Expression of Adipose Tissue Extracellular Matrix-Related Genes Predicts Weight Loss after Bariatric Surgery
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Anthropometric and Clinical Assessments
2.3. Tissue Processing and Histology
2.4. RNA Extraction and Quantitative Real-Time PCR
2.5. Statistics
3. Results
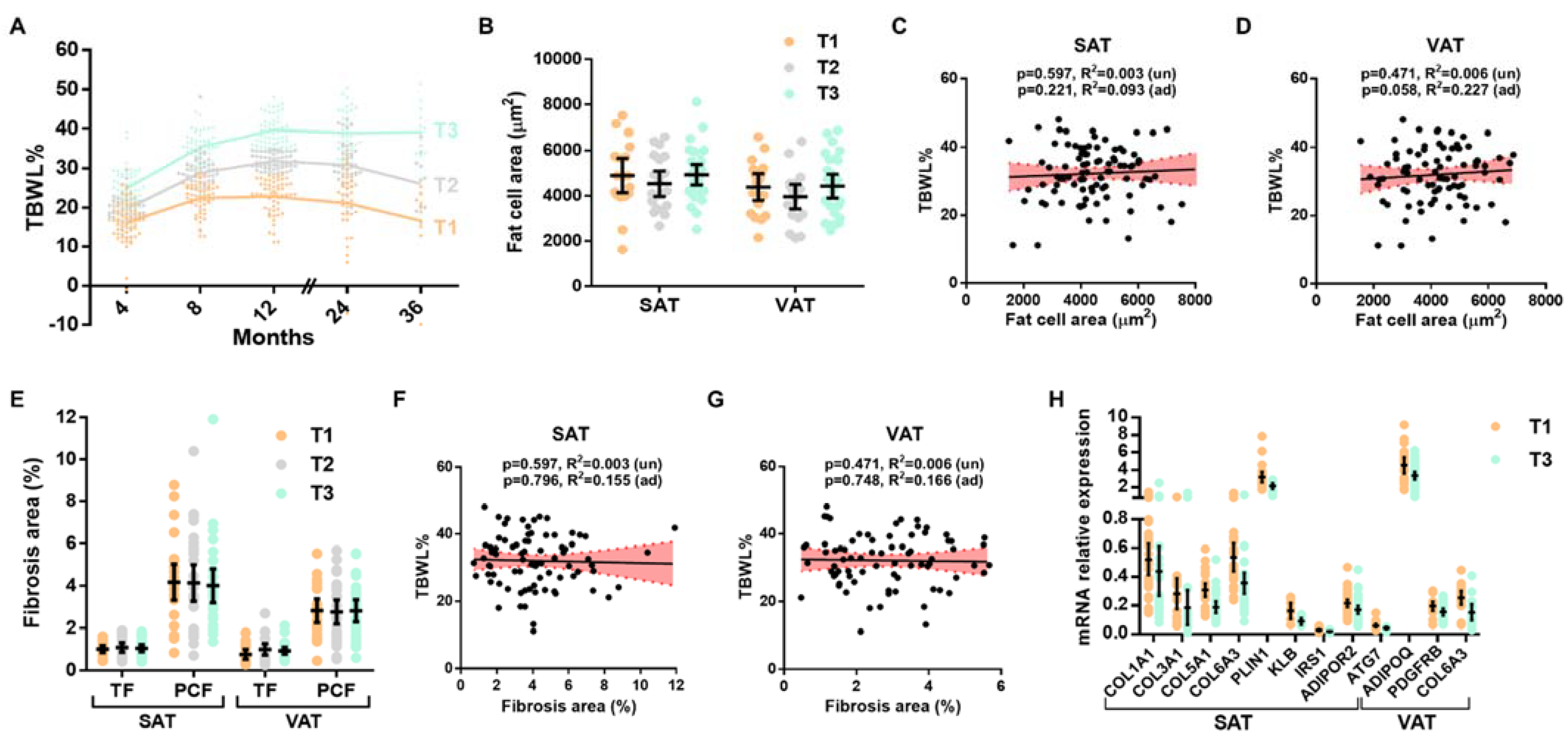
3.1. White Adipose Tissue Parameters and Weight Loss after BS
3.2. Expression of ECM Components and Associations with Weight Loss
3.3. Predictive Value of ECM Components on Insufficient Weight Loss
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sjöström, L. Review of the Key Results from the Swedish Obese Subjects (SOS) Trial—A Prospective Controlled Intervention Study of Bariatric Surgery. J. Intern. Med. 2013, 273, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Mingrone, G.; Panunzi, S.; De Gaetano, A.; Guidone, C.; Iaconelli, A.; Capristo, E.; Chamseddine, G.; Bornstein, S.R.; Rubino, F. Metabolic Surgery versus Conventional Medical Therapy in Patients with Type 2 Diabetes: 10-Year Follow-up of an Open-Label, Single-Centre, Randomised Controlled Trial. Lancet 2021, 397, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Courcoulas, A.P.; Christian, N.J.; Belle, S.H.; Berk, P.D.; Flum, D.R.; Garcia, L.; Horlick, M.; Kalarchian, M.A.; King, W.C.; Mitchell, J.E.; et al. Weight Change and Health Outcomes at 3 Years after Bariatric Surgery among Individuals with Severe Obesity. JAMA 2013, 310, 2416–2425. [Google Scholar] [CrossRef] [PubMed]
- de Hollanda, A.; Ruiz, T.; Jiménez, A.; Flores, L.; Lacy, A.; Vidal, J. Patterns of Weight Loss Response Following Gastric Bypass and Sleeve Gastrectomy. Obes. Surg. 2015, 25, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Velapati, S.R.; Shah, M.; Kuchkuntla, A.R.; Abu-dayyeh, B.; Grothe, K.; Hurt, R.T.; Mundi, M.S. Weight Regain After Bariatric Surgery: Prevalence, Etiology, and Treatment. Curr. Nutr. Rep. 2018, 7, 329–334. [Google Scholar] [CrossRef]
- Osorio-Conles, Ó.; Vidal, J.; de Hollanda, A. Impact of Bariatric Surgery on Adipose Tissue Biology. J. Clin. Med. 2021, 10, 5516. [Google Scholar] [CrossRef]
- Debari, M.K.; Abbott, R.D. Adipose Tissue Fibrosis: Mechanisms, Models, and Importance. Int. J. Mol. Sci. 2020, 21, 6030. [Google Scholar] [CrossRef]
- Adami, G.F.; Carbone, F.; Montecucco, F.; Camerini, G.; Cordera, R. Adipose Tissue Composition in Obesity and After Bariatric Surgery. Obes. Surg. 2019, 29, 3030–3038. [Google Scholar] [CrossRef]
- van Baak, M.A.; Mariman, E.C.M. Mechanisms of Weight Regain after Weight Loss—The Role of Adipose Tissue. Nat. Rev. Endocrinol. 2019, 15, 274–287. [Google Scholar] [CrossRef]
- Pellegrinelli, V.; Carobbio, S.; Vidal-Puig, A. Adipose Tissue Plasticity: How Fat Depots Respond Differently to Pathophysiological Cues. Diabetologia 2016, 59, 1075–1088. [Google Scholar] [CrossRef]
- Fried, M.; Yumuk, V.; Oppert, J.M.; Scopinaro, N.; Torres, A.; Weiner, R.; Yashkov, Y.; Frühbeck, G. Interdisciplinary European Guidelines on Metabolic and Bariatric Surgery. Obes. Surg. 2014, 24, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Morínigo, R.; Vidal, J.; Lacy, A.M.; Delgado, S.; Casamitjana, R.; Gomis, R. Circulating Peptide YY, Weight Loss, and Glucose Homeostasis after Gastric Bypass Surgery in Morbidly Obese Subjects. Ann. Surg. 2008, 247, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Vidal, J.; Ibarzabal, A.; Romero, F.; Delgado, S.; Momblán, D.; Flores, L.; Lacy, A. Type 2 Diabetes Mellitus and the Metabolic Syndrome Following Sleeve Gastrectomy in Severely Obese Subjects. Obes. Surg. 2008, 18, 1077–1082. [Google Scholar] [CrossRef]
- Jiménez, A.; Pané, A.; Ibarzábal, A.; de Hollanda, A.; Tundidor, D.; Balibrea, J.M.; Andreu, A.; Molero, J.; Cañizares, S.; Obach, A.; et al. Weight-Loss Thresholds after Bariatric Surgery and Cardiovascular Outcomes: More Is Better. Int. J. Obes. 2022, 46, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Divoux, A.; Tordjman, J.; Lacasa, D.; Veyrie, N.; Hugol, D.; Aissat, A.; Basdevant, A.; Guerre-Millo, M.; Poitou, C.; Zucker, J.D.; et al. Fibrosis in Human Adipose Tissue: Composition, Distribution, and Link with Lipid Metabolism and Fat Mass Loss. Diabetes 2010, 59, 2817–2825. [Google Scholar] [CrossRef]
- Hanley, J.A.; McNeil, B.J. A Method of Comparing the Areas under Receiver Operating Characteristic Curves Derived from the Same Cases. Radiology 1983, 148, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Muir, L.A.; Baker, N.A.; Washabaugh, A.R.; Neeley, C.K.; Flesher, C.G.; DelProposto, J.B.; Geletka, L.M.; Ghaferi, A.A.; Finks, J.F.; Singer, K.; et al. Adipocyte Hypertrophy-Hyperplasia Balance Contributes to Weight Loss after Bariatric Surgery. Adipocyte 2017, 6, 134–140. [Google Scholar] [CrossRef]
- Abdennour, M.; Reggio, S.; Le Naour, G.; Liu, Y.; Poitou, C.; Aron-Wisnewsky, J.; Charlotte, F.; Bouillot, J.L.; Torcivia, A.; Sasso, M.; et al. Association of Adipose Tissue and Liver Fibrosis with Tissue Stiffness in Morbid Obesity: Links with Diabetes and BMI Loss after Gastric Bypass. J. Clin. Endocrinol. Metab. 2014, 99, 898–907. [Google Scholar] [CrossRef]
- Lassen, P.B.; Charlotte, F.; Liu, Y.; Bedossa, P.; Le Naour, G.; Tordjman, J.; Poitou, C.; Bouillot, J.L.; Genser, L.; Zucker, J.D.; et al. The FAT Score, a Fibrosis Score of Adipose Tissue: Predicting Weight-Loss Outcome After Gastric Bypass. J. Clin. Endocrinol. Metab. 2017, 102, 2443–2453. [Google Scholar] [CrossRef]
- Roumans, N.J.T.; Vink, R.G.; Bouwman, F.G.; Fazelzadeh, P.; Van Baak, M.A.; Mariman, E.C.M. Weight Loss-Induced Cellular Stress in Subcutaneous Adipose Tissue and the Risk for Weight Regain in Overweight and Obese Adults. Int. J. Obes. 2017, 41, 894–901. [Google Scholar] [CrossRef]
- Roumans, N.J.T.; Camps, S.G.; Renes, J.; Bouwman, F.G.; Westerterp, K.R.; Mariman, E.C.M. Weight Loss-Induced Stress in Subcutaneous Adipose Tissue Is Related to Weight Regain. Br. J. Nutr. 2016, 115, 913–920. [Google Scholar] [CrossRef]
- Márquez-Quiñones, A.; Mutch, D.M.; Debard, C.; Wang, P.; Combes, M.; Roussel, B.; Holst, C.; Martinez, J.A.; Handjieva-Darlenska, T.; Kalouskova, P.; et al. Adipose Tissue Transcriptome Reflects Variations between Subjects with Continued Weight Loss and Subjects Regaining Weight 6 Mo after Caloric Restriction Independent of Energy Intake. Am. J. Clin. Nutr. 2010, 92, 975–984. [Google Scholar] [CrossRef]
- Mutch, D.M.; Pers, T.H.; Temanni, M.R.; Pelloux, V.; Marquez-Quinõnes, A.; Holst, C.; Martinez, J.A.; Babalis, D.; Van Baak, M.A.; Handjieva-Darlenska, T.; et al. A Distinct Adipose Tissue Gene Expression Response to Caloric Restriction Predicts 6-Mo Weight Maintenance in Obese Subjects. Am. J. Clin. Nutr. 2011, 94, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- Bollepalli, S.; Kaye, S.; Heinonen, S.; Kaprio, J.; Rissanen, A.; Virtanen, K.A.; Pietiläinen, K.H.; Ollikainen, M. Subcutaneous Adipose Tissue Gene Expression and DNA Methylation Respond to Both Short- and Long-Term Weight Loss. Int. J. Obes. 2018, 42, 412–423. [Google Scholar] [CrossRef]
- Kim, K.; Perroud, B.; Espinal, G.; Kachinskas, D.; Austrheim-Smith, I.; Wolfe, B.M.; Warden, C.H. Genes and Networks Expressed in Perioperative Omental Adipose Tissue Are Correlated with Weight Loss from Roux-En-Y Gastric Bypass. Int. J. Obes. 2008, 32, 1395–1406. [Google Scholar] [CrossRef] [PubMed]
- Lasselin, J.; Magne, E.; Beau, C.; Ledaguenel, P.; Dexpert, S.; Aubert, A.; Layé, S.; Capuron, L. Adipose Inflammation in Obesity: Relationship with Circulating Levels of Inflammatory Markers and Association with Surgery-Induced Weight Loss. J. Clin. Endocrinol. Metab. 2014, 99, E53–E61. [Google Scholar] [CrossRef]
- Serrano, J.C.E.; Baena-Fustegueras, J.A.; Martin-Gari, M.; Rassendren, H.; Cassanye, A.; Naudí, A.; López-Cano, C.; Sánchez, E.; de la Fuente-Juárez, M.C.; Herrerías González, F.; et al. Adipose Tissue Protein Glycoxidation Is Associated with Weight-Loss Potential. Obesity 2019, 27, 1133–1140. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, B.A.P.; de Souza Pinhel, M.A.; Nicoletti, C.F.; de Oliveira, C.C.; Quinhoneiro, D.C.G.; Noronha, N.Y.; Fassini, P.G.; da Silva Júnior, W.A.; Junior, W.S.; Nonino, C.B. UCP2 and PLIN1 Expression Affects the Resting Metabolic Rate and Weight Loss on Obese Patients. Obes. Surg. 2017, 27, 343–348. [Google Scholar] [CrossRef]
- Yeh, J.K.; Chen, C.C.; Liu, K.H.; Peng, C.C.; Lin, T.A.; Chang, Y.S.; Wen, M.S.; Yeh, T.S.; Wang, C.Y. Serum MicroRNA Panels Predict Bariatric Surgery Outcomes. Obesity 2022, 30, 389–399. [Google Scholar] [CrossRef]
- Chabot, K.; Gauthier, M.-S.S.; Garneau, P.Y.Y.; Rabasa-Lhoret, R. Evolution of Subcutaneous Adipose Tissue Fibrosis after Bariatric Surgery. Diabetes Metab. 2017, 43, 125–133. [Google Scholar] [CrossRef]
- Liu, Y.; Aron-Wisnewsky, J.; Marcelin, G.; Genser, L.; Le Naour, G.; Torcivia, A.; Bauvois, B.; Bouchet, S.; Pelloux, V.; Sasso, M.; et al. Accumulation and Changes in Composition of Collagens in Subcutaneous Adipose Tissue After Bariatric Surgery. J. Clin. Endocrinol. Metab. 2016, 101, 293–304. [Google Scholar] [CrossRef]
- Dankel, S.N.; Fadnes, D.J.; Stavrum, A.K.; Stansberg, C.; Holdhus, R.; Hoang, T.; Veum, V.L.; Christensen, B.J.; Våge, V.; Sagen, J.V.; et al. Switch from Stress Response to Homeobox Transcription Factors in Adipose Tissue after Profound Fat Loss. PLoS ONE 2010, 5, e11033. [Google Scholar] [CrossRef]
- Mariman, E.C.M. Human Biology of Weight Maintenance after Weight Loss. J. Nutrigenet. Nutr. 2012, 5, 13–25. [Google Scholar] [CrossRef]
- You, T.; Murphy, K.M.; Lyles, M.F.; Demons, J.L.; Lenchik, L.; Nicklas, B.J. Addition of Aerobic Exercise to Dietary Weight Loss Preferentially Reduces Abdominal Adipocyte Size. Int. J. Obes. 2006, 30, 1211–1216. [Google Scholar] [CrossRef]
- Pasarica, M.; Tchoukalova, Y.D.; Heilbronn, L.K.; Fang, X.; Albu, J.B.; Kelley, D.E.; Smith, S.R.; Ravussin, E. Differential Effect of Weight Loss on Adipocyte Size Subfractions in Patients with Type 2 Diabetes. Obesity 2009, 17, 1976–1978. [Google Scholar] [CrossRef]
- Eastman, Q. Very Low Calorie Diet Makes Adipocytes “Scream”. J. Proteome Res. 2009, 8, 5408. [Google Scholar] [CrossRef]
- Aumailley, M.; Gayraud, B. Structure and Biological Activity of the Extracellular Matrix. J. Mol. Med. 1998, 76, 253–265. [Google Scholar] [CrossRef]
- Plant, A.L.; Bhadriraju, K.; Spurlin, T.A.; Elliott, J.T. Cell Response to Matrix Mechanics: Focus on Collagen. Biochim. Biophys. Acta 2009, 1793, 893–902. [Google Scholar] [CrossRef]
- Scherer, P.E.; Bickel, P.E.; Kotler, M.; Lodish, H.F. Cloning of Cell-Specific Secreted and Surface Proteins by Subtractive Antibody Screening. Nat. Biotechnol. 1998, 16, 581–586. [Google Scholar] [CrossRef]
- Pasarica, M.; Gowronska-Kozak, B.; Burk, D.; Remedios, I.; Hymel, D.; Gimble, J.; Ravussin, E.; Bray, G.A.; Smith, S.R. Adipose Tissue Collagen VI in Obesity. J. Clin. Endocrinol. Metab. 2009, 94, 5155–5162. [Google Scholar] [CrossRef]
- Henegar, C.; Tordjman, J.; Achard, V.; Lacasa, D.; Cremer, I.; Guerre-Millo, M.; Poitou, C.; Basdevant, A.; Stich, V.; Viguerie, N.; et al. Adipose Tissue Transcriptomic Signature Highlights the Pathological Relevance of Extracellular Matrix in Human Obesity. Genome Biol. 2008, 9, R14. [Google Scholar] [CrossRef]
- Adapala, V.J.; Adedokun, S.A.; Considine, R.V.; Ajuwon, K.M. Acute Inflammation Plays a Limited Role in the Regulation of Adipose Tissue COL1A1 Protein Abundance. J. Nutr. Biochem. 2012, 23, 567–572. [Google Scholar] [CrossRef]
- Soták, M.; Rajan, M.R.; Clark, M.; Biörserud, C.; Wallenius, V.; Hagberg, C.E.; Börgeson, E. Healthy Subcutaneous and Omental Adipose Tissue Is Associated with High Expression of Extracellular Matrix Components. Int. J. Mol. Sci. 2022, 23, 520. [Google Scholar] [CrossRef]
- Khan, T.; Muise, E.S.; Iyengar, P.; Wang, Z.V.; Chandalia, M.; Abate, N.; Zhang, B.B.; Bonaldo, P.; Chua, S.; Scherer, P.E. Metabolic Dysregulation and Adipose Tissue Fibrosis: Role of Collagen VI. Mol. Cell. Biol. 2009, 29, 1575–1591. [Google Scholar] [CrossRef]
- Dankel, S.N.; Grytten, E.; Bjune, J.I.; Nielsen, H.J.; Dietrich, A.; Blüher, M.; Sagen, J.V.; Mellgren, G. COL6A3 Expression in Adipose Tissue Cells Is Associated with Levels of the Homeobox Transcription Factor PRRX1. Sci. Rep. 2020, 10, 20164. [Google Scholar] [CrossRef]
- McCulloch, L.J.; Rawling, T.J.; Sjöholm, K.; Franck, N.; Dankel, S.N.; Price, E.J.; Knight, B.; Liversedge, N.H.; Mellgren, G.; Nystrom, F.; et al. COL6A3 Is Regulated by Leptin in Human Adipose Tissue and Reduced in Obesity. Endocrinology 2015, 156, 134–146. [Google Scholar] [CrossRef]
- Staunstrup, L.M.; Bager, C.L.; Frederiksen, P.; Helge, J.W.; Brunak, S.; Christiansen, C.; Karsdal, M. Endotrophin Is Associated with Chronic Multimorbidity and All-Cause Mortality in a Cohort of Elderly Women. EBioMedicine 2021, 68, 103391. [Google Scholar] [CrossRef]
- Scherer, P.E.; Gupta, O.T. Endotrophin: Nominated for Best Supporting Actor in the Fibro-Inflammatory Saga. EBioMedicine 2021, 69, 103447. [Google Scholar] [CrossRef]
- Eruzun, H.; Toprak, İ.D.; Arman, Y.; Yılmaz, U.; Özcan, M.; Kutlu, Y.; Irmak, S.; Kutlu, O.; Yoldemir, Ş.A.; Altun, Ö.; et al. Serum Endotrophin Levels in Patients with Heart Failure with Reduced and Mid-Range Ejection Fraction. Eur. J. Intern. Med. 2019, 64, 29–32. [Google Scholar] [CrossRef]
- Sun, K.; Park, J.; Kim, M.; Scherer, P.E. Endotrophin, a Multifaceted Player in Metabolic Dysregulation and Cancer Progression, Is a Predictive Biomarker for the Response to PPARγ Agonist Treatment. Diabetologia 2017, 60, 24–29. [Google Scholar] [CrossRef]
- Sun, K.; Park, J.; Gupta, O.T.; Holland, W.L.; Auerbach, P.; Zhang, N.; Goncalves Marangoni, R.; Nicoloro, S.M.; Czech, M.P.; Varga, J.; et al. Endotrophin Triggers Adipose Tissue Fibrosis and Metabolic Dysfunction. Nat. Commun. 2014, 5, 3485. [Google Scholar] [CrossRef]
- Adikaram, S.G.S.; Chathurangana, P.W.; Perera, T.M.R. Classic Ehlers-Danlos Syndrome. Sri Lanka J. Child Health 1993, 43, 61–62. [Google Scholar] [CrossRef]
- Wenstrup, R.J.; Florer, J.B.; Willing, M.C.; Giunta, C.; Steinmann, B.; Young, F.; Susic, M.; Cole, W.G. COL5A1 Haploinsufficiency Is a Common Molecular Mechanism Underlying the Classical Form of EDS. Am. J. Hum. Genet. 2000, 66, 1766–1776. [Google Scholar] [CrossRef]
- Lim, S.-T.; Kim, C.-S.; Kim, W.-N.; Min, S.-K. The COL5A1 Genotype Is Associated with Range of Motion. J. Exerc. Nutr. Biochem. 2015, 19, 49–53. [Google Scholar] [CrossRef]
- Pabalan, N.; Tharabenjasin, P.; Phababpha, S.; Jarjanazi, H. Association of COL5A1 Gene Polymorphisms and Risk of Tendon-Ligament Injuries among Caucasians: A Meta-Analysis. Sports Med. Open 2018, 4, 46. [Google Scholar] [CrossRef]
- Gu, S.; Peng, Z.; Wu, Y.; Wang, Y.; Lei, D.; Jiang, X.; Zhao, H.; Fu, P. COL5A1 Serves as a Biomarker of Tumor Progression and Poor Prognosis and May Be a Potential Therapeutic Target in Gliomas. Front. Oncol. 2021, 11, 752694. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, X.; Feng, S.; Jian, Z.; Xu, X.; Gu, L.; Xiong, X. The Hypoxia-Related Gene COL5A1 Is a Prognostic and Immunological Biomarker for Multiple Human Tumors. Oxid. Med. Cell. Longev. 2022, 2022, 6419695. [Google Scholar] [CrossRef]
- Zhang, Y.; Jing, Y.; Wang, Y.; Tang, J.; Zhu, X.; Jin, W.L.; Wang, Y.; Yuan, W.; Li, X.; Li, X. NAT10 Promotes Gastric Cancer Metastasis via N4-Acetylated COL5A1. Signal Transduct. Target. Ther. 2021, 6, 173. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, J.; Wang, F.; Xu, X.; Li, X.; Guan, W.; Men, T.; Xu, G. Overexpressed COL5A1 Is Correlated with Tumor Progression, Paclitaxel Resistance, and Tumor-Infiltrating Immune Cells in Ovarian Cancer. J. Cell. Physiol. 2021, 236, 6907–6919. [Google Scholar] [CrossRef]
- Wu, M.; Sun, Q.; Mo, C.H.; Pang, J.S.; Hou, J.Y.; Pang, L.L.; Lu, H.P.; Dang, Y.W.; Fang, S.J.; Tang, D.; et al. Prospective Molecular Mechanism of COL5A1 in Breast Cancer Based on a Microarray, RNA Sequencing and Immunohistochemistry. Oncol. Rep. 2019, 42, 151–175. [Google Scholar] [CrossRef]
- Spencer, M.; Unal, R.; Zhu, B.; Rasouli, N.; McGehee, R.E.; Peterson, C.A.; Kern, P.A. Adipose Tissue Extracellular Matrix and Vascular Abnormalities in Obesity and Insulin Resistance. J. Clin. Endocrinol. Metab. 2011, 96, E1990–E1998. [Google Scholar] [CrossRef] [PubMed]
- Kotzé-Hörstmann, L.M.; Keswell, D.; Adams, K.; Dlamini, T.; Goedecke, J.H. Hypoxia and Extra-Cellular Matrix Gene Expression in Adipose Tissue Associates with Reduced Insulin Sensitivity in Black South African Women. Endocrine 2017, 55, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Hocking, S.L.; Wu, L.E.; Guilhaus, M.; Chisholm, D.J.; James, D.E. Intrinsic Depot-Specific Differences in the Secretome of Adipose Tissue, Preadipocytes, and Adipose Tissue-Derived Microvascular Endothelial Cells. Diabetes 2010, 59, 3008–3016. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, I.; Muroya, S.; Tanabe, R.I.; Chikuni, K. Positive Effect of Collagen V and VI on Triglyceride Accumulation during Differentiation in Cultures of Bovine Intramuscular Adipocytes. Differentiation 2002, 70, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, I.; Muroya, S.; Tanabe, R.I.; Chikuni, K. Extracellular Matrix Development during Differentiation into Adipocytes with a Unique Increase in Type V and VI Collagen. Biol. Cell 2002, 94, 197–203. [Google Scholar] [CrossRef]


| Baseline (n = 144) | |
|---|---|
| Type of Surgery (RYGB) | 88 (61.1%) |
| Sex (Female) | 124 (86.1%) |
| Age (years) | 51.8 ± 10.9 |
| Weight (kg) | 113.8 ± 25.7 |
| BMI (kg/m2) | 43.1 (42.1, 44.5) |
| T2D (yes/no) | 49 (34.0%) |
| T2D treatment (yes/no) | 48 (33.3%) |
| T2D medications (number) | 0 (0–0) |
| Insulin treatment (yes/no) | 19 (13.2%) |
| Hypertension (yes/no) | 47.9 (32%) |
| Statins (yes/no) | 49 (34.0%) |
| FPG (mg/dL) | 99 (96, 102) |
| HbA1c (%) | 5.7 (5.6, 5.8) |
| Total cholesterol (mg/dL) | 184.6 ± 36.18 |
| HDL-cholesterol (mg/dL) | |
| Male | 38.5 (37, 44) |
| Female | 50.55 ± 10.65 |
| LDL-cholesterol (mg/dL) | |
| Male | 108.9 ± 33.8 |
| Female | 111.7 ± 27.08 |
| Tryglycerides (mg/dL) | |
| Male | 130.5 ± 55.9 |
| Female | 116 (106, 130) |
| hs-CRP (mg/L) | 0.66 (0.58, 0.85) |
| AST (UI/L) | 20 (19, 22) |
| ALT (UI/L) | 23 (21, 25) |
| GGT (UI/L) | 26 (24, 28) |
| Exploratory | Validation | Pooled | ||||
|---|---|---|---|---|---|---|
| β (95% CI) | p | β (95% CI) | p | β (95% CI) | p | |
| PLIN1 | ||||||
| Unadjusted | −3.44 (−7.12, 0.29) | 0.069 | −0.791 (−2.33, 0.75) | 0.305 | −1.37 (−2.8, 0.07) | 0.061 |
| Adjusted 1 | −3.52 (−6.94, −0.11) | 0.044 | −0.72 (−2.14, 0.71) | 0.312 | −1.35 (−2.69, −0.01) | 0.049 |
| ADIPOR2 | ||||||
| Unadjusted | −21.71 (−42.91, −0.50) | 0.045 | −21.02 (−49.59, 7.55) | 0.145 | −22.34 (−39.01, −5.48) | 0.010 |
| Adjusted 1 | −16.86 (−36.62, 2.89) | 0.093 | −25.01 (−50.05, 0.019) | 0.050 | −20.02 (−35.76, −4.27) | 0.013 |
| COL1A1 | ||||||
| Unadjusted | −4.31 (−9.29, 0.67) | 0.089 | −8.51 (−18.46, 1.25) | 0.085 | −3.65 (−7.77, 0.47) | 0.082 |
| Adjusted 1 | −5.87 (−10.44, −1.30) | 0.013 | −8.93 (−17.91, 0.045) | 0.049 | −5.25 (−8.99, −1.51) | <0.001 |
| COL6A3 | ||||||
| Unadjusted | −11.87 (−21.05, −2.70) | 0.012 | −8.75 (−16.48, −1.01) | 0.028 | −11.40 (−17.37, −5.43) | <0.001 |
| Adjusted 1 | −13.23 (−20.73, −3.73) | 0.006 | −8.06 (−15.81, −0.31) | 0.042 | −12.25 (−17.68, −6.82) | <0.001 |
| COL5A1 | ||||||
| Unadjusted | −24.61 (−41.31, −7.92) | 0.005 | −21.01 (−35.44, −6.58) | 0.005 | −23.59 (−34.18, −13.01) | <0.001 |
| Adjusted 1 | −24.79 (−43, −6.59) | 0.009 | −16.74 (−30.94, −2.53) | 0.022 | −23.78 (−34.7, −12.86) | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osorio-Conles, Ó.; Olbeyra, R.; Vidal, J.; Ibarzabal, A.; Balibrea, J.M.; de Hollanda, A. Expression of Adipose Tissue Extracellular Matrix-Related Genes Predicts Weight Loss after Bariatric Surgery. Cells 2023, 12, 1262. https://doi.org/10.3390/cells12091262
Osorio-Conles Ó, Olbeyra R, Vidal J, Ibarzabal A, Balibrea JM, de Hollanda A. Expression of Adipose Tissue Extracellular Matrix-Related Genes Predicts Weight Loss after Bariatric Surgery. Cells. 2023; 12(9):1262. https://doi.org/10.3390/cells12091262
Chicago/Turabian StyleOsorio-Conles, Óscar, Romina Olbeyra, Josep Vidal, Ainitze Ibarzabal, José María Balibrea, and Ana de Hollanda. 2023. "Expression of Adipose Tissue Extracellular Matrix-Related Genes Predicts Weight Loss after Bariatric Surgery" Cells 12, no. 9: 1262. https://doi.org/10.3390/cells12091262
APA StyleOsorio-Conles, Ó., Olbeyra, R., Vidal, J., Ibarzabal, A., Balibrea, J. M., & de Hollanda, A. (2023). Expression of Adipose Tissue Extracellular Matrix-Related Genes Predicts Weight Loss after Bariatric Surgery. Cells, 12(9), 1262. https://doi.org/10.3390/cells12091262








