Characterization of Two Parabacteroides distasonis Candidate Strains as New Live Biotherapeutics against Obesity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Measurement of SCFA Production
2.3. STC1 Enteroendocrine Cell Culture and Induction of GLP-1 Secretion
2.4. Murine Model of Diet-Induced Obesity
2.5. Histological Analysis of the Adipose Tissue
2.6. Analysis of Metabolic Parameters
2.7. Gene Expression Quantification by Real-Time RT-PCR
2.8. Caecal Microbiota Analysis
2.9. Statistical Analysis
3. Results
3.1. In Vitro Functional Characteristics of the Strains
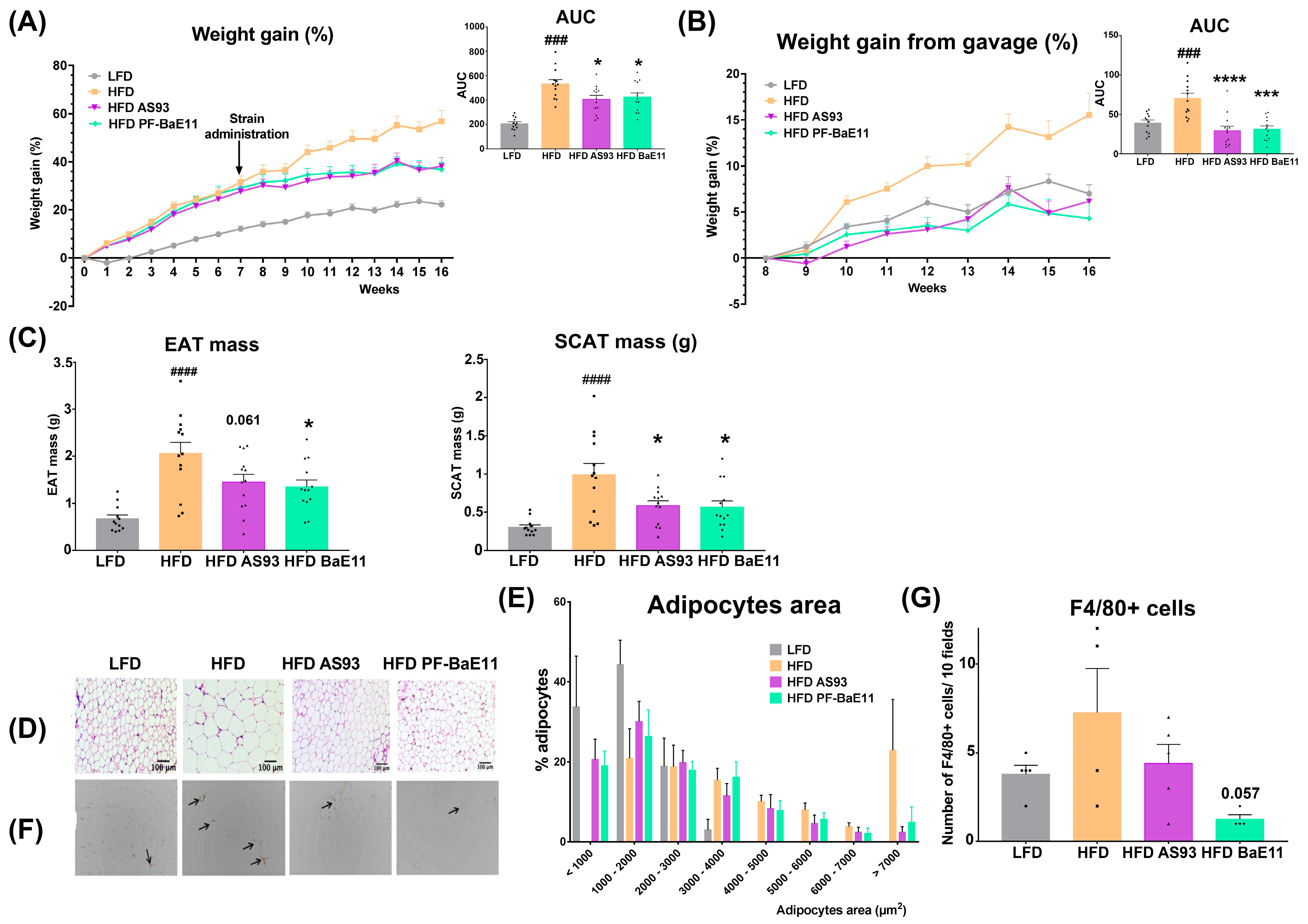
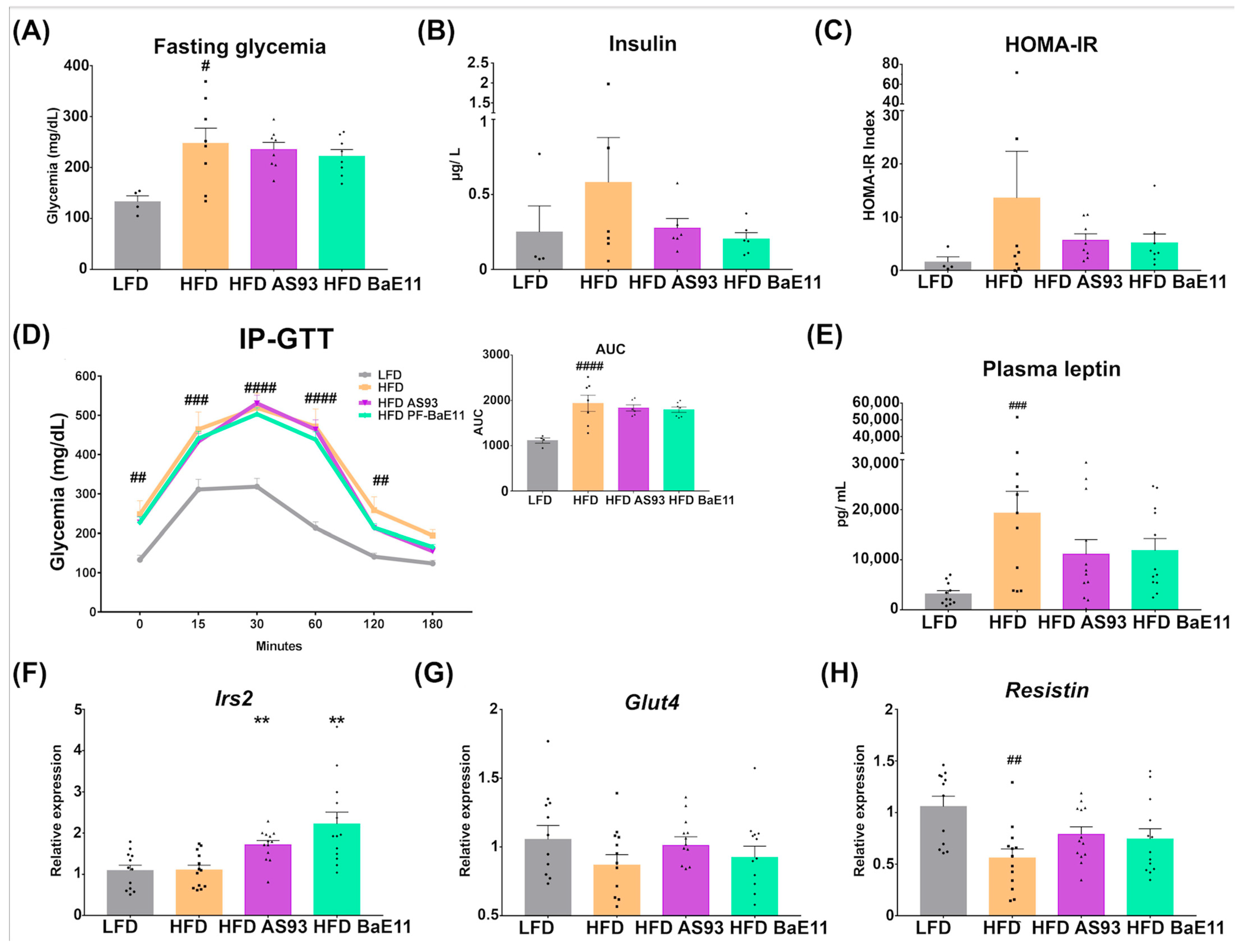
3.2. The Two P. distasonis Strains Counteract HFD-Induced Obesity and Adiposity and Improve Glucose Homeostasis
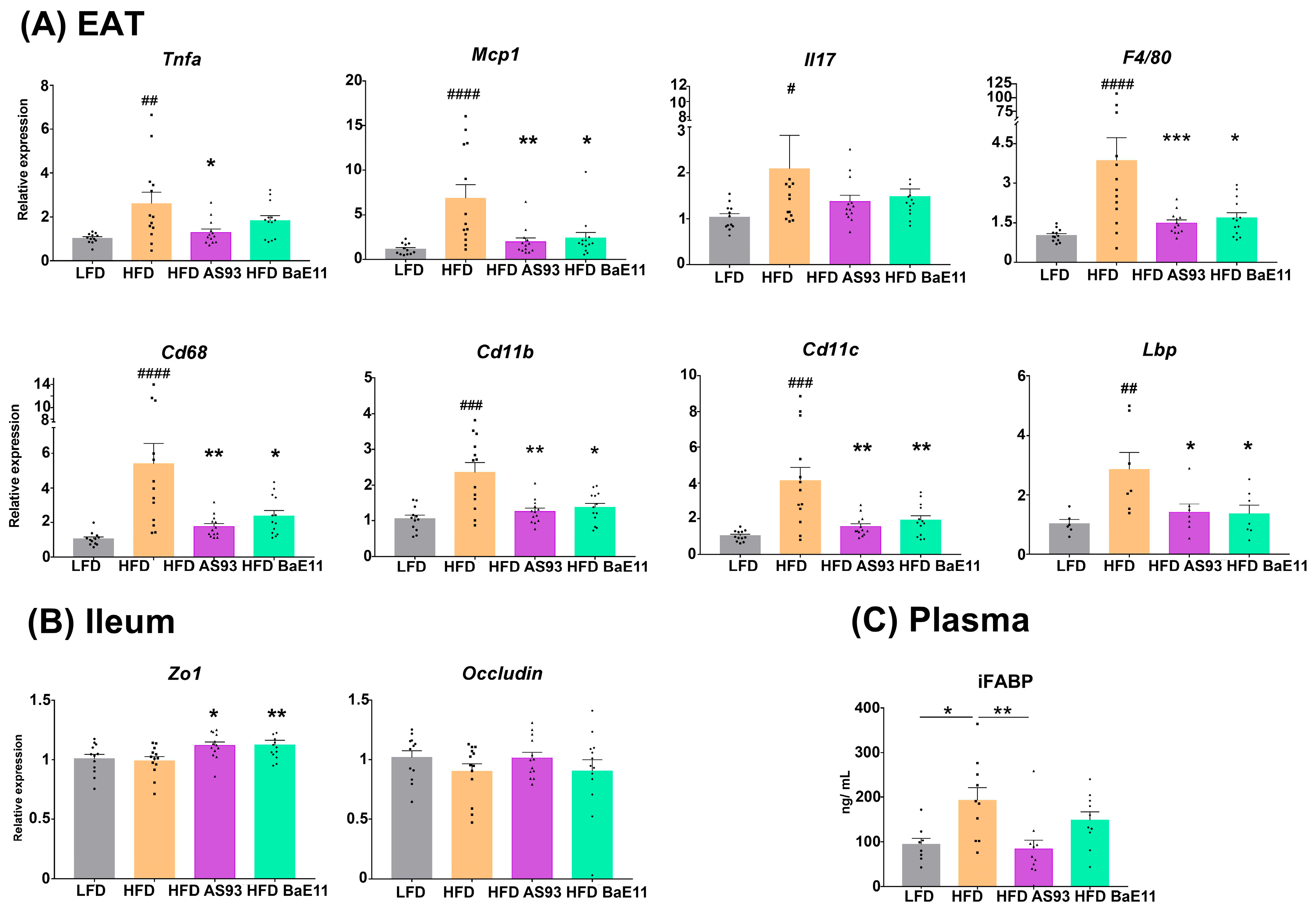
3.3. Supplementation by the Two P. distasonis Strains Decreases Adipose Tissue Inflammation and Improves Markers of Gut Permeability
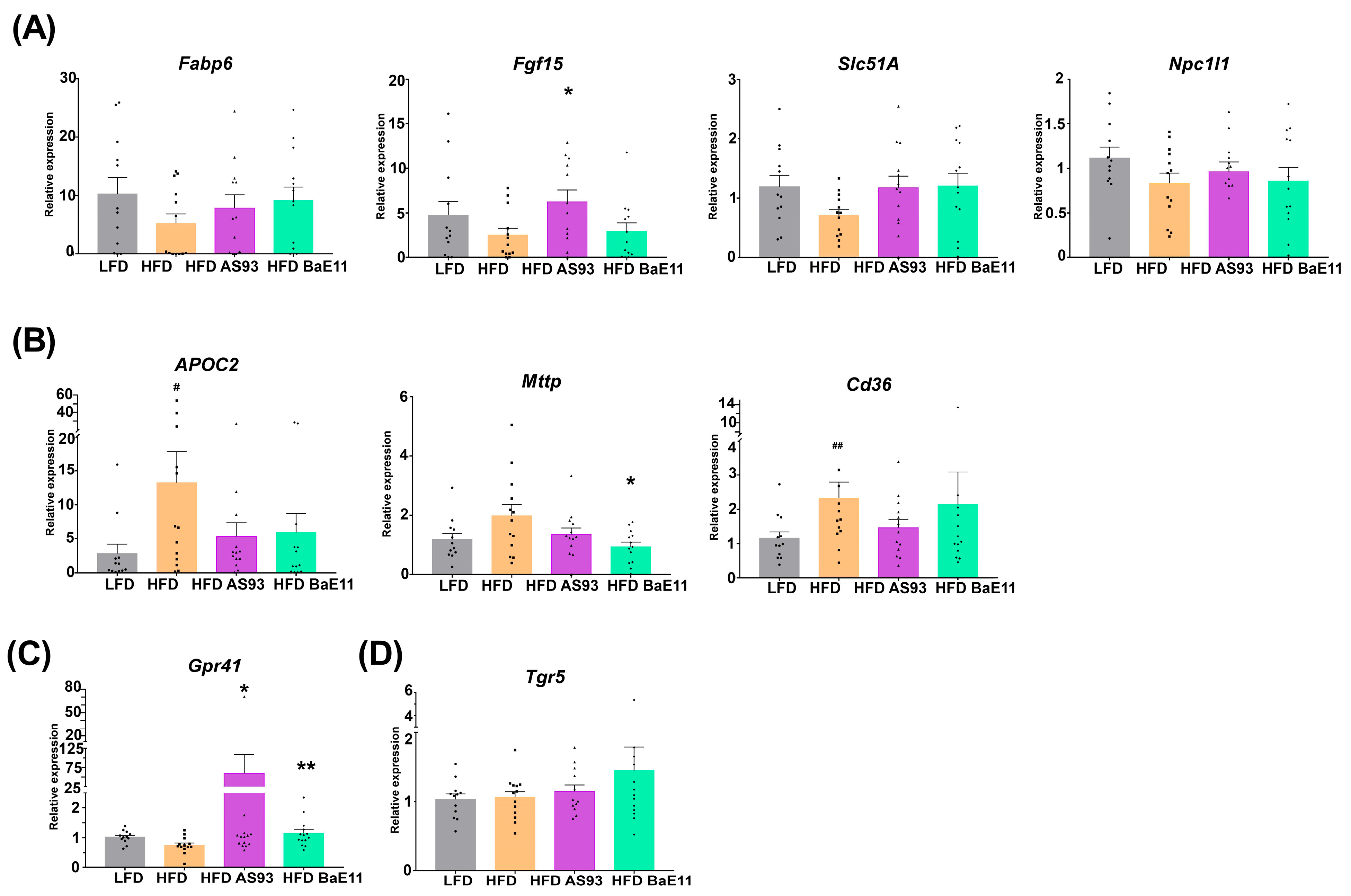
3.4. P. distasonis Strains Restore Cholesterol and Bile Acids Metabolism and Limit Lipid Metabolism in the Gut
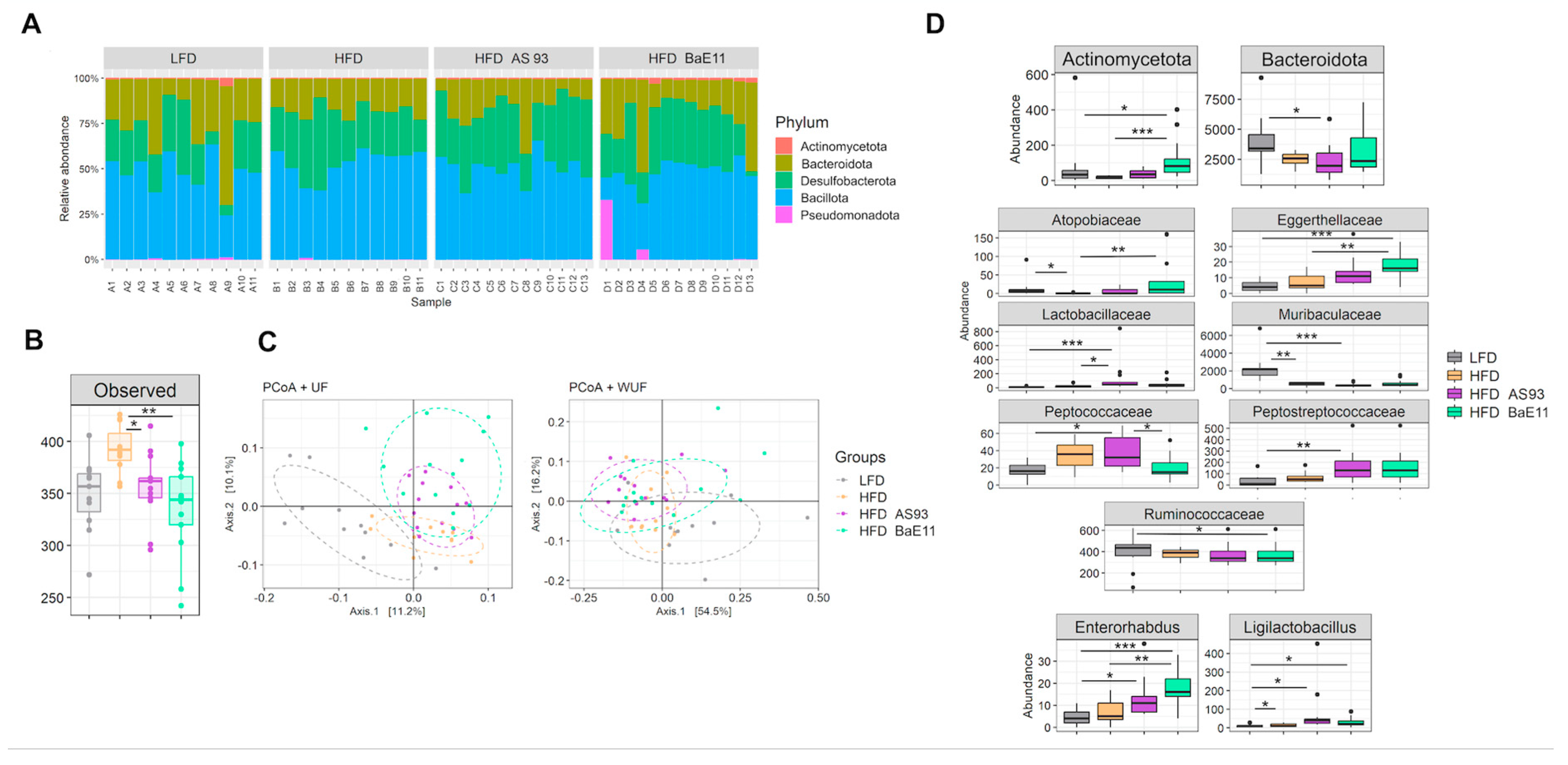
3.5. P. distasonis Strains Differentially Modulate the Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malard, F.; Doré, J.; Gaugler, B.; Mohty, M. Introduction to Host Microbiome Symbiosis in Health and Disease. Mucosal. Immunol. 2021, 14, 547–554. [Google Scholar] [CrossRef]
- Koh, A.; Bäckhed, F. From Association to Causality: The Role of the Gut Microbiota and Its Functional Products on Host Metabolism. Mol. Cell 2020, 78, 584–596. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An Obesity-Associated Gut Microbiome with Increased Capacity for Energy Harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut Microbiota from Twins Discordant for Obesity Modulate Metabolism in Mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Hong, J.; Xu, X.; Feng, Q.; Zhang, D.; Gu, Y.; Shi, J.; Zhao, S.; Liu, W.; Wang, X.; et al. Gut Microbiome and Serum Metabolome Alterations in Obesity and after Weight-Loss Intervention. Nat. Med. 2017, 23, 859–868. [Google Scholar] [CrossRef]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic Endotoxemia Initiates Obesity and Insulin Resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef]
- Kawai, T.; Autieri, M.V.; Scalia, R. Adipose Tissue Inflammation and Metabolic Dysfunction in Obesity. Am. J. Physiol. Cell Physiol. 2021, 320, C375–C391. [Google Scholar] [CrossRef] [PubMed]
- Frost, G.; Sleeth, M.L.; Sahuri-Arisoylu, M.; Lizarbe, B.; Cerdan, S.; Brody, L.; Anastasovska, J.; Ghourab, S.; Hankir, M.; Zhang, S.; et al. The Short-Chain Fatty Acid Acetate Reduces Appetite via a Central Homeostatic Mechanism. Nat. Commun. 2014, 5, 3611. [Google Scholar] [CrossRef]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E. Short-Chain Fatty Acids in Control of Body Weight and Insulin Sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577–591. [Google Scholar] [CrossRef]
- Sanz, Y.; Rastmanesh, R.; Agostoni, C.; Agostonic, C. Understanding the Role of Gut Microbes and Probiotics in Obesity: How Far Are We? Pharmacol. Res. 2013, 69, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Alard, J.; Lehrter, V.; Rhimi, M.; Mangin, I.; Peucelle, V.; Abraham, A.-L.; Mariadassou, M.; Maguin, E.; Waligora-Dupriet, A.-J.; Pot, B.; et al. Beneficial Metabolic Effects of Selected Probiotics on Diet-Induced Obesity and Insulin Resistance in Mice Are Associated with Improvement of Dysbiotic Gut Microbiota. Environ. Microbiol. 2016, 18, 1484–1497. [Google Scholar] [CrossRef]
- Alard, J.; Cudennec, B.; Boutillier, D.; Peucelle, V.; Descat, A.; Decoin, R.; Kuylle, S.; Jablaoui, A.; Rhimi, M.; Wolowczuk, I.; et al. Multiple Selection Criteria for Probiotic Strains with High Potential for Obesity Management. Nutrients 2021, 13, 713. [Google Scholar] [CrossRef] [PubMed]
- Aoki, R.; Kamikado, K.; Suda, W.; Takii, H.; Mikami, Y.; Suganuma, N.; Hattori, M.; Koga, Y. A Proliferative Probiotic Bifidobacterium Strain in the Gut Ameliorates Progression of Metabolic Disorders via Microbiota Modulation and Acetate Elevation. Sci. Rep. 2017, 7, 43522. [Google Scholar] [CrossRef]
- Balcells, F.; Martínez Monteros, M.J.; Gómez, A.L.; Cazorla, S.I.; Perdigón, G.; Maldonado-Galdeano, C. Probiotic Consumption Boosts Thymus in Obesity and Senescence Mouse Models. Nutrients 2022, 14, 616. [Google Scholar] [CrossRef]
- Wang, J.; Tang, H.; Zhang, C.; Zhao, Y.; Derrien, M.; Rocher, E.; van-Hylckama Vlieg, J.E.T.; Strissel, K.; Zhao, L.; Obin, M.; et al. Modulation of Gut Microbiota during Probiotic-Mediated Attenuation of Metabolic Syndrome in High Fat Diet-Fed Mice. ISME J. 2015, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yadav, H.; Lee, J.-H.; Lloyd, J.; Walter, P.; Rane, S.G. Beneficial Metabolic Effects of a Probiotic via Butyrate-Induced GLP-1 Hormone Secretion. J. Biol. Chem. 2013, 288, 25088–25097. [Google Scholar] [CrossRef] [PubMed]
- Koutnikova, H.; Genser, B.; Monteiro-Sepulveda, M.; Faurie, J.-M.; Rizkalla, S.; Schrezenmeir, J.; Clément, K. Impact of Bacterial Probiotics on Obesity, Diabetes and Non-Alcoholic Fatty Liver Disease Related Variables: A Systematic Review and Meta-Analysis of Randomised Controlled Trials. BMJ Open 2019, 9, e017995. [Google Scholar] [CrossRef] [PubMed]
- Crovesy, L.; Ostrowski, M.; Ferreira, D.M.T.P.; Rosado, E.L.; Soares-Mota, M. Effect of Lactobacillus on Body Weight and Body Fat in Overweight Subjects: A Systematic Review of Randomized Controlled Clinical Trials. Int. J. Obes. 2017, 41, 1607–1614. [Google Scholar] [CrossRef]
- Gauffin Cano, P.; Santacruz, A.; Moya, Á.; Sanz, Y. Bacteroides Uniformis CECT 7771 Ameliorates Metabolic and Immunological Dysfunction in Mice with High-Fat-Diet Induced Obesity. PLoS ONE 2012, 7, e41079. [Google Scholar] [CrossRef]
- Yoshida, N.; Yamashita, T.; Osone, T.; Hosooka, T.; Shinohara, M.; Kitahama, S.; Sasaki, K.; Sasaki, D.; Yoneshiro, T.; Suzuki, T.; et al. Bacteroides Spp. Promotes Branched-Chain Amino Acid Catabolism in Brown Fat and Inhibits Obesity. iScience 2021, 24, 103342. [Google Scholar] [CrossRef]
- Munukka, E.; Rintala, A.; Toivonen, R.; Nylund, M.; Yang, B.; Takanen, A.; Hänninen, A.; Vuopio, J.; Huovinen, P.; Jalkanen, S.; et al. Faecalibacterium Prausnitzii Treatment Improves Hepatic Health and Reduces Adipose Tissue Inflammation in High-Fat Fed Mice. ISME J. 2017, 11, 1667–1679. [Google Scholar] [CrossRef]
- Udayappan, S.; Manneras-Holm, L.; Chaplin-Scott, A.; Belzer, C.; Herrema, H.; Dallinga-Thie, G.M.; Duncan, S.H.; Stroes, E.S.G.; Groen, A.K.; Flint, H.J.; et al. Oral Treatment with Eubacterium Hallii Improves Insulin Sensitivity in Db/Db Mice. NPJ Biofilms Microbiomes 2016, 2, 16009. [Google Scholar] [CrossRef]
- Lucas, N.; Legrand, R.; Deroissart, C.; Dominique, M.; Azhar, S.; Le Solliec, M.-A.; Léon, F.; do Rego, J.-C.; Déchelotte, P.; Fetissov, S.O.; et al. Hafnia Alvei HA4597 Strain Reduces Food Intake and Body Weight Gain and Improves Body Composition, Glucose, and Lipid Metabolism in a Mouse Model of Hyperphagic Obesity. Microorganisms 2019, 8, E35. [Google Scholar] [CrossRef] [PubMed]
- Paquet, J.-C.; Claus, S.P.; Cordaillat-Simmons, M.; Mazier, W.; Rawadi, G.; Rinaldi, L.; Elustondo, F.; Rouanet, A. Entering First-in-Human Clinical Study with a Single-Strain Live Biotherapeutic Product: Input and Feedback Gained from the EMA and the FDA. Front. Med. 2021, 8, 716266. [Google Scholar] [CrossRef] [PubMed]
- YSOPIA Bioscience. Xla1 Christensenella Minuta, Phase I, Randomized, Partially Placebo-Controlled Double-Blind Protocol, Evaluating Safety, Tolerability and Impact on the Gut Microbiota in Healthy Volunteers, Overweight and Obese Adults; clinicaltrials.gov; YSOPIA Bioscience: Bordeaux, France, 2021. [Google Scholar]
- Gilijamse, P.W.; Hartstra, A.V.; Levin, E.; Wortelboer, K.; Serlie, M.J.; Ackermans, M.T.; Herrema, H.; Nederveen, A.J.; Imangaliyev, S.; Aalvink, S.; et al. Treatment with Anaerobutyricum Soehngenii: A Pilot Study of Safety and Dose-Response Effects on Glucose Metabolism in Human Subjects with Metabolic Syndrome. NPJ Biofilms Microbiomes 2020, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Atlantia Food Clinical Trials. A Double-Blind, Randomized, Placebo-Controlled Trial to Assess the Efficacy and Safety of 12-Weeks Supplementation of Eubacterium Hallii on Insulin Sensitivity and Markers of Glycaemic Control in Healthy Hyperglycaemic Males; clinicaltrials.gov; Atlantia Clinical Trials: Cork, Ireland, 2020. [Google Scholar]
- Déchelotte, P.; Breton, J.; Trotin-Picolo, C.; Grube, B.; Erlenbeck, C.; Bothe, G.; Fetissov, S.O.; Lambert, G. The Probiotic Strain H. Alvei HA4597® Improves Weight Loss in Overweight Subjects under Moderate Hypocaloric Diet: A Proof-of-Concept, Multicenter Randomized, Double-Blind Placebo-Controlled Study. Nutrients 2021, 13, 1902. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia Muciniphila in Overweight and Obese Human Volunteers: A Proof-of-Concept Exploratory Study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Depommier, C.; Vitale, R.M.; Iannotti, F.A.; Silvestri, C.; Flamand, N.; Druart, C.; Everard, A.; Pelicaen, R.; Maiter, D.; Thissen, J.-P.; et al. Beneficial Effects of Akkermansia Muciniphila Are Not Associated with Major Changes in the Circulating Endocannabinoidome but Linked to Higher Mono-Palmitoyl-Glycerol Levels as New PPARα Agonists. Cells 2021, 10, 185. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Nutrition, Novel Foods and Food Allergens (NDA); Turck, D.; Bohn, T.; Castenmiller, J.; De Henauw, S.; Hirsch-Ernst, K.I.; Maciuk, A.; Mangelsdorf, I.; McArdle, H.J.; Naska, A.; et al. Safety of Pasteurised Akkermansia Muciniphila as a Novel Food Pursuant to Regulation (EU) 2015/2283. EFSA J. 2021, 19, e06780. [Google Scholar] [CrossRef] [PubMed]
- López-Moreno, A.; Acuña, I.; Torres-Sánchez, A.; Ruiz-Moreno, Á.; Cerk, K.; Rivas, A.; Suárez, A.; Monteoliva-Sánchez, M.; Aguilera, M. Next Generation Probiotics for Neutralizing Obesogenic Effects: Taxa Culturing Searching Strategies. Nutrients 2021, 13, 1617. [Google Scholar] [CrossRef] [PubMed]
- Cuffaro, B.; Assohoun, A.L.W.; Boutillier, D.; Peucelle, V.; Desramaut, J.; Boudebbouze, S.; Croyal, M.; Waligora-Dupriet, A.-J.; Rhimi, M.; Grangette, C.; et al. Identification of New Potential Biotherapeutics from Human Gut Microbiota-Derived Bacteria. Microorganisms 2021, 9, 565. [Google Scholar] [CrossRef]
- Cuffaro, B.; Assohoun, A.L.W.; Boutillier, D.; Súkeníková, L.; Desramaut, J.; Boudebbouze, S.; Salomé-Desnoulez, S.; Hrdý, J.; Waligora-Dupriet, A.-J.; Maguin, E.; et al. In Vitro Characterization of Gut Microbiota-Derived Commensal Strains: Selection of Parabacteroides Distasonis Strains Alleviating TNBS-Induced Colitis in Mice. Cells 2020, 9, 2104. [Google Scholar] [CrossRef] [PubMed]
- Klen, J.; Dolžan, V. Glucagon-like Peptide-1 Receptor Agonists in the Management of Type 2 Diabetes Mellitus and Obesity: The Impact of Pharmacological Properties and Genetic Factors. Int. J. Mol. Sci. 2022, 23, 3451. [Google Scholar] [CrossRef] [PubMed]
- Laurans, L.; Venteclef, N.; Haddad, Y.; Chajadine, M.; Alzaid, F.; Metghalchi, S.; Sovran, B.; Denis, R.G.P.; Dairou, J.; Cardellini, M.; et al. Genetic Deficiency of Indoleamine 2,3-Dioxygenase Promotes Gut Microbiota-Mediated Metabolic Health. Nat. Med. 2018, 24, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Godon, J.J.; Zumstein, E.; Dabert, P.; Habouzit, F.; Moletta, R. Molecular Microbial Diversity of an Anaerobic Digestor as Determined by Small-Subunit RDNA Sequence Analysis. Appl. Environ. Microbiol. 1997, 63, 2802–2813. [Google Scholar] [CrossRef]
- Escudié, F.; Auer, L.; Bernard, M.; Mariadassou, M.; Cauquil, L.; Vidal, K.; Maman, S.; Hernandez-Raquet, G.; Combes, S.; Pascal, G. FROGS: Find, Rapidly, OTUs with Galaxy Solution. Bioinformatics 2018, 34, 1287–1294. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic. Acids. Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Hochberg, Y.; Benjamini, Y. More Powerful Procedures for Multiple Significance Testing. Stat. Med. 1990, 9, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Kuhre, R.E.; Albrechtsen, N.J.W.; Deacon, C.F.; Balk-Møller, E.; Rehfeld, J.F.; Reimann, F.; Gribble, F.M.; Holst, J.J. Peptide Production and Secretion in GLUTag, NCI-H716, and STC-1 Cells: A Comparison to Native L-Cells. J. Mol. Endocrinol. 2016, 56, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Rindi, G.; Grant, S.G.; Yiangou, Y.; Ghatei, M.A.; Bloom, S.R.; Bautch, V.L.; Solcia, E.; Polak, J.M. Development of Neuroendocrine Tumors in the Gastrointestinal Tract of Transgenic Mice. Heterogeneity of Hormone Expression. Am. J. Pathol. 1990, 136, 1349–1363. [Google Scholar] [PubMed]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W. Obesity Is Associated with Macrophage Accumulation in Adipose Tissue. J. Clin Investig. 2003, 112, 1796–1808. [Google Scholar] [CrossRef] [PubMed]
- Zatterale, F.; Longo, M.; Naderi, J.; Raciti, G.A.; Desiderio, A.; Miele, C.; Beguinot, F. Chronic Adipose Tissue Inflammation Linking Obesity to Insulin Resistance and Type 2 Diabetes. Front. Physiol. 2020, 10, 1607. [Google Scholar] [CrossRef] [PubMed]
- Hersoug, L.-G.; Møller, P.; Loft, S. Role of Microbiota-Derived Lipopolysaccharide in Adipose Tissue Inflammation, Adipocyte Size and Pyroptosis during Obesity. Nutr. Res. Rev. 2018, 31, 153–163. [Google Scholar] [CrossRef]
- Fang, S.; Suh, J.M.; Reilly, S.M.; Yu, E.; Osborn, O.; Lackey, D.; Yoshihara, E.; Perino, A.; Jacinto, S.; Lukasheva, Y.; et al. Intestinal FXR Agonism Promotes Adipose Tissue Browning and Reduces Obesity and Insulin Resistance. Nat. Med. 2015, 21, 159–165. [Google Scholar] [CrossRef]
- Samuel, B.S.; Shaito, A.; Motoike, T.; Rey, F.E.; Backhed, F.; Manchester, J.K.; Hammer, R.E.; Williams, S.C.; Crowley, J.; Yanagisawa, M.; et al. Effects of the Gut Microbiota on Host Adiposity Are Modulated by the Short-Chain Fatty-Acid Binding G Protein-Coupled Receptor, Gpr41. Proc. Natl. Acad. Sci. USA 2008, 105, 16767–16772. [Google Scholar] [CrossRef]
- Lu, Y.; Fan, C.; Li, P.; Lu, Y.; Chang, X.; Qi, K. Short Chain Fatty Acids Prevent High-Fat-Diet-Induced Obesity in Mice by Regulating G Protein-Coupled Receptors and Gut Microbiota. Sci. Rep. 2016, 6, 37589. [Google Scholar] [CrossRef]
- Guo, C.; Chen, W.-D.; Wang, Y.-D. TGR5, Not Only a Metabolic Regulator. Front. Physiol. 2016, 7, 646. [Google Scholar] [CrossRef]
- Suez, J.; Zmora, N.; Elinav, E. Probiotics in the Next-Generation Sequencing Era. Gut. Microbes. 2020, 11, 77–93. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Gordon, J.I. The Core Gut Microbiome, Energy Balance and Obesity. J. Physiol. 2009, 587, 4153–4158. [Google Scholar] [CrossRef] [PubMed]
- Ezeji, J.C.; Sarikonda, D.K.; Hopperton, A.; Erkkila, H.L.; Cohen, D.E.; Martinez, S.P.; Cominelli, F.; Kuwahara, T.; Dichosa, A.E.K.; Good, C.E.; et al. Parabacteroides Distasonis: Intriguing Aerotolerant Gut Anaerobe with Emerging Antimicrobial Resistance and Pathogenic and Probiotic Roles in Human Health. Gut. Microbes. 2021, 13, 1922241. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Li, D.; He, Y.; Li, Y.; Yang, Z.; Zhao, X.; Liu, Y.; Wang, Y.; Sun, J.; Feng, X.; et al. Discrepant Gut Microbiota Markers for the Classification of Obesity-Related Metabolic Abnormalities. Sci. Rep. 2019, 9, 13424. [Google Scholar] [CrossRef] [PubMed]
- Verdam, F.J.; Fuentes, S.; de Jonge, C.; Zoetendal, E.G.; Erbil, R.; Greve, J.W.; Buurman, W.A.; de Vos, W.M.; Rensen, S.S. Human Intestinal Microbiota Composition Is Associated with Local and Systemic Inflammation in Obesity. Obesity 2013, 21, E607–E615. [Google Scholar] [CrossRef]
- Kuang, Y.-S.; Lu, J.-H.; Li, S.-H.; Li, J.-H.; Yuan, M.-Y.; He, J.-R.; Chen, N.-N.; Xiao, W.-Q.; Shen, S.-Y.; Qiu, L.; et al. Connections between the Human Gut Microbiome and Gestational Diabetes Mellitus. Gigascience 2017, 6, gix058. [Google Scholar] [CrossRef] [PubMed]
- Hasain, Z.; Mokhtar, N.M.; Kamaruddin, N.A.; Mohamed Ismail, N.A.; Razalli, N.H.; Gnanou, J.V.; Raja Ali, R.A. Gut Microbiota and Gestational Diabetes Mellitus: A Review of Host-Gut Microbiota Interactions and Their Therapeutic Potential. Front. Cell Infect. Microbiol. 2020, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, J.; Liu, H.; Tang, Y.; Zhan, Q.; Lai, W.; Ao, L.; Meng, X.; Ren, H.; Xu, D.; et al. The Intestinal Microbiota Associated with Cardiac Valve Calcification Differs from That of Coronary Artery Disease. Atherosclerosis 2019, 284, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Bank, N.C.; Singh, V.; Rodriguez-Palacios, A. Classification of Parabacteroides Distasonis and Other Bacteroidetes Using O- Antigen Virulence Gene: RfbA-Typing and Hypothesis for Pathogenic vs. Probiotic Strain Differentiation. Gut Microbes 2022, 14, 1997293. [Google Scholar] [CrossRef]
- Haro, C.; Montes-Borrego, M.; Rangel-Zúñiga, O.A.; Alcalá-Díaz, J.F.; Gómez-Delgado, F.; Pérez-Martínez, P.; Delgado-Lista, J.; Quintana-Navarro, G.M.; Tinahones, F.J.; Landa, B.B.; et al. Two Healthy Diets Modulate Gut Microbial Community Improving Insulin Sensitivity in a Human Obese Population. J. Clin. Endocrinol. Metab. 2016, 101, 233–242. [Google Scholar] [CrossRef]
- Palacios, T.; Vitetta, L.; Coulson, S.; Madigan, C.D.; Lam, Y.Y.; Manuel, R.; Briskey, D.; Hendy, C.; Kim, J.-N.; Ishoey, T.; et al. Targeting the Intestinal Microbiota to Prevent Type 2 Diabetes and Enhance the Effect of Metformin on Glycaemia: A Randomised Controlled Pilot Study. Nutrients 2020, 12, 2041. [Google Scholar] [CrossRef]
- Elbere, I.; Silamikelis, I.; Dindune, I.I.; Kalnina, I.; Ustinova, M.; Zaharenko, L.; Silamikele, L.; Rovite, V.; Gudra, D.; Konrade, I.; et al. Baseline Gut Microbiome Composition Predicts Metformin Therapy Short-Term Efficacy in Newly Diagnosed Type 2 Diabetes Patients. PLoS ONE 2020, 15, e0241338. [Google Scholar] [CrossRef]
- Ranaivo, H.; Thirion, F.; Béra-Maillet, C.; Guilly, S.; Simon, C.; Sothier, M.; Van Den Berghe, L.; Feugier-Favier, N.; Lambert-Porcheron, S.; Dussous, I.; et al. Increasing the Diversity of Dietary Fibers in a Daily-Consumed Bread Modifies Gut Microbiota and Metabolic Profile in Subjects at Cardiometabolic Risk. Gut. Microbes 2022, 14, 2044722. [Google Scholar] [CrossRef] [PubMed]
- Thirion, F.; Da Silva, K.; Plaza Oñate, F.; Alvarez, A.-S.; Thabuis, C.; Pons, N.; Berland, M.; Le Chatelier, E.; Galleron, N.; Levenez, F.; et al. Diet Supplementation with NUTRIOSE, a Resistant Dextrin, Increases the Abundance of Parabacteroides Distasonis in the Human Gut. Mol. Nutr. Food Res. 2022, 66, e2101091. [Google Scholar] [CrossRef]
- Włodarczyk, M.; Śliżewska, K. Efficiency of Resistant Starch and Dextrins as Prebiotics: A Review of the Existing Evidence and Clinical Trials. Nutrients 2021, 13, 3808. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Chen, B.; Luo, L.; Zhang, X.; Dai, X.; Gong, S. Chemical Compositions and Antioxidant Activities of Water Extracts of Chinese Propolis. J. Agric. Food Chem. 2011, 59, 12610–12616. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Xu, J.; Li, G.; Liu, T.; Guo, X.; Wang, H.; Luo, L. Ethanol Extract of Propolis Prevents High-Fat Diet-Induced Insulin Resistance and Obesity in Association with Modulation of Gut Microbiota in Mice. Food Res. Int. 2020, 130, 108939. [Google Scholar] [CrossRef]
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides Distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019, 26, 222–235.e5. [Google Scholar] [CrossRef]
- Wu, T.-R.; Lin, C.-S.; Chang, C.-J.; Lin, T.-L.; Martel, J.; Ko, Y.-F.; Ojcius, D.M.; Lu, C.-C.; Young, J.D.; Lai, H.-C. Gut Commensal Parabacteroides Goldsteinii Plays a Predominant Role in the Anti-Obesity Effects of Polysaccharides Isolated from Hirsutella Sinensis. Gut 2019, 68, 248–262. [Google Scholar] [CrossRef]
- Chamarande, J.; Cunat, L.; Pavlov, N.; Alauzet, C.; Cailliez-Grimal, C. Parabacteroides Distasonis Properties Linked to the Selection of New Biotherapeutics. Nutrients 2022, 14, 4176. [Google Scholar] [CrossRef]
- Müller, T.D.; Finan, B.; Bloom, S.R.; D’Alessio, D.; Drucker, D.J.; Flatt, P.R.; Fritsche, A.; Gribble, F.; Grill, H.J.; Habener, J.F.; et al. Glucagon-like Peptide 1 (GLP-1). Mol. Metab. 2019, 30, 72–130. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Bao, L.; Zhou, N.; Zhang, J.; Liao, M.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, J.; Wang, L.; et al. Structural Modification of Natural Product Ganomycin I Leading to Discovery of a α-Glucosidase and HMG-CoA Reductase Dual Inhibitor Improving Obesity and Metabolic Dysfunction in Vivo. J. Med. Chem. 2018, 61, 3609–3625. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuffaro, B.; Boutillier, D.; Desramaut, J.; Jablaoui, A.; Werkmeister, E.; Trottein, F.; Waligora-Dupriet, A.-J.; Rhimi, M.; Maguin, E.; Grangette, C. Characterization of Two Parabacteroides distasonis Candidate Strains as New Live Biotherapeutics against Obesity. Cells 2023, 12, 1260. https://doi.org/10.3390/cells12091260
Cuffaro B, Boutillier D, Desramaut J, Jablaoui A, Werkmeister E, Trottein F, Waligora-Dupriet A-J, Rhimi M, Maguin E, Grangette C. Characterization of Two Parabacteroides distasonis Candidate Strains as New Live Biotherapeutics against Obesity. Cells. 2023; 12(9):1260. https://doi.org/10.3390/cells12091260
Chicago/Turabian StyleCuffaro, Bernardo, Denise Boutillier, Jérémy Desramaut, Amin Jablaoui, Elisabeth Werkmeister, François Trottein, Anne-Judith Waligora-Dupriet, Moez Rhimi, Emmanuelle Maguin, and Corinne Grangette. 2023. "Characterization of Two Parabacteroides distasonis Candidate Strains as New Live Biotherapeutics against Obesity" Cells 12, no. 9: 1260. https://doi.org/10.3390/cells12091260




