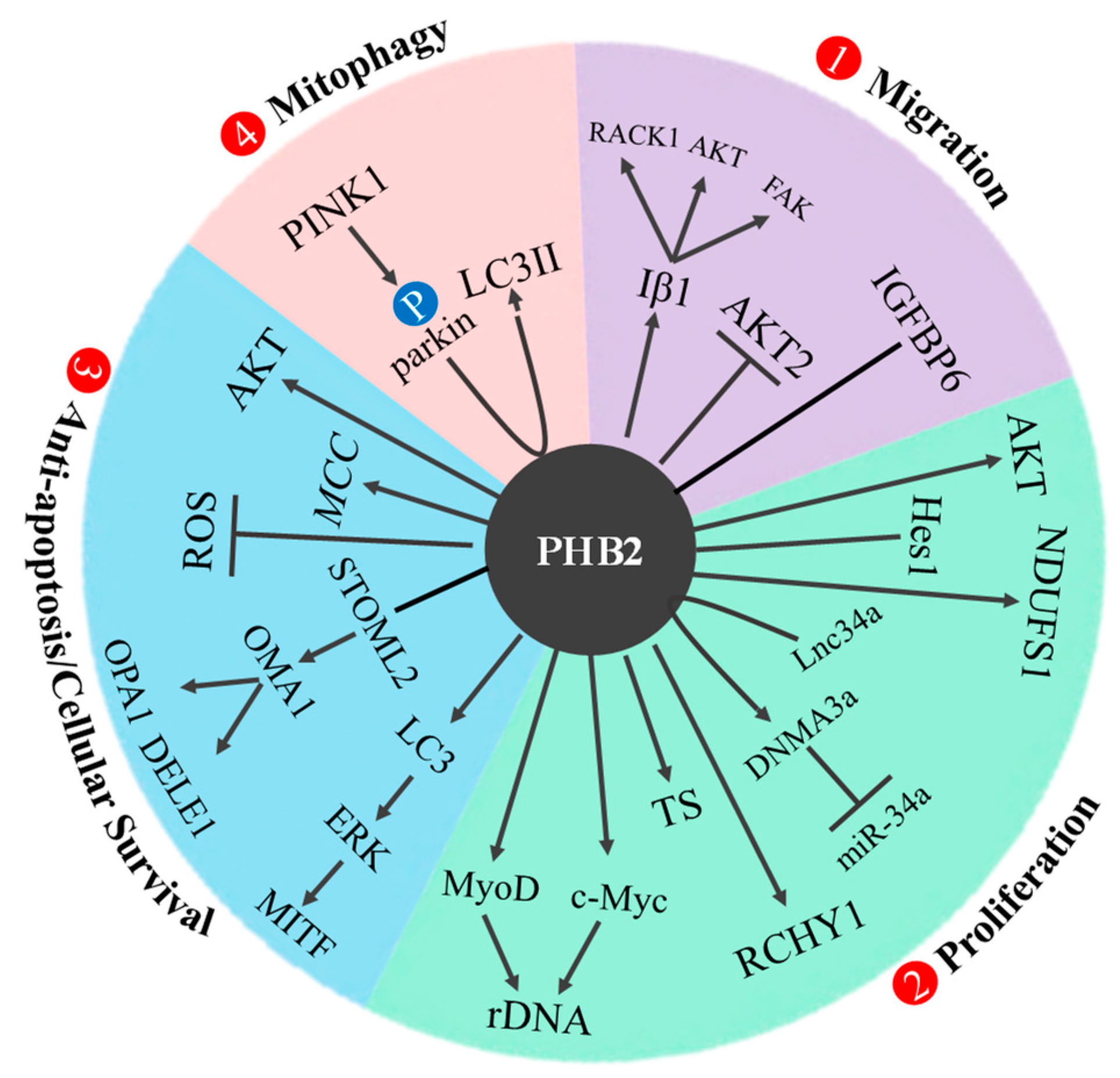
2. Critical Functions of PHB2
2.1. Conditional Depletion of PHB2
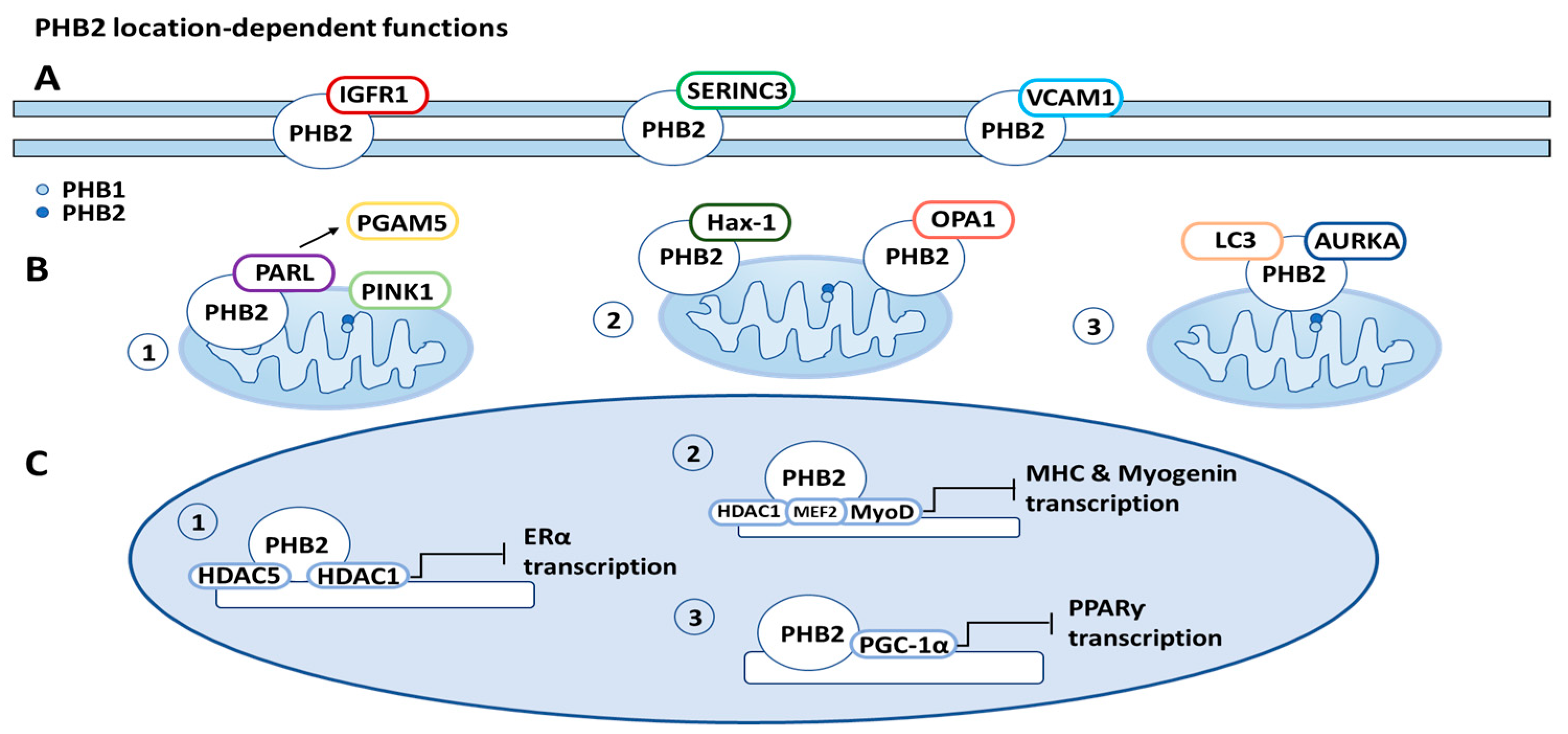
2.2. Location-Dependent PHB2 Function
2.2.1. PHB2 Function on Cell Membrane
2.2.2. PHB2 Function in Mitochondria
2.2.3. PHB2 Function in Nucleus
3. PHB2 Tumor Suppressor Function
3.1. Breast Cancer (BC)
3.2. Osteosarcoma (OS)
3.3. Head and Neck Squamous Cancer Cells
4. PHB2 Oncogenic Function
4.1. Prostate Cancer (PCa)
4.2. Non-Small Cell Lung Cancer (NSCLC)
4.3. Colon Cancer (CRC)
4.4. Hepatocellular (HCC)
4.5. Leukemia
4.6. Ovarian Cancer
4.7. Rhabdomyosarcoma (RMS)
4.8. Multiple Myeloma (MM)
4.9. Esophageal Squamous Cell Carcinoma (ESCC)
4.10. Hematologic: Lymphoid and Myeloid Tumor
4.11. Melanoma
4.12. Pancreatic Cancer
5. Reported PHB2 Modulators
6. Discussion
6.1. Dual Function of PHB2 in Different Cancers
6.2. Shared Molecular Mechanism in Different Cancers
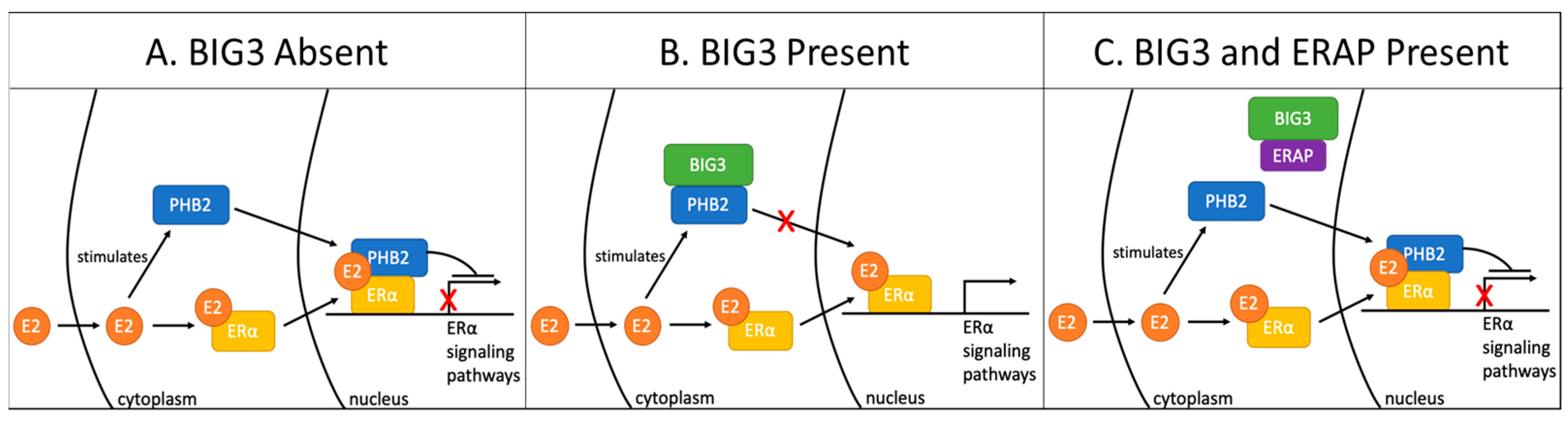
6.2.1. BIG3-PHB2
6.2.2. Inhibit miR-34a
6.2.3. Crosstalk with AKT
6.3. PHB2 Location-Dependent Functions (Figure 3)
6.4. PHB2 Modulators Block Translation Initiation
6.5. Rational Design of Targeting PHB2
7. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PHB1 | prohibitin 1 |
| PHB2 | prohibitin 2 |
| IMM | inner mitochondrial membrane |
| OMM | outer mitochondrial membrane |
| CPT1b | carnitine palmitoyltransferase1b |
| MAPLC3/LC3 | gene for microtubule-associated proteins 1A/1B light chain 3B |
| PINK1 | PTEN induced putative kinase 1 |
| PARL | presenilin-associated rhomboid-like protein |
| OXPHOS | oxidative phosphorylation system |
| OPA1 | optic atrophy 1 protein |
| Hax-1 | HCLS1-associated protein X-1 |
| ROS | reactive oxygen species |
| AURKA | aurora kinase A |
| MAP1LC3 | gene for microtubule-associated proteins 1A/1B light chain 3B |
| AKT2 | AKT serine/threonine kinase 2 |
| PPARγ | peroxisome proliferator-activated receptor |
| PGC-1α | a co-transcriptional activator of PPARγ |
| BIG3 | brefeldin A-inhibited guanine nucleotide-exchange protein 3 |
| ERAP | ERα activity-regulator synthetic peptide |
| PGRMC1 | progesterone receptor membrane component 1 PGRMC1 |
| GGCT | γ-glutamylcyclotransferases |
| PARP-1 | Poly [ADP-ribose] polymerase 1 |
| RACK1 | activated C kinase 1 |
| ROS | reactive oxygen species |
| TS | thymidylate synthase |
| RCHY1 | RING finger and CHY zinc finger domain-containing protein 1 |
| NDUFS1 | NADH: ubiquinone oxidoreductase core subunit S1 |
| FOXM1 | forkhead box protein M1 |
| BAX | Bcl-2-associated X protein |
| BAK1 | Bcl-2 homologous antagonist killer |
| DELE1 | DAP3 Binding Cell Death Enhancer 1 |
| EIF2AK1 | eukaryotic translation initiation factor 2 alpha kinase 1 |
| MCL1 | induced myeloid leukemia cell differentiation protein |
| IGFBP-6 | Insulin Like Growth Factor Binding Protein 6 |
| IGF-II | Insulin Like Growth Factor 2 |
| PBMCs | Peripheral Blood Mononuclear Cells |
| MITF | microphthalmia associated transcription factor |
References
- McClung, J.K.; Danner, D.B.; Stewart, D.A.; Smith, J.R.; Schneider, E.L.; Lumpkin, C.K.; Dell’Orco, R.T.; Nuell, M.J. Isolation of a cDNA that hybrid selects antiproliferative mRNA from rat liver. Biochem. Biophys. Res. Commun. 1989, 164, 1316–1322. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Saito, H.; Swensen, J.; Olifant, A.; Wood, C.; Danner, D.; Sakamoto, T.; Takita, K.; Kasumi, F.; Miki, Y.; et al. The human prohibitin gene located on chromosome 17q21 is mutated in sporadic breast cancer. Cancer Res. 1992, 52, 1643–1646. [Google Scholar] [PubMed]
- Nuell, M.J.; Stewart, D.A.; Walker, L.; Friedman, V.; Wood, C.M.; Owens, G.A.; Smith, J.R.; Schneider, E.L.; Dell’Orco, R.; Lumpkin, C.K.; et al. Prohibitin, an evolutionarily conserved intracellular protein that blocks DNA synthesis in normal fibroblasts and HeLa cells. Mol. Cell. Biol. 1991, 11, 1372–1381. [Google Scholar] [CrossRef] [PubMed]
- Terashima, M.; Kim, K.M.; Adachi, T.; Nielsen, P.J.; Reth, M.; Kohler, G.; Lamers, M.C. The IgM antigen receptor of B lymphocytes is associated with prohibitin and a prohibitin-related protein. EMBO J. 1994, 13, 3782–3792. [Google Scholar] [CrossRef]
- Montano, M.M.; Ekena, K.; Delage-Mourroux, R.; Chang, W.; Martini, P.; Katzenellenbogen, B.S. An estrogen receptor-selective coregulator that potentiates the effectiveness of antiestrogens and represses the activity of estrogens. Proc. Natl. Acad. Sci. USA 1999, 96, 6947–6952. [Google Scholar] [CrossRef] [PubMed]
- Browman, D.T.; Hoegg, M.B.; Robbins, S.M. The SPFH domain-containing proteins: More than lipid raft markers. Trends Cell. Biol. 2007, 17, 394–402. [Google Scholar] [CrossRef]
- Signorile, A.; Sgaramella, G.; Bellomo, F.; De Rasmo, D. Prohibitins: A Critical Role in Mitochondrial Functions and Implication in Diseases. Cells 2019, 8, 71. [Google Scholar] [CrossRef]
- Tatsuta, T.; Model, K.; Langer, T. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell 2005, 16, 248–259. [Google Scholar] [CrossRef]
- Bavelloni, A.; Piazzi, M.; Raffini, M.; Faenza, I.; Blalock, W.L. Prohibitin 2: At a communications crossroads. IUBMB Life 2015, 67, 239–254. [Google Scholar] [CrossRef]
- Funk, L.; Su, K.C.; Ly, J.; Feldman, D.; Singh, A.; Moodie, B.; Blainey, P.C.; Cheeseman, I.M. The phenotypic landscape of essential human genes. Cell 2022, 185, 4634–4653 e4622. [Google Scholar] [CrossRef]
- Yang, J.; Li, B.; He, Q.Y. Significance of prohibitin domain family in tumorigenesis and its implication in cancer diagnosis and treatment. Cell Death Dis. 2018, 9, 580. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.T.; Chen, P.; Ouyang, R.Y.; Song, L. Multifaceted role of prohibitin in cell survival and apoptosis. Apoptosis 2015, 20, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Ande, S.R.; Xu, Y.X.Z.; Mishra, S. Prohibitin: A potential therapeutic target in tyrosine kinase signaling. Signal Transduct. Target. Ther. 2017, 2, 17059. [Google Scholar] [CrossRef] [PubMed]
- Supale, S.; Thorel, F.; Merkwirth, C.; Gjinovci, A.; Herrera, P.L.; Scorrano, L.; Meda, P.; Langer, T.; Maechler, P. Loss of prohibitin induces mitochondrial damages altering beta-cell function and survival and is responsible for gradual diabetes development. Diabetes 2013, 62, 3488–3499. [Google Scholar] [CrossRef]
- Wu, D.; Jian, C.; Peng, Q.; Hou, T.; Wu, K.; Shang, B.; Zhao, M.; Wang, Y.; Zheng, W.; Ma, Q.; et al. Prohibitin 2 deficiency impairs cardiac fatty acid oxidation and causes heart failure. Cell Death Dis. 2020, 11, 181. [Google Scholar] [CrossRef]
- Merkwirth, C.; Martinelli, P.; Korwitz, A.; Morbin, M.; Bronneke, H.S.; Jordan, S.D.; Rugarli, E.I.; Langer, T. Loss of prohibitin membrane scaffolds impairs mitochondrial architecture and leads to tau hyperphosphorylation and neurodegeneration. PLoS Genet. 2012, 8, e1003021. [Google Scholar] [CrossRef]
- Li, L.; Martin-Levilain, J.; Jimenez-Sanchez, C.; Karaca, M.; Foti, M.; Martinou, J.C.; Maechler, P. In vivo stabilization of OPA1 in hepatocytes potentiates mitochondrial respiration and gluconeogenesis in a prohibitin-dependent way. J. Biol. Chem. 2019, 294, 12581–12598. [Google Scholar] [CrossRef]
- Chowdhury, I.; Thompson, W.E.; Thomas, K. Prohibitins role in cellular survival through Ras-Raf-MEK-ERK pathway. J. Cell Physiol. 2014, 229, 998–1004. [Google Scholar] [CrossRef]
- Osman, C.; Merkwirth, C.; Langer, T. Prohibitins and the functional compartmentalization of mitochondrial membranes. J. Cell Sci. 2009, 122, 3823–3830. [Google Scholar] [CrossRef]
- Zhou, Z.; Ai, H.; Li, K.; Yao, X.; Zhu, W.; Liu, L.; Yu, C.; Song, Z.; Bao, Y.; Huang, Y.; et al. Prohibitin 2 localizes in nucleolus to regulate ribosomal RNA transcription and facilitate cell proliferation in RD cells. Sci. Rep. 2018, 8, 1479. [Google Scholar] [CrossRef]
- Ding, W.X.; Yin, X.M. Mitophagy: Mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012, 393, 547–564. [Google Scholar] [CrossRef]
- Ma, K.; Chen, G.; Li, W.; Kepp, O.; Zhu, Y.; Chen, Q. Mitophagy, Mitochondrial Homeostasis, and Cell Fate. Front. Cell Dev. Biol. 2020, 8, 467. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Chiang, W.C.; Sumpter, R., Jr.; Mishra, P.; Levine, B. Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell 2017, 168, 224–238.e210. [Google Scholar] [CrossRef]
- Lahiri, V.; Klionsky, D.J. PHB2/prohibitin 2: An inner membrane mitophagy receptor. Cell Res. 2017, 27, 311–312. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Gong, L.; Chen, L.; Xu, M.; Abou-Hamdan, H.; Tang, M.; Desaubry, L.; Song, Z. PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy 2020, 16, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Badrinath, N.; Yoo, S.Y. Mitochondria in cancer: In the aspects of tumorigenesis and targeted therapy. Carcinogenesis 2018, 39, 1419–1430. [Google Scholar] [CrossRef]
- Merkwirth, C.; Dargazanli, S.; Tatsuta, T.; Geimer, S.; Lower, B.; Wunderlich, F.T.; von Kleist-Retzow, J.C.; Waisman, A.; Westermann, B.; Langer, T. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008, 22, 476–488. [Google Scholar] [CrossRef]
- Estaquier, J.; Vallette, F.; Vayssiere, J.L.; Mignotte, B. The mitochondrial pathways of apoptosis. Adv. Exp. Med. Biol. 2012, 942, 157–183. [Google Scholar] [CrossRef] [PubMed]
- Bertolin, G.; Alves-Guerra, M.C.; Cheron, A.; Burel, A.; Prigent, C.; Le Borgne, R.; Tramier, M. Mitochondrial Aurora kinase A induces mitophagy by interacting with MAP1LC3 and Prohibitin 2. Life Sci. Alliance 2021, 4, e202000806. [Google Scholar] [CrossRef]
- Wang, S.; Faller, D.V. Roles of prohibitin in growth control and tumor suppression in human cancers. Transl. Oncogenomics 2008, 3, 23–37. [Google Scholar]
- Thuaud, F.; Ribeiro, N.; Nebigil, C.G.; Desaubry, L. Prohibitin ligands in cell death and survival: Mode of action and therapeutic potential. Chem. Biol. 2013, 20, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Kasashima, K.; Ohta, E.; Kagawa, Y.; Endo, H. Mitochondrial functions and estrogen receptor-dependent nuclear translocation of pleiotropic human prohibitin 2. J. Biol. Chem. 2006, 281, 36401–36410. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Liu, L.; Yang, X.J.; Wu, Z. Akt binds prohibitin 2 and relieves its repression of MyoD and muscle differentiation. J. Cell Sci. 2004, 117, 3021–3029. [Google Scholar] [CrossRef] [PubMed]
- Kurtev, V.; Margueron, R.; Kroboth, K.; Ogris, E.; Cavailles, V.; Seiser, C. Transcriptional regulation by the repressor of estrogen receptor activity via recruitment of histone deacetylases. J. Biol. Chem. 2004, 279, 24834–24843. [Google Scholar] [CrossRef] [PubMed]
- Chigira, T.; Nagatoishi, S.; Takeda, H.; Yoshimaru, T.; Katagiri, T.; Tsumoto, K. Biophysical characterization of the breast cancer-related BIG3-PHB2 interaction: Effect of non-conserved loop region of BIG3 on the structure and the interaction. Biochem. Biophys. Res. Commun. 2019, 518, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Yoshimaru, T.; Komatsu, M.; Matsuo, T.; Chen, Y.A.; Murakami, Y.; Mizuguchi, K.; Mizohata, E.; Inoue, T.; Akiyama, M.; Yamaguchi, R.; et al. Targeting BIG3-PHB2 interaction to overcome tamoxifen resistance in breast cancer cells. Nat. Commun. 2013, 4, 2443. [Google Scholar] [CrossRef][Green Version]
- Yoshimaru, T.; Aihara, K.; Komatsu, M.; Matsushita, Y.; Okazaki, Y.; Toyokuni, S.; Honda, J.; Sasa, M.; Miyoshi, Y.; Otaka, A.; et al. Stapled BIG3 helical peptide ERAP potentiates anti-tumour activity for breast cancer therapeutics. Sci. Rep. 2017, 7, 1821. [Google Scholar] [CrossRef]
- Bai, Y.; Ludescher, M.; Poschmann, G.; Stuhler, K.; Wyrich, M.; Oles, J.; Franken, A.; Rivandi, M.; Abramova, A.; Reinhardt, F.; et al. PGRMC1 Promotes Progestin-Dependent Proliferation of Breast Cancer Cells by Binding Prohibitins Resulting in Activation of ERalpha Signaling. Cancers 2021, 13, 5635. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Z.; Li, S.; Deng, Y.; He, N. Identification of PHB2 as a Potential Biomarker of Luminal A Breast Cancer Cells Using a Cell-Specific Aptamer. ACS Appl. Mater. Interfaces 2022, 14, 51593–51601. [Google Scholar] [CrossRef]
- Takagi, H.; Moyama, C.; Taniguchi, K.; Ando, K.; Matsuda, R.; Ando, S.; Ii, H.; Kageyama, S.; Kawauchi, A.; Chouha, N.; et al. Fluorizoline Blocks the Interaction between Prohibitin-2 and gamma-Glutamylcyclotransferase and Induces p21(Waf1/Cip1) Expression in MCF7 Breast Cancer Cells. Mol. Pharmacol. 2022, 101, 78–86. [Google Scholar] [CrossRef]
- Perez-Perarnau, A.; Preciado, S.; Palmeri, C.M.; Moncunill-Massaguer, C.; Iglesias-Serret, D.; Gonzalez-Girones, D.M.; Miguel, M.; Karasawa, S.; Sakamoto, S.; Cosialls, A.M.; et al. A trifluorinated thiazoline scaffold leading to pro-apoptotic agents targeting prohibitins. Angew. Chem. Int. Ed. Engl. 2014, 53, 10150–10154. [Google Scholar] [CrossRef]
- Nunez-Vazquez, S.; Saura-Esteller, J.; Sanchez-Vera, I.; Guilbaud, E.; Cosialls, A.M.; Pons, G.; Ricci, J.E.; Iglesias-Serret, D.; Marchetti, S.; Gil, J. The prohibitin-binding compound fluorizoline inhibits mitophagy in cancer cells. Oncogenesis 2021, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Toki, S.; Yoshimaru, T.; Matsushita, Y.; Aihara, H.; Ono, M.; Tsuneyama, K.; Sairyo, K.; Katagiri, T. The survival and proliferation of osteosarcoma cells are dependent on the mitochondrial BIG3-PHB2 complex formation. Cancer Sci. 2021, 112, 4208–4219. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Liu, Z.; Wang, L.; Johnson, D.G.; Wei, Q. Identification of prohibitin and prohibiton as novel factors binding to the p53 induced gene 3 (PIG3) promoter (TGYCC)(15) motif. Biochem. Biophys. Res. Commun. 2014, 443, 1239–1244. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shen, Y.; Gao, Y.; Yuan, H.; Cao, J.; Jia, B.; Li, M.; Peng, Y.; Du, X.; Zhang, J.; Shi, J. Prohibitin-2 negatively regulates AKT2 expression to promote prostate cancer cell migration. Int. J. Mol. Med. 2018, 41, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Chang, N.; Xi, H.; Xiong, J.; Zhou, Y.; Wu, Y.; Wu, S.; Wang, N.; Yi, H.; Song, Y.; et al. PHB2 promotes tumorigenesis via RACK1 in non-small cell lung cancer. Theranostics 2021, 11, 3150–3166. [Google Scholar] [CrossRef]
- Zhang, H.; Yin, C.; Liu, X.; Bai, X.; Wang, L.; Xu, H.; Ju, J.; Zhang, L. Prohibitin 2/PHB2 in Parkin-Mediated Mitophagy: A Potential Therapeutic Target for Non-Small Cell Lung Carcinoma. Med. Sci. Monit. 2020, 26, e923227. [Google Scholar] [CrossRef]
- Watanabe, M.; Yamada, Y.; Kurumida, Y.; Kameda, T.; Sukeno, M.; Iizuka-Ohashi, M.; Sowa, Y.; Iizumi, Y.; Takakura, H.; Miyamoto, S.; et al. Rabdosianone I, a Bitter Diterpene from an Oriental Herb, Suppresses Thymidylate Synthase Expression by Directly Binding to ANT2 and PHB2. Cancers 2021, 13, 982. [Google Scholar] [CrossRef]
- Wang, X.; Zheng, Y.; Chai, Z.; Li, J.; Zhu, C.; Peng, Y.; Qiu, J.; Xu, J.; Liu, C. Dihydroartemisinin synergistically enhances the cytotoxic effects of oxaliplatin in colon cancer by targeting the PHB2-RCHY1 mediated signaling pathway. Mol. Carcinog. 2023, 62, 293–302. [Google Scholar] [CrossRef]
- Ren, L.; Meng, L.; Gao, J.; Lu, M.; Guo, C.; Li, Y.; Rong, Z.; Ye, Y. PHB2 promotes colorectal cancer cell proliferation and tumorigenesis through NDUFS1-mediated oxidative phosphorylation. Cell Death Dis. 2023, 14, 44. [Google Scholar] [CrossRef]
- Wang, L.; Bu, P.; Ai, Y.; Srinivasan, T.; Chen, H.J.; Xiang, K.; Lipkin, S.M.; Shen, X. A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. Elife 2016, 5, e14620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Niu, H.; Ma, J.; Yuan, B.Y.; Chen, Y.H.; Zhuang, Y.; Chen, G.W.; Zeng, Z.C.; Xiang, Z.L. The molecular mechanism of LncRNA34a-mediated regulation of bone metastasis in hepatocellular carcinoma. Mol. Cancer 2019, 18, 120. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Fu, Y.; Zhao, K.; Miao, R.; Zhang, X.; Ma, X.; Liu, C.; Zhang, N.; Qu, K. Cellular senescence in hepatocellular carcinoma induced by a long non-coding RNA-encoded peptide PINT87aa by blocking FOXM1-mediated PHB2. Theranostics 2021, 11, 4929–4944. [Google Scholar] [CrossRef] [PubMed]
- Kuramori, C.; Azuma, M.; Kume, K.; Kaneko, Y.; Inoue, A.; Yamaguchi, Y.; Kabe, Y.; Hosoya, T.; Kizaki, M.; Suematsu, M.; et al. Capsaicin binds to prohibitin 2 and displaces it from the mitochondria to the nucleus. Biochem. Biophys. Res. Commun. 2009, 379, 519–525. [Google Scholar] [CrossRef]
- Belser, M.; Walker, D.W. Role of Prohibitins in Aging and Therapeutic Potential Against Age-Related Diseases. Front. Genet. 2021, 12, 714228. [Google Scholar] [CrossRef]
- Bavelloni, A.; Piazzi, M.; Faenza, I.; Raffini, M.; D’Angelo, A.; Cattini, L.; Cocco, L.; Blalock, W.L. Prohibitin 2 represents a novel nuclear AKT substrate during all-trans retinoic acid-induced differentiation of acute promyelocytic leukemia cells. FASEB J. 2014, 28, 2009–2019. [Google Scholar] [CrossRef]
- Cheng, M.; Yu, H.; Kong, Q.; Wang, B.; Shen, L.; Dong, D.; Sun, L. The Mitochondrial PHB2/OMA1/DELE1 Pathway Cooperates with Endoplasmic Reticulum Stress to Facilitate the Response to Chemotherapeutics in Ovarian Cancer. Int. J. Mol. Sci. 2022, 23, 1320. [Google Scholar] [CrossRef]
- Fu, P.; Yang, Z.; Bach, L.A. Prohibitin-2 binding modulates insulin-like growth factor-binding protein-6 (IGFBP-6)-induced rhabdomyosarcoma cell migration. J. Biol. Chem. 2013, 288, 29890–29900. [Google Scholar] [CrossRef]
- Edwards, S.K.; Baron, J.; Moore, C.R.; Liu, Y.; Perlman, D.H.; Hart, R.P.; Xie, P. Mutated in colorectal cancer (MCC) is a novel oncogene in B lymphocytes. J. Hematol. Oncol. 2014, 7, 56. [Google Scholar] [CrossRef]
- Cai, X.W.; Yu, W.W.; Yu, W.; Zhang, Q.; Feng, W.; Liu, M.N.; Sun, M.H.; Xiang, J.Q.; Zhang, Y.W.; Fu, X.L. Tissue-based quantitative proteomics to screen and identify the potential biomarkers for early recurrence/metastasis of esophageal squamous cell carcinoma. Cancer Med. 2018, 7, 2504–2517. [Google Scholar] [CrossRef]
- Ross, J.A.; Robles-Escajeda, E.; Oaxaca, D.M.; Padilla, D.L.; Kirken, R.A. The prohibitin protein complex promotes mitochondrial stabilization and cell survival in hematologic malignancies. Oncotarget 2017, 8, 65445–65456. [Google Scholar] [CrossRef] [PubMed]
- Djehal, A.; Krayem, M.; Najem, A.; Hammoud, H.; Cresteil, T.; Nebigil, C.G.; Wang, D.; Yu, P.; Bentouhami, E.; Ghanem, G.E.; et al. Targeting prohibitin with small molecules to promote melanogenesis and apoptosis in melanoma cells. Eur. J. Med. Chem. 2018, 155, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Perron, A.; Nishikawa, Y.; Iwata, J.; Shimojo, H.; Takaya, J.; Kobayashi, K.; Imayoshi, I.; Mbenza, N.M.; Takenoya, M.; Kageyama, R.; et al. Small-molecule screening yields a compound that inhibits the cancer-associated transcription factor Hes1 via the PHB2 chaperone. J. Biol. Chem. 2018, 293, 8285–8294. [Google Scholar] [CrossRef] [PubMed]
- Virtakoivu, R.; Pellinen, T.; Rantala, J.K.; Perala, M.; Ivaska, J. Distinct roles of AKT isoforms in regulating beta1-integrin activity, migration, and invasion in prostate cancer. Mol. Biol. Cell 2012, 23, 3357–3369. [Google Scholar] [CrossRef]
- Dillon, R.L.; Marcotte, R.; Hennessy, B.T.; Woodgett, J.R.; Mills, G.B.; Muller, W.J. Akt1 and akt2 play distinct roles in the initiation and metastatic phases of mammary tumor progression. Cancer Res. 2009, 69, 5057–5064. [Google Scholar] [CrossRef]
- Dai, X.; Zhang, X.; Chen, W.; Chen, Y.; Zhang, Q.; Mo, S.; Lu, J. Dihydroartemisinin: A Potential Natural Anticancer Drug. Int. J. Biol. Sci. 2021, 17, 603–622. [Google Scholar] [CrossRef]
- Zhang, L.; Liao, Y.; Tang, L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019, 38, 53. [Google Scholar] [CrossRef]
- Lee, Y.E.; Lim, H.J.; Park, J.H.; Kim, H.R.; Kang, M.G.; Cho, Y.K.; Shin, J.H.; Shin, M.G. Overexpression of Prohibitin 2 Protein is Associated with Adverse Prognosis in Cytogenetically Normal Acute Myeloid Leukemia. Ann. Lab. Med. 2022, 42, 585–589. [Google Scholar] [CrossRef]
- Wang, D.; Tabti, R.; Elderwish, S.; Abou-Hamdan, H.; Djehal, A.; Yu, P.; Yurugi, H.; Rajalingam, K.; Nebigil, C.G.; Desaubry, L. Prohibitin ligands: A growing armamentarium to tackle cancers, osteoporosis, inflammatory, cardiac and neurological diseases. Cell. Mol. Life Sci. 2020, 77, 3525–3546. [Google Scholar] [CrossRef]
- Cosialls, A.M.; Pomares, H.; Iglesias-Serret, D.; Saura-Esteller, J.; Nunez-Vazquez, S.; Gonzalez-Girones, D.M.; de la Banda, E.; Preciado, S.; Albericio, F.; Lavilla, R.; et al. The prohibitin-binding compound fluorizoline induces apoptosis in chronic lymphocytic leukemia cells through the upregulation of NOXA and synergizes with ibrutinib, 5-aminoimidazole-4-carboxamide riboside or venetoclax. Haematologica 2017, 102, 1587–1593. [Google Scholar] [CrossRef]
- Choi, S.; Bhagwat, A.M.; Al Mismar, R.; Goswami, N.; Ben Hamidane, H.; Sun, L.; Graumann, J. Proteomic profiling of human cancer pseudopodia for the identification of anti-metastatic drug candidates. Sci. Rep. 2018, 8, 5858. [Google Scholar] [CrossRef]
- Polier, G.; Neumann, J.; Thuaud, F.; Ribeiro, N.; Gelhaus, C.; Schmidt, H.; Giaisi, M.; Kohler, R.; Muller, W.W.; Proksch, P.; et al. The natural anticancer compounds rocaglamides inhibit the Raf-MEK-ERK pathway by targeting prohibitin 1 and 2. Chem. Biol. 2012, 19, 1093–1104. [Google Scholar] [CrossRef]
- Doudican, N.A.; Orlow, S.J. Inhibition of the CRAF/prohibitin interaction reverses CRAF-dependent resistance to vemurafenib. Oncogene 2017, 36, 423–428. [Google Scholar] [CrossRef]
- Yurugi, H.; Marini, F.; Weber, C.; David, K.; Zhao, Q.; Binder, H.; Desaubry, L.; Rajalingam, K. Targeting prohibitins with chemical ligands inhibits KRAS-mediated lung tumours. Oncogene 2017, 36, 4778–4789. [Google Scholar] [CrossRef] [PubMed]
- Thuaud, F.; Bernard, Y.; Turkeri, G.; Dirr, R.; Aubert, G.; Cresteil, T.; Baguet, A.; Tomasetto, C.; Svitkin, Y.; Sonenberg, N.; et al. Synthetic analogue of rocaglaol displays a potent and selective cytotoxicity in cancer cells: Involvement of apoptosis inducing factor and caspase-12. J. Med. Chem. 2009, 52, 5176–5187. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Murata, A.; Orihara, T.; Shirakawa, T.; Suenaga, K.; Kigoshi, H.; Uesugi, M. Marine natural product aurilide activates the OPA1-mediated apoptosis by binding to prohibitin. Chem. Biol. 2011, 18, 131–139. [Google Scholar] [CrossRef]
- Santhanam, S.K.; Dutta, D.; Parween, F.; Qadri, A. The virulence polysaccharide Vi released by Salmonella Typhi targets membrane prohibitin to inhibit T-cell activation. J. Infect. Dis. 2014, 210, 79–88. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Kruse, F.E.; Bacher, S.; Schmitz, M.L. Lipoteichoic acid selectively induces the ERK signaling pathway in the cornea. Investig. Ophthalmol. Vis. Sci. 2002, 43, 2272–2277. [Google Scholar]
- Asselin, E.; Bazer, F.W.; Fortier, M.A. Recombinant ovine and bovine interferons tau regulate prostaglandin production and oxytocin response in cultured bovine endometrial cells. Biol. Reprod. 1997, 56, 402–408. [Google Scholar] [CrossRef][Green Version]
- Yoshimaru, T.; Komatsu, M.; Tashiro, E.; Imoto, M.; Osada, H.; Miyoshi, Y.; Honda, J.; Sasa, M.; Katagiri, T. Xanthohumol suppresses oestrogen-signalling in breast cancer through the inhibition of BIG3-PHB2 interactions. Sci. Rep. 2014, 4, 7355. [Google Scholar] [CrossRef]
- Vanrenterghem, Y.; Waer, M.; Ang, K.K.; Lerut, T.; Bouillon, R.; Vandeputte, I.; Vandeputte, M.; van der Schueren, E.; Gruwez, J.; Michielsen, P. Renal cadaveric transplantation after total lymphoid irradiation in patients with diabetes. Kidney Int. Suppl. 1983, 14, S69–S73. [Google Scholar]
- San, T.B. A case of benign reticuloendothelial tumor of the placenta. Pediatr. Akus. Ginekol. 1966, 4, 64. [Google Scholar] [PubMed]
- Slabakova, E.; Culig, Z.; Remsik, J.; Soucek, K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017, 8, e3100. [Google Scholar] [CrossRef] [PubMed]
- Degan, S.E.; Gelman, I.H. Emerging Roles for AKT Isoform Preference in Cancer Progression Pathways. Mol. Cancer Res. 2021, 19, 1251–1257. [Google Scholar] [CrossRef]
- Bleumink, M.; Kohler, R.; Giaisi, M.; Proksch, P.; Krammer, P.H.; Li-Weber, M. Rocaglamide breaks TRAIL resistance in HTLV-1-associated adult T-cell leukemia/lymphoma by translational suppression of c-FLIP expression. Cell Death Differ. 2011, 18, 362–370. [Google Scholar] [CrossRef]
- Li-Weber, M. Molecular mechanisms and anti-cancer aspects of the medicinal phytochemicals rocaglamides (=flavaglines). Int. J. Cancer 2015, 137, 1791–1799. [Google Scholar] [CrossRef]
- Truitt, M.L.; Conn, C.S.; Shi, Z.; Pang, X.; Tokuyasu, T.; Coady, A.M.; Seo, Y.; Barna, M.; Ruggero, D. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell 2015, 162, 59–71. [Google Scholar] [CrossRef]


| (A) | ||
| Cancer Type | Mechanisms and References | |
| 1 | Breast cancer | |
| 2 | Osteosarcoma |
|
| 3 | Head and neck squamous cell |
|
| (B) | ||
| Cancer Type | Mechanisms and References | |
| 1 | Prostate | |
| 2 | Non-small cell lung | |
| 3 | Colon |
|
| 4 | Hepatocellular | |
| 5 | Leukemia |
|
| 6 | Ovarian |
|
| 7 | Rhabdomyosarcoma | |
| 8 | Multiple myeloma |
|
| 9 | Esophageal squamous cell |
|
| 10 | Hematologic: lymphoid and myeloid |
|
| 11 | Melanoma | |
| 12 | Pancreatic |
|
| Ligand Name | Description | Target | Kd Potency | System | Effect of Binding | Mechanisms | |
|---|---|---|---|---|---|---|---|
| 1 | Fluorizoline (FLZ) | A cytotoxic trifluorothiazoline | PHBs | 1 µM | Lymphocytic leukemia cells | Inhibits epidermal growth factor (EGF)/RAS-induced C-RAF activation |
|
| 2 | Rocaglamide (RocA) | Flavagline | PHBs | 50 nM |
|
|
|
| 3 | FL3 | Flavagline | PHBs | 50 nM |
|
|
|
| 4 | Mel9 and Mel41 | Melanogenin analogs | PHBs | 0.1 µM | Melanoma cancer cells | Triggers a cascade of events involving LC3, the kinase ERK and the transcription factor MITF | Mel9 and Mel41 binds PHB2 and inhibits p-ERK and p-AKT [62] |
| 5 | JI051 | Synthesized Hes1 inhibiting compound | PHB2 | EC50 of 0.3 μm | Pancreatic cancer | G2/M cell cycle arrest is promoted | JI051 binds to PHB2 to stabilize the interaction between Hes1 and PHB2 outside the nucleus [63] |
| 6 | Aurilide | Cyclic depsipeptide isolated from a marine mollusk | PHBs | 3–6 nM | HeLa cells | Disintegration of mitochondria and apoptosis, leading to an extreme toxicity | Blocks the PHB-dependent inhibition of the proteolytic processing of the dynamin-like GTPase optic atrophy 1 (OPA1) [76] |
| 7 | Vi capsular polysaccharide (Vi) | Of Salmonella typhi, causative agent of typhoid fever | PHBs | 1–5 µg/mL | Human intestinal epithelial cells | Blocks IL-2 secretion from T cells stimulated through the T-cell receptor (TCR) but not Protein Kinase C | Interacts with cell surface PHBs to inhibit ERK activity and interleukin-8 secretion in human intestinal epithelial cells [77] |
| 8 | Lipoteichoic acid | Polyanionic bacterial lipid, part of gram-positive bacteria cell walls | PHBs | Unknown | Human corneal keratocytes | Highlights the implication of PHBs in the immune response to infections | Lipoteichoic acid has been shown to activate RAF in the cornea, but whether this involves PHBs has not been documented yet [78] |
| 9 | Capsaicin | A component of hot chili peppers | PHB2 | Too high to be pertinent for treatment | Leukemia | Initiates mitochondrial apoptosis in human myeloid leukemia cells | Promotes PHB2’s translocation from mitochondria to the nucleus, dissociating it from the Adenine Nucleotide Translocator 2 (ANT2) [79] |
| 10 | Xanthohumol | Polyphenol chalcone found in hops | PHB2 | 50 µM | Breast cancer cells | Block ERα-positive breast cancer cell growth in vitro and in vivo | Binds to PHB2 inhibiting its interaction with BIG3, which suppresses the inhibitory activity PHB2 has on the ERα in breast cancer cells [80] |
| 11 | PDD005 | Purine derivative drug | PHBs | 1.29 ± 1.16 µM for PHB2 | Nervous system | Inhibits neuro-inflammation, levels of PHBs increase in mice when treated with PDD005 and decreases levels of the cytokine IL-1β | Enhances expression of PHBs in the hippocampal dentate gyrus of aged mouse and increases the phosphorylation of GSK-3β in organotypic hippocampal slice cultures, suggesting that this signaling protein is involved, at least partially, in the mechanism of neuroprotection [81,82] |
| 12 | DHA | Derivative of the antimalaria drug, Artemisinin | PHB2 | 20 µM | Colon cancer | Synergistic anticancer effects when with oxaliplatin by the promotion of PHB2 degradation in colon cancer cells | Downregulates PHB2 expression in a ubiquitylation-dependent manner, blocking PHB2-induced RCHY1 upregulation and the downregulation of p53 and p51 [49] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, A.; Lamont, L.; Liu, E.; Murray, S.D.; Meng, X.; Yang, S. Essential Protein PHB2 and Its Regulatory Mechanisms in Cancer. Cells 2023, 12, 1211. https://doi.org/10.3390/cells12081211
Qi A, Lamont L, Liu E, Murray SD, Meng X, Yang S. Essential Protein PHB2 and Its Regulatory Mechanisms in Cancer. Cells. 2023; 12(8):1211. https://doi.org/10.3390/cells12081211
Chicago/Turabian StyleQi, Amanda, Lillie Lamont, Evelyn Liu, Sarina D. Murray, Xiangbing Meng, and Shujie Yang. 2023. "Essential Protein PHB2 and Its Regulatory Mechanisms in Cancer" Cells 12, no. 8: 1211. https://doi.org/10.3390/cells12081211
APA StyleQi, A., Lamont, L., Liu, E., Murray, S. D., Meng, X., & Yang, S. (2023). Essential Protein PHB2 and Its Regulatory Mechanisms in Cancer. Cells, 12(8), 1211. https://doi.org/10.3390/cells12081211






