Molecular and Functional Characterization of Different BrainSphere Models for Use in Neurotoxicity Testing on Microelectrode Arrays
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation of hiPSCs
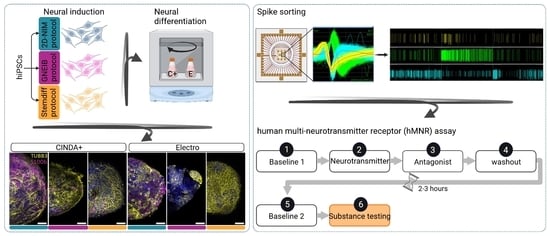
2.2. Neural Induction of hiPSCs into Human-Induced Neural Progenitor Cells (hiNPCs)
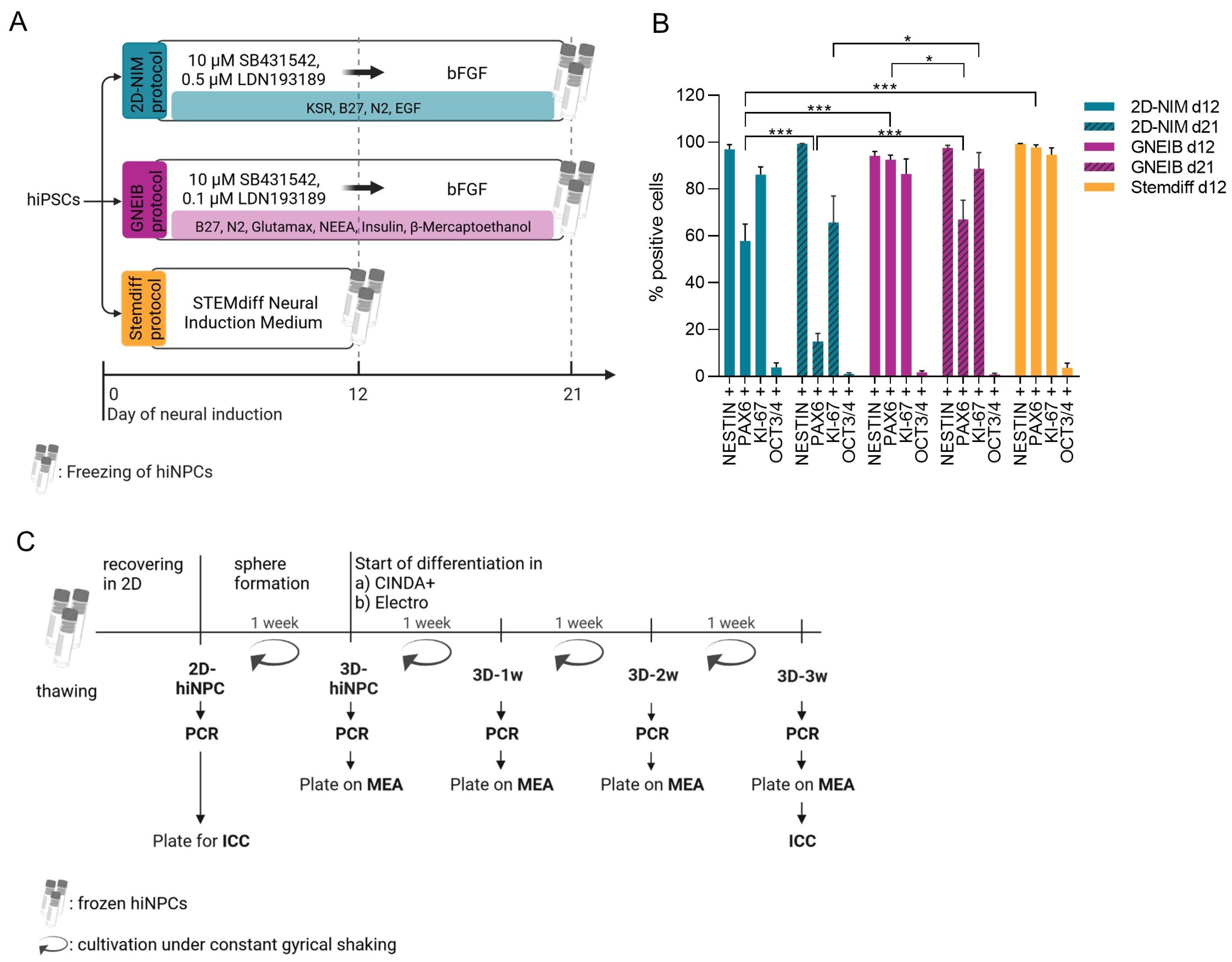
2.2.1. 2D-NIM Protocol
2.2.2. GNEIB Protocol
2.2.3. Stemdiff Protocol
2.3. Thawing of hiNPCs
2.4. Formation of BrainSpheres
2.5. Neural Differentiation
2.6. Neural Differentiation on Microelectrode Arrays (MEA)
2.6.1. Recording and Data Analysis of MEA Neuronal Electrical Activity
2.6.2. Spike Sorting
2.7. Cytotoxicity Assessment
2.8. Cultivation of SynFire Cells
2.9. Flow Cytometry
2.10. Quantitative Polymerase Chain Reaction (qPCR)
2.11. Immunocytochemistry (ICC) of Adherent hiNPCs
2.12. ICC of BrainSpheres
2.13. Statistical Analysis
3. Results
3.1. All Three Neural Induction Protocols Successfully Induce hiPSCs into the Neural Lineage
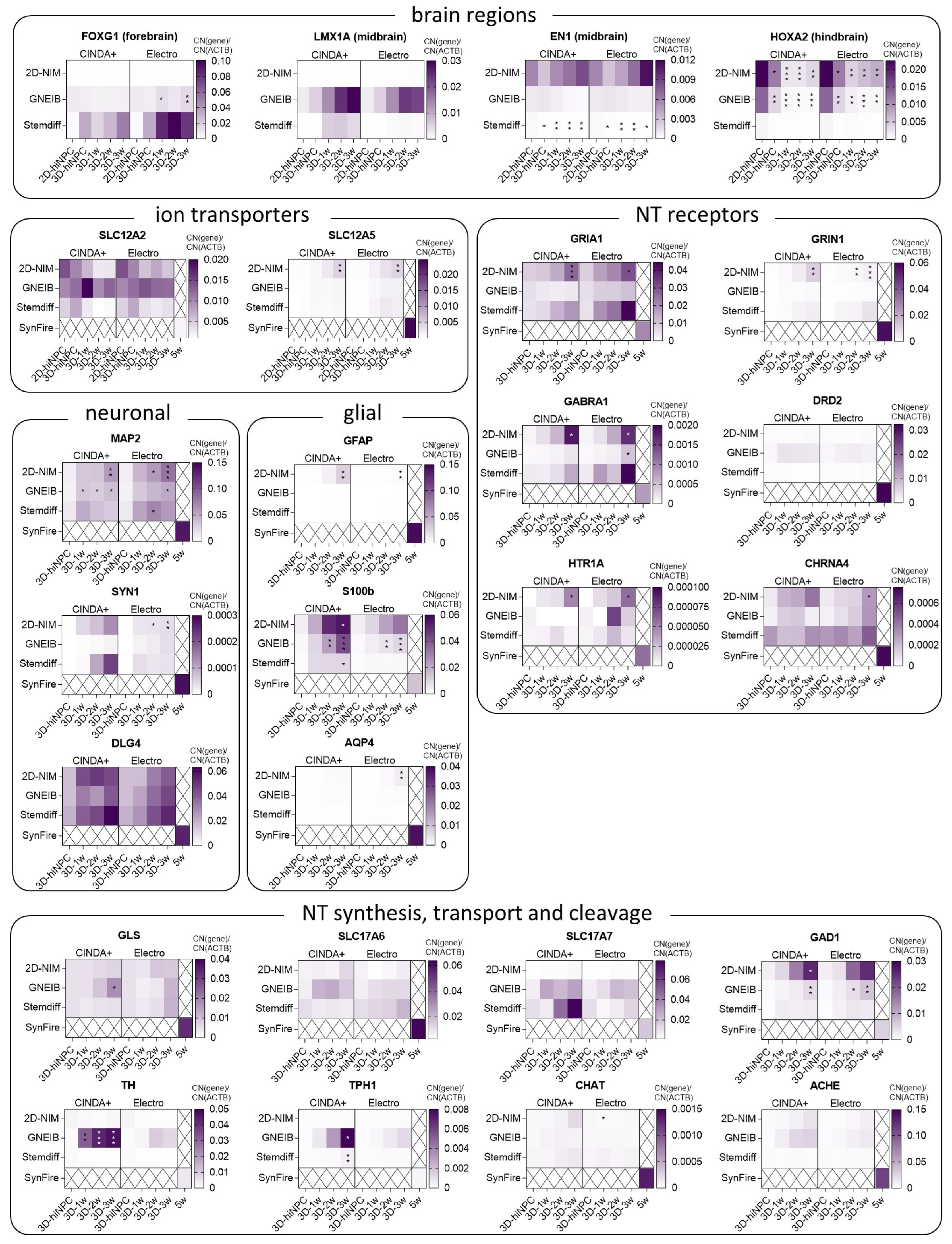
3.2. BrainSpheres Differ in Neural Marker Gene Expression Depending on the Applied Protocol
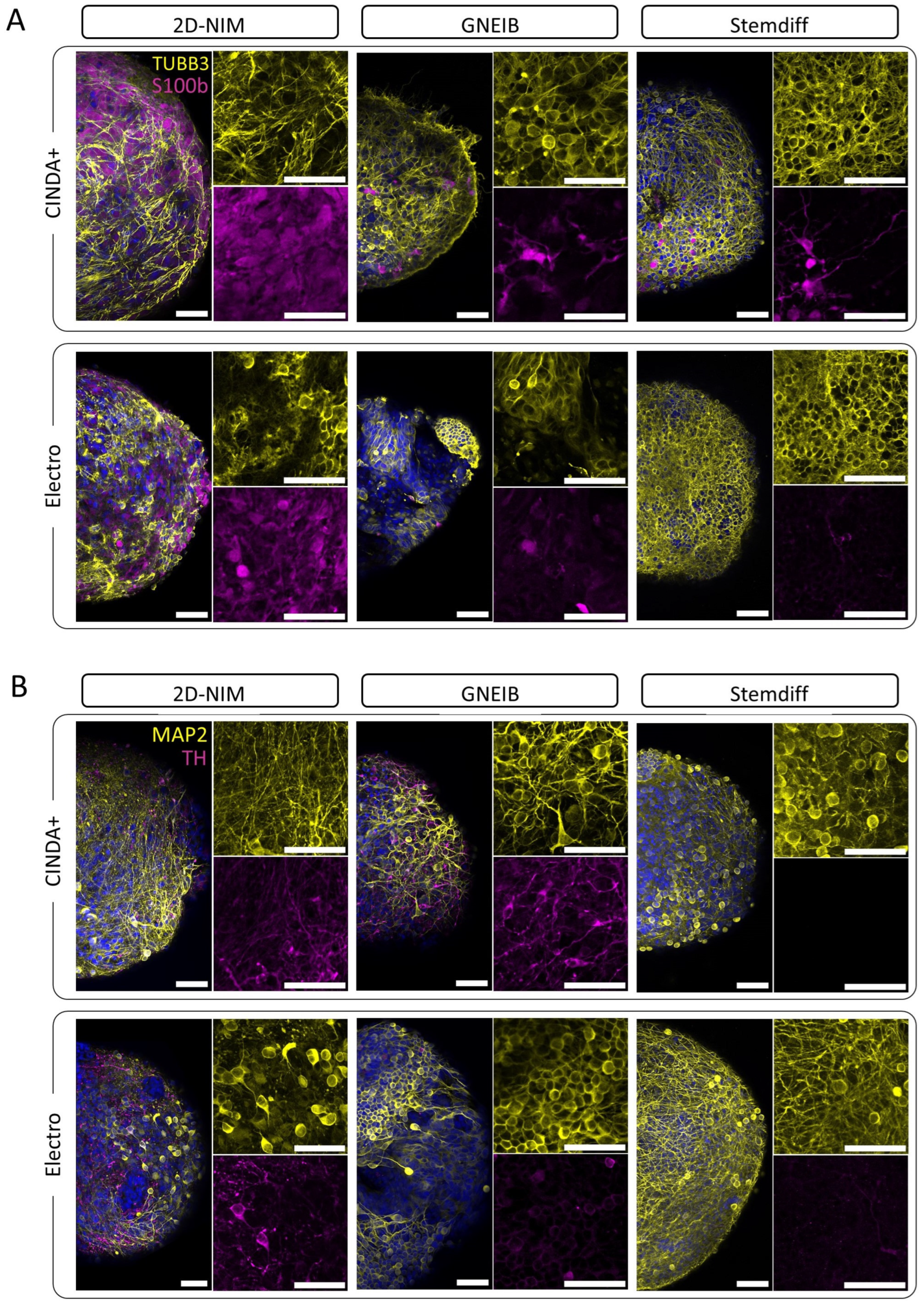
3.3. Neural Induction and Differentiation Protocols Determine the Potential of BrainSpheres to Differentiate into Astrocytes and Dopaminergic Neurons
3.4. Neural Induction and Differentiation Media Determine Neuronal Activity and Neural Network Function of BrainSpheres on MEAs
3.5. Neural Induction and Differentiation Media Determine BrainSpheres’ Neuronal Subtype Differentiation
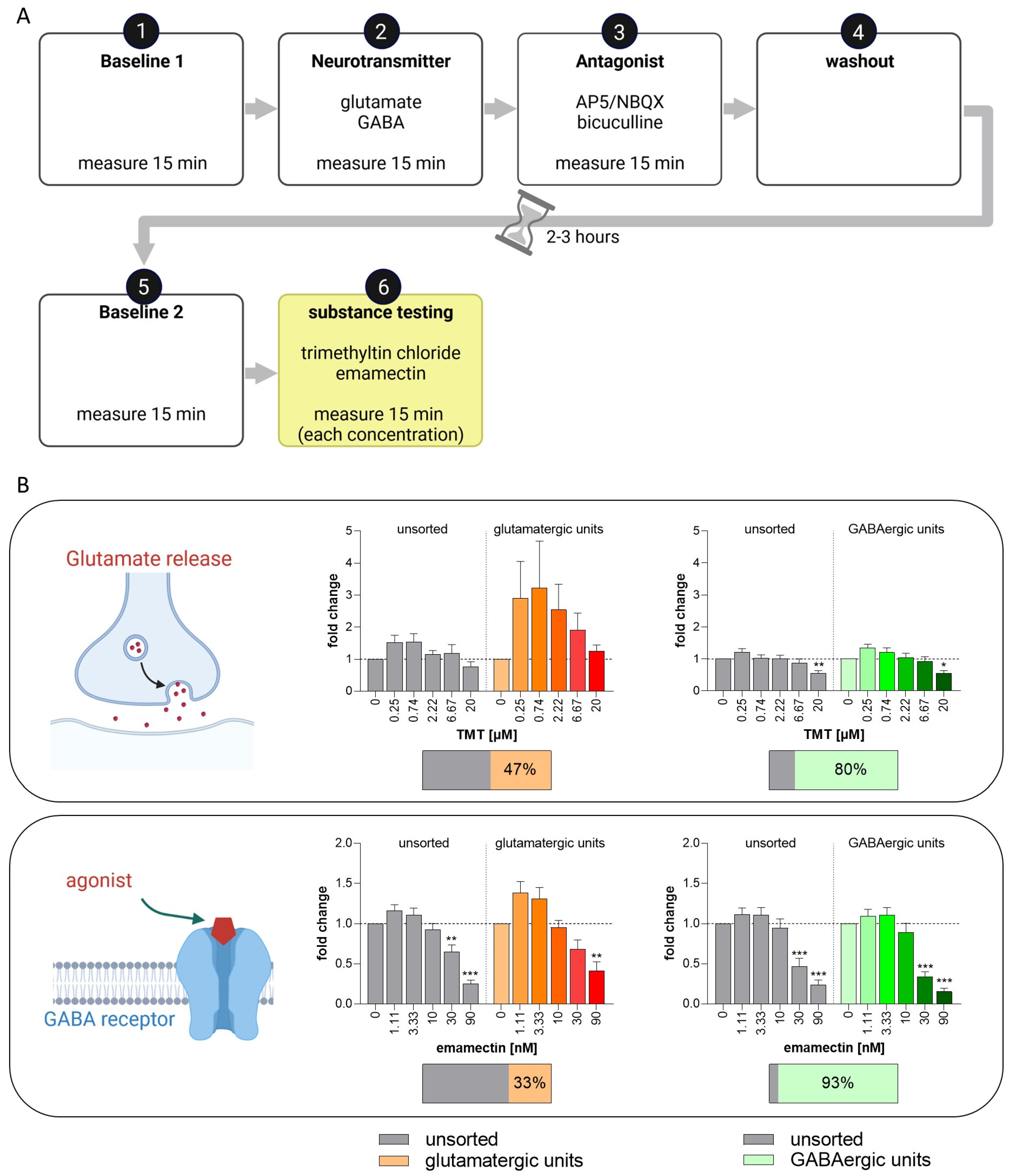
3.6. Set-Up of a New NAM for Acute Neurotoxicity Testing Using MEAs and Spike Sorting, the Human Multi-Neurotransmitter Receptor (hMNR) Assay
4. Discussion
5. Summary and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- OECD. OECD Guideline for the Testing of Chemicals 419 Delayed Neurotoxicity of Organophosphorus Substances: 28-Day Repeated Dose Study; OECD: Paris, France, 1995; pp. 1–7. [Google Scholar]
- OECD. OECD Guideline for the Testing of Chemicals 418 Delayed Neurotoxicity of Organophosphorus Substances Following Acute Exposure; OECD: Paris, France, 1995; pp. 1–8. [Google Scholar]
- OECD. OECD guideline for the testing of Chemicals Test No. 424: Neurotoxicity Study in Rodents. In OECD Guideline for the Testing of Chemicals; Section 4; OECD: Paris, France, 1997; pp. 1–15. ISBN 9789264071025. [Google Scholar]
- Buschmann, J. The OECD Guidelines for the Testing of Chemicals and Pesticides. Methods Mol. Biol. 2013, 947, 37–56. [Google Scholar] [CrossRef]
- Bal-Price, A.K.; Hogberg, H.T.; Buzanska, L.; Coecke, S. Relevance of in Vitro Neurotoxicity Testing for Regulatory Requirements: Challenges to Be Considered. Neurotoxicol. Teratol. 2010, 32, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Somel, M.; Liu, X.; Tang, L.; Yan, Z.; Hu, H.; Guo, S.; Jiang, X.; Zhang, X.; Xu, G.; Xie, G.; et al. MicroRNA-Driven Developmental Remodeling in the Brain Distinguishes Humans from Other Primates. PLoS Biol. 2011, 9, e1001214. [Google Scholar] [CrossRef] [PubMed]
- Pollen, A.A.; Nowakowski, T.J.; Chen, J.; Retallack, H.; Sandoval-Espinosa, C.; Nicholas, C.R.; Shuga, J.; Liu, S.J.; Oldham, M.C.; Diaz, A.; et al. Molecular Identity of Human Outer Radial Glia during Cortical Development. Cell 2015, 163, 55–67. [Google Scholar] [CrossRef]
- Waring, M.J.; Arrowsmith, J.; Leach, A.R.; Leeson, P.D.; Mandrell, S.; Owen, R.M.; Pairaudeau, G.; Pennie, W.D.; Pickett, S.D.; Wang, J.; et al. An Analysis of the Attrition of Drug Candidates from Four Major Pharmaceutical Companies. Nat. Rev. Drug Discov. 2015, 14, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Mohs, R.C.; Greig, N.H. Drug Discovery and Development: Role of Basic Biological Research. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2017, 3, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Toutain, P.-L.; Ferran, A.; Bousquet-Melou, A. Species Differences in Pharmacokinetics and Pharmacodynamics. In Handbook of Experimental Pharmacology; Springer: Cham, Switzerland, 2010; pp. 19–48. [Google Scholar]
- Marshall, L.J.; Bailey, J.; Cassotta, M.; Herrmann, K.; Pistollato, F. Poor Translatability of Biomedical Research Using Animals—A Narrative Review. Altern. Lab. Anim. 2023, 51, 102–135. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Walker, G.W.; Muir, D.C.G.; Nagatani-Yoshida, K. Toward a Global Understanding of Chemical Pollution: A First Comprehensive Analysis of National and Regional Chemical Inventories. Environ. Sci. Technol. 2020, 54, 2575–2584. [Google Scholar] [CrossRef] [PubMed]
- Pallocca, G.; Moné, M.J.; Kamp, H.; Luijten, M.; Van de Water, B.; Leist, M. Next-Generation Risk Assessment of Chemicals—Rolling out a Human-Centric Testing Strategy to Drive 3R Implementation: The RISK-HUNT3R Project Perspective. ALTEX 2022, 39, 419–426. [Google Scholar] [CrossRef]
- Dent, M.P.; Vaillancourt, E.; Thomas, R.S.; Carmichael, P.L.; Ouedraogo, G.; Kojima, H.; Barroso, J.; Ansell, J.; Barton-Maclaren, T.S.; Bennekou, S.H.; et al. Paving the Way for Application of next Generation Risk Assessment to Safety Decision-Making for Cosmetic Ingredients. Regul. Toxicol. Pharmacol. 2021, 125, 105026. [Google Scholar] [CrossRef]
- Krewski, D.; Andersen, M.E.; Tyshenko, M.G.; Krishnan, K.; Hartung, T.; Boekelheide, K.; Wambaugh, J.F.; Jones, D.; Whelan, M.; Thomas, R.; et al. Toxicity Testing in the 21st Century: Progress in the Past Decade and Future Perspectives; Springer Berlin/Heidelberg: Heidelberg, Germany, 2020; Volume 94, ISBN 0123456789. [Google Scholar]
- Kavlock, R.J.; Bahadori, T.; Barton-Maclaren, T.S.; Gwinn, M.R.; Rasenberg, M.; Thomas, R.S. Accelerating the Pace of Chemical Risk Assessment. Chem. Res. Toxicol. 2018, 31, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Vinken, M.; Benfenati, E.; Busquet, F.; Castell, J.; Clevert, D.A.; de Kok, T.M.; Dirven, H.; Fritsche, E.; Geris, L.; Gozalbes, R.; et al. Safer Chemicals Using Less Animals: Kick-off of the European ONTOX Project. Toxicology 2021, 458, 152846. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, P.L.; Baltazar, M.T.; Cable, S.; Cochrane, S.; Dent, M.; Li, H.; Middleton, A.; Muller, I.; Reynolds, G.; Westmoreland, C.; et al. Ready for Regulatory Use: NAMs and NGRA for Chemical Safety Assurance. ALTEX 2022, 39, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Maertens, A.; Golden, E.; Luechtefeld, T.H.; Hoffmann, S.; Tsaioun, K.; Hartung, T. Probabilistic Risk Assessment—The Keystone for the Future of Toxicology. ALTEX 2022, 39, 3–29. [Google Scholar] [CrossRef]
- Mahony, C.; Ashton, R.S.; Birk, B.; Boobis, A.R.; Cull, T.; Daston, G.P.; Ewart, L.; Knudsen, T.B.; Manou, I.; Maurer-Stroh, S.; et al. New Ideas for Non-Animal Approaches to Predict Repeated-Dose Systemic Toxicity: Report from an EPAA Blue Sky Workshop. Regul. Toxicol. Pharmacol. 2020, 114, 104668. [Google Scholar] [CrossRef] [PubMed]
- van der Stel, W.; Carta, G.; Eakins, J.; Delp, J.; Suciu, I.; Forsby, A.; Cediel-Ulloa, A.; Attoff, K.; Troger, F.; Kamp, H.; et al. New Approach Methods (NAMs) Supporting Read-Across: Two Neurotoxicity AOP-Based IATA Case Studies. ALTEX 2021, 38, 615–635. [Google Scholar] [CrossRef] [PubMed]
- Masjosthusmann, S.; Barenys, M.; El-Gamal, M.; Geerts, L.; Gerosa, L.; Gorreja, A.; Kühne, B.; Marchetti, N.; Tigges, J.; Viviani, B.; et al. Literature Review and Appraisal on Alternative Neurotoxicity Testing Methods. EFSA Support. Publ. 2018, 15, 1410E. [Google Scholar] [CrossRef]
- Harry, G.J.; Tiffany-Castiglioni, E. Evaluation of Neurotoxic Potential by Use of in Vitro Systems. Expert Opin. Drug Metab. Toxicol. 2005, 1, 701–713. [Google Scholar] [CrossRef]
- Lisek, M.; Boczek, T.; Stragierowicz, J.; Wawrzyniak, J.; Guo, F.; Klimczak, M.; Kilanowicz, A.; Zylinska, L. Hexachloronaphthalene (HxCN) Impairs the Dopamine Pathway in an in Vitro Model of PC12 Cells. Chemosphere 2022, 287, 132284. [Google Scholar] [CrossRef]
- Schultz, L.; Zurich, M.G.; Culot, M.; da Costa, A.; Landry, C.; Bellwon, P.; Kristl, T.; Hörmann, K.; Ruzek, S.; Aiche, S.; et al. Evaluation of Drug-Induced Neurotoxicity Based on Metabolomics, Proteomics and Electrical Activity Measurements in Complementary CNS in Vitro Models. Toxicol. Vitr. 2015, 30, 138–165. [Google Scholar] [CrossRef] [PubMed]
- Hausherr, V.; van Thriel, C.; Krug, A.; Leist, M.; Schöbel, N. Impairment of Glutamate Signaling in Mouse Central Nervous System Neurons in Vitro by Tri-Ortho-Cresyl Phosphate at Noncytotoxic Concentrations. Toxicol. Sci. 2014, 142, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Loser, D.; Schaefer, J.; Danker, T.; Möller, C.; Brüll, M.; Suciu, I.; Ückert, A.K.; Klima, S.; Leist, M.; Kraushaar, U. Human Neuronal Signaling and Communication Assays to Assess Functional Neurotoxicity. Arch. Toxicol. 2021, 95, 229–252. [Google Scholar] [CrossRef] [PubMed]
- Kosnik, M.B.; Strickland, J.D.; Marvel, S.W.; Wallis, D.J.; Wallace, K.; Richard, A.M.; Reif, D.M.; Shafer, T.J. Concentration–Response Evaluation of ToxCast Compounds for Multivariate Activity Patterns of Neural Network Function. Arch. Toxicol. 2020, 94, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, A.F.M.; Gross, G.W.; Weiss, D.G.; Schroeder, O.H.-U.; Gramowski, A.; Shafer, T.J. Microelectrode Arrays: A Physiologically Based Neurotoxicity Testing Platform for the 21st Century. Neurotoxicology 2010, 31, 331–350. [Google Scholar] [CrossRef] [PubMed]
- Vassallo, A.; Chiappalone, M.; De Camargos Lopes, R.; Scelfo, B.; Novellino, A.; Defranchi, E.; Palosaari, T.; Weisschu, T.; Ramirez, T.; Martinoia, S.; et al. A Multi-Laboratory Evaluation of Microelectrode Array-Based Measurements of Neural Network Activity for Acute Neurotoxicity Testing. Neurotoxicology 2017, 60, 280–292. [Google Scholar] [CrossRef]
- Saavedra, L.; Wallace, K.; Freudenrich, T.F.; Mall, M.; Mundy, W.R.; Davila, J.; Shafer, T.J.; Wernig, M.; Haag, D. Comparison of Acute Effects of Neurotoxic Compounds on Network Activity in Human and Rodent Neural Cultures. Toxicol. Sci. 2021, 180, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Tukker, A.M.; De Groot, M.W.G.D.M.; Wijnolts, F.M.J.; Kasteel, E.E.J.J.; Hondebrink, L.; Westerink, R.H.S. Is the Time Right for in Vitro Neurotoxicity Testing Using Human IPSC-Derived Neurons? ALTEX 2016, 33, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Tukker, A.M.; van Kleef, R.G.D.M.; Wijnolts, F.M.J.; de Groot, A.; Westerink, R.H.S. Towards Animal-Free Neurotoxicity Screening: Applicability of HiPSC-Derived Neuronal Models for in Vitro Seizure Liability Assessment. ALTEX 2019, 37, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Tukker, A.M.; Wijnolts, F.M.J.J.; de Groot, A.; Westerink, R.H.S. Human IPSC-Derived Neuronal Models for in Vitro Neurotoxicity Assessment. Neurotoxicology 2018, 67, 215–225. [Google Scholar] [CrossRef]
- Mack, C.M.; Lin, B.J.; Turner, J.D.; Johnstone, A.F.M.; Burgoon, L.D.; Shafer, T.J. Burst and Principal Components Analyses of MEA Data for 16 Chemicals Describe at Least Three Effects Classes. Neurotoxicology 2014, 40, 75–85. [Google Scholar] [CrossRef]
- Brown, J.P.; Hall, D.; Frank, C.L.; Wallace, K.; Mundy, W.R.; Shafer, T.J. Editor’s Highlight: Evaluation of a Microelectrode Array-Based Assay for Neural Network Ontogeny Using Training Set Chemicals. Toxicol. Sci. 2016, 154, 126–139. [Google Scholar] [CrossRef]
- Tukker, A.M.; Wijnolts, F.M.J.; de Groot, A.; Westerink, R.H.S. Applicability of HiPSC-Derived Neuronal Cocultures and Rodent Primary Cortical Cultures for In Vitro Seizure Liability Assessment. Toxicol. Sci. 2020, 178, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Balestrino, R.; Schapira, A.H.V. Parkinson Disease. Eur. J. Neurol. 2020, 27, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Nopoulos, P.C. Huntington Disease: A Single-Gene Degenerative Disorder of the Striatum. Dialogues Clin. Neurosci. 2016, 18, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced Pluripotent Stem Cell Technology: A Decade of Progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef]
- Galiakberova, A.A.; Dashinimaev, E.B. Neural Stem Cells and Methods for Their Generation From Induced Pluripotent Stem Cells in Vitro. Front. Cell Dev. Biol. 2020, 8, 815. [Google Scholar] [CrossRef]
- Li, W.; Sun, W.; Zhang, Y.; Wei, W.; Ambasudhan, R.; Xia, P.; Talantova, M.; Lin, T.; Kim, J.; Wang, X.; et al. Rapid Induction and Long-Term Self-Renewal of Primitive Neural Precursors from Human Embryonic Stem Cells by Small Molecule Inhibitors. Proc. Natl. Acad. Sci. USA 2011, 108, 8299–8304. [Google Scholar] [CrossRef]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly Efficient Neural Conversion of Human ES and IPS Cells by Dual Inhibition of SMAD Signaling. Nat. Biotechnol. 2009, 27, 275–280. [Google Scholar] [CrossRef]
- Shi, Y.; Kirwan, P.; Smith, J.; Robinson, H.P.C.; Livesey, F.J. Human Cerebral Cortex Development from Pluripotent Stem Cells to Functional Excitatory Synapses. Nat. Neurosci. 2012, 15, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Izsak, J.; Seth, H.; Andersson, M.; Vizlin-hodzic, D.; Theiss, S.; Hanse, E.; Ågren, H.; Funa, K.; Illes, S. Robust Generation of Person-Specific, Synchronously Active Neuronal Networks Using Purely Isogenic Human IPSC-3D Neural Aggregate Cultures. Front. Neurosci. 2019, 13, 351. [Google Scholar] [CrossRef] [PubMed]
- Hyvärinen, T.; Hyysalo, A.; Kapucu, F.E.; Aarnos, L.; Vinogradov, A.; Eglen, S.J.; Ylä-Outinen, L.; Narkilahti, S. Functional Characterization of Human Pluripotent Stem Cell-Derived Cortical Networks Differentiated on Laminin-521 Substrate: Comparison to Rat Cortical Cultures. Sci. Rep. 2019, 9, 17125. [Google Scholar] [CrossRef] [PubMed]
- Paavilainen, T.; Pelkonen, A.; Mäkinen, M.E.; Peltola, M.; Huhtala, H.; Fayuk, D.; Narkilahti, S. Effect of Prolonged Differentiation on Functional Maturation of Human Pluripotent Stem Cell-Derived Neuronal Cultures. Stem Cell Res. 2018, 27, 151–161. [Google Scholar] [CrossRef]
- Gunhanlar, N.; Shpak, G.; van der Kroeg, M.; Gouty-Colomer, L.A.; Munshi, S.T.; Lendemeijer, B.; Ghazvini, M.; Dupont, C.; Hoogendijk, W.J.G.; Gribnau, J.; et al. A Simplified Protocol for Differentiation of Electrophysiologically Mature Neuronal Networks from Human Induced Pluripotent Stem Cells. Mol. Psychiatry 2017, 23, 1336–1344. [Google Scholar] [CrossRef]
- de Leeuw, V.C.; van Oostrom, C.T.M.; Wackers, P.F.K.; Pennings, J.L.A.; Hodemaekers, H.M.; Piersma, A.H.; Hessel, E.V.S. Neuronal Differentiation Pathways and Compound-Induced Developmental Neurotoxicity in the Human Neural Progenitor Cell Test (HNPT) Revealed by RNA-Seq. Chemosphere 2022, 304, 135298. [Google Scholar] [CrossRef]
- Pistollato, F.; Canovas-Jorda, D.; Zagoura, D.; Price, A. Protocol for the Differentiation of Human Induced Pluripotent Stem Cells into Mixed Cultures of Neurons and Glia for Neurotoxicity Testing. J. Vis. Exp. 2017, 2017, 55702. [Google Scholar] [CrossRef]
- Schenke, M.; Schjeide, B.M.; Püschel, G.P.; Seeger, B. Analysis of Motor Neurons Differentiated from Human Induced Pluripotent Stem Cells for the Use in Cell-Based Botulinum Neurotoxin Activity Assays. Toxins 2020, 12, 276. [Google Scholar] [CrossRef]
- De Leeuw, V.C.; Van Oostrom, C.T.M.; Zwart, E.P.; Heusinkveld, H.J.; Hessel, E.V.S. Prolonged Differentiation of Neuron-Astrocyte Co-Cultures Results in Emergence of Dopaminergic Neurons. Int. J. Mol. Sci. 2023, 24, 3608. [Google Scholar] [CrossRef]
- Suzuki, I.K.; Vanderhaeghen, P. Is This a Brain Which I See before Me? Modeling Human Neural Development with Pluripotent Stem Cells. Development 2015, 142, 3138–3150. [Google Scholar] [CrossRef]
- Nimtz, L.; Hartmann, J.; Tigges, J.; Masjosthusmann, S.; Schmuck, M.; Keßel, E.; Theiss, S.; Köhrer, K.; Petzsch, P.; Adjaye, J.; et al. Characterization and Application of Electrically Active Neuronal Networks Established from Human Induced Pluripotent Stem Cell-Derived Neural Progenitor Cells for Neurotoxicity Evaluation. Stem Cell Res. 2020, 45, 101761. [Google Scholar] [CrossRef]
- Pamies, D.; Barreras, P.; Block, K.; Makri, G.; Kumar, A.; Wiersma, D.; Smirnova, L.; Zhang, C.; Bressler, J.; Christian, K.M.; et al. A Human Brain Microphysiological System Derived from Induced Pluripotent Stem Cells to Study Neurological Diseases and Toxicity. ALTEX 2017, 34, 362–376. [Google Scholar] [CrossRef]
- Pistollato, F.; Louisse, J.; Scelfo, B.; Mennecozzi, M.; Accordi, B.; Basso, G.; Gaspar, J.A.; Zagoura, D.; Barilari, M.; Palosaari, T.; et al. Development of a Pluripotent Stem Cell Derived Neuronal Model to Identify Chemically Induced Pathway Perturbations in Relation to Neurotoxicity: Effects of CREB Pathway Inhibition. Toxicol. Appl. Pharmacol. 2014, 280, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, E.; Tigges, J.; Hartmann, J.; Kapr, J.; Serafini, M.M.; Viviani, B. Neural In Vitro Models for Studying Substances Acting on the Central Nervous System. In Handbook of Experimental Pharmacology; Springer: Cham, Switzerland, 2020. [Google Scholar] [CrossRef]
- Logan, S.; Arzua, T.; Canfield, S.G.; Seminary, E.R.; Sison, S.L.; Ebert, A.D.; Bai, X. Studying Human Neurological Disorders Using Induced Pluripotent Stem Cells: From 2D Monolayer to 3D Organoid and Blood Brain Barrier Models. Compr. Physiol. 2019, 9, 565–611. [Google Scholar]
- Abreu, C.M.; Gama, L.; Krasemann, S.; Chesnut, M.; Odwin-Dacosta, S.; Hogberg, H.T.; Hartung, T.; Pamies, D. Microglia Increase Inflammatory Responses in IPSC-Derived Human BrainSpheres. Front. Microbiol. 2018, 9, 2766. [Google Scholar] [CrossRef] [PubMed]
- Pamies, D.; Wiersma, D.; Katt, M.E.; Zhao, L.; Burtscher, J.; Harris, G.; Smirnova, L.; Searson, P.C.; Hartung, T.; Hogberg, H.T. Human IPSC 3D Brain Model as a Tool to Study Chemical-Induced Dopaminergic Neuronal Toxicity. Neurobiol. Dis. 2022, 169, 105719. [Google Scholar] [CrossRef] [PubMed]
- Pamies, D.; Block, K.; Lau, P.; Gribaldo, L.; Pardo, C.A.; Barreras, P.; Smirnova, L.; Wiersma, D.; Zhao, L.; Harris, G.; et al. Rotenone Exerts Developmental Neurotoxicity in a Human Brain Spheroid Model. Toxicol. Appl. Pharmacol. 2018, 354, 101–114. [Google Scholar] [CrossRef]
- Nunes, C.; Gorczyca, G.; Mendoza-deGyves, E.; Ponti, J.; Bogni, A.; Carpi, D.; Bal-Price, A.; Pistollato, F. Upscaling Biological Complexity to Boost Neuronal and Oligodendroglia Maturation and Improve in Vitro Developmental Neurotoxicity (DNT) Evaluation. Reprod. Toxicol. 2022, 110, 124–140. [Google Scholar] [CrossRef]
- Kobolak, J.; Teglasi, A.; Bellak, T.; Janstova, Z.; Molnar, K.; Zana, M.; Bock, I.; Laszlo, L.; Dinnyes, A. Human Induced Pluripotent Stem Cell-Derived 3D-Neurospheres Are Suitable for Neurotoxicity Screening. Cells 2020, 9, 1122. [Google Scholar] [CrossRef]
- Harris, G.; Hogberg, H.; Hartung, T.; Smirnova, L. 3D Differentiation of LUHMES Cell Line to Study Recovery and Delayed Neurotoxic Effects. Curr. Protoc. Toxicol. 2017, 73, 11–23. [Google Scholar] [CrossRef]
- Smirnova, L.; Harris, G.; Delp, J.; Valadares, M.; Pamies, D.; Hogberg, H.T.; Waldmann, T.; Leist, M.; Hartung, T. A LUHMES 3D Dopaminergic Neuronal Model for Neurotoxicity Testing Allowing Long-Term Exposure and Cellular Resilience Analysis. Arch. Toxicol. 2016, 90, 2725–2743. [Google Scholar] [CrossRef] [PubMed]
- Leite, P.E.C.; Pereira, M.R.; Harris, G.; Pamies, D.; Dos Santos, L.M.G.; Granjeiro, J.M.; Hogberg, H.T.; Hartung, T.; Smirnova, L. Suitability of 3D Human Brain Spheroid Models to Distinguish Toxic Effects of Gold and Poly-Lactic Acid Nanoparticles to Assess Biocompatibility for Brain Drug Delivery. Part. Fibre Toxicol. 2019, 16, 22. [Google Scholar] [CrossRef] [PubMed]
- Tigges, J.; Bielec, K.; Brockerhoff, G.; Hildebrandt, B.; Hübenthal, U.; Kapr, J.; Koch, K.; Teichweyde, N.; Wieczorek, D.; Rossi, A.; et al. Academic Application of Good Cell Culture Practice for Induced Pluripotent Stem Cells. ALTEX 2021, 38, 595–614. [Google Scholar] [CrossRef] [PubMed]
- Bartmann, K.; Hartmann, J.; Kapr, J.; Fritsche, E. Measurement of of Differentiated Human IPSC-Derived Neurospheres Recorded by Microelectrode Arrays (MEA). In Experimental Neurotoxicology Methods; Humana: New York, NY, USA, 2021; pp. 473–488. ISBN 978-1-0716-1637-6. [Google Scholar]
- Koch, K.; Bartmann, K.; Hartmann, J.; Kapr, J.; Klose, J.; Kuchovská, E.; Pahl, M.; Schlüppmann, K.; Zühr, E.; Fritsche, E. Scientific Validation of Human Neurosphere Assays for Developmental Neurotoxicity Evaluation. Front. Toxicol. 2022, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Bartmann, K.; Bendt, F.; Dönmez, A.; Haag, D.; Keßel, E.; Masjosthusmann, S.; Noel, C.; Wu, J.; Zhou, P.; Fritsche, E. A Human IPSC-Based in Vitro Neural Network Formation Assay to Investigate Neurodevelopmental Toxicity of Pesticides. bioRxiv 2023, 1–41. [Google Scholar] [CrossRef]
- Walter, K.M.; Dach, K.; Hayakawa, K.; Giersiefer, S.; Heuer, H.; Lein, P.J.; Fritsche, E. Ontogenetic Expression of Thyroid Hormone Signaling Genes: An in Vitro and in Vivo Species Comparison. PLoS ONE 2019, 14, e0221230. [Google Scholar] [CrossRef]
- Park, D.; Xiang, A.P.; Mao, F.F.; Zhang, L.; Di, C.G.; Liu, X.M.; Shao, Y.; Ma, B.F.; Lee, J.H.; Ha, K.S.; et al. Nestin Is Required for the Proper Self-Renewal of Neural Stem Cells. Stem Cells 2010, 28, 2162–2171. [Google Scholar] [CrossRef]
- Sansom, S.N.; Griffiths, D.S.; Faedo, A.; Kleinjan, D.J.; Ruan, Y.; Smith, J.; Van Heyningen, V.; Rubenstein, J.L.; Livesey, F.J. The Level of the Transcription Factor Pax6 Is Essential for Controlling the Balance between Neural Stem Cell Self-Renewal and Neurogenesis. PLoS Genet. 2009, 5, e1000511. [Google Scholar] [CrossRef]
- Honegger, P.; Lenoir, D.; Favrod, P. Growth and Differentiation of Aggregating Fetal Brain Cells in a Serum-Free Defined Medium. Nature 1979, 282, 305–308. [Google Scholar] [CrossRef]
- Tao, Y.; Zhang, S.C. Neural Subtype Specification from Human Pluripotent Stem Cells. Cell Stem Cell 2016, 19, 573–586. [Google Scholar] [CrossRef]
- Leonzino, M.; Busnelli, M.; Antonucci, F.; Verderio, C.; Mazzanti, M.; Chini, B. The Timing of the Excitatory-to-Inhibitory GABA Switch Is Regulated by the Oxytocin Receptor via KCC2. Cell Rep. 2016, 15, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Maccioni, R.B.; Cambiazo, V. Role of Microtubule-Associated Proteins in the Control of Microtubule Assembly. Physiol. Rev. 1995, 75, 835–864. [Google Scholar] [CrossRef]
- Holst, C.B.; Brøchner, C.B.; Vitting-Seerup, K.; Møllgård, K. Astrogliogenesis in Human Fetal Brain: Complex Spatiotemporal Immunoreactivity Patterns of GFAP, S100, AQP4 and YKL-40. J. Anat. 2019, 235, 590–615. [Google Scholar] [CrossRef]
- Rowley, N.M.; Madsen, K.K.; Schousboe, A.; White, H.S. Glutamate and GABA Synthesis, Release, Transport and Metabolism as Targets for Seizure Control. Neurochem. Int. 2012, 61, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.B.; Nilsen, L.H.; Eyjolfsson, E.M.; Vestergaard, H.T.; Hansen, S.L.; Schousboe, A.; Sonnewald, U.; Waagepetersen, H.S. GAD65 Is Essential for Synthesis of GABA Destined for Tonic Inhibition Regulating Epileptiform Activity. J. Neurochem. 2010, 115, 1398–1408. [Google Scholar] [CrossRef]
- Niciu, M.J.; Kelmendi, B.; Sanacora, G. Overview of Glutamatergic Neurotransmission in the Nervous System. Pharmacol. Biochem. Behav. 2012, 100, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Bormann, J. The “ABC” of GABA Receptors. Trends Pharmacol. Sci. 2000, 21, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Popova, E. Ionotropic GABA Receptors and Distal Retinal ON and OFF Responses. Scientifica 2014, 2014, 149187. [Google Scholar] [CrossRef]
- Fan, L.; Tan, L.; Chen, Z.; Qi, J.; Nie, F.; Luo, Z.; Cheng, J.; Wang, S. Haloperidol Bound D2 Dopamine Receptor Structure Inspired the Discovery of Subtype Selective Ligands. Nat. Commun. 2020, 11, 1074. [Google Scholar] [CrossRef]
- Martel, J.C.; McArthur, S.G. Dopamine Receptor Subtypes, Physiology and Pharmacology: New Ligands and Concepts in Schizophrenia. Front. Pharmacol. 2020, 11, 1003. [Google Scholar] [CrossRef]
- Sagarduy, A.; Llorente, J.; Miguelez, C.; Morera-Herreras, T.; Ruiz-Ortega, J.A.; Ugedo, L. Buspirone Requires the Intact Nigrostriatal Pathway to Reduce the Activity of the Subthalamic Nucleus via 5-HT1A Receptors. Exp. Neurol. 2016, 277, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Qujeq, D.; Roushan, T.; Norouzy, A.; Habibi-Rezaei, M.; Mehdinejad-Shani, M. Effects of Dichlorvos and Carbaryl on the Activity of Free and Immobilized Acetylcholinesterase. Toxicol. Ind. Health 2012, 28, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Smulders, C.J.G.M.; Bueters, T.J.H.; Van Kleef, R.G.D.M.; Vijverberg, H.P.M. Selective Effects of Carbamate Pesticides on Rat Neuronal Nicotinic Acetylcholine Receptors and Rat Brain Acetylcholinesterase. Toxicol. Appl. Pharmacol. 2003, 193, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Huang, C.S.; Song, J.H.; Narahashi, T. Direct Actions of Anticholinesterases on the Neuronal Nicotinic Acetylcholine Receptor Channels. Brain Res. 1997, 769, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Sepich, C.; Lukas, R.J.; Zhu, G.; Chang, Y. Emamectin Is a Non-Selective Allosteric Activator of Nicotinic Acetylcholine Receptors and GABAA/C Receptors. Biochem. Biophys. Res. Commun. 2016, 473, 795–800. [Google Scholar] [CrossRef]
- Patterson, T.A.; Eppler, B.; Dawson, R. Attenuation of Trimethyltin-Evoked Glutamate (GLU) Efflux from Rat Cortical and Hippocampal Slices. Neurotoxicol. Teratol. 1996, 18, 697–702. [Google Scholar] [CrossRef]
- Bal-Price, A.K.; Suñol, C.; Weiss, D.G.; van Vliet, E.; Westerink, R.H.S.; Costa, L.G. Application of in Vitro Neurotoxicity Testing for Regulatory Purposes: Symposium III Summary and Research Needs. Neurotoxicology 2008, 29, 520–531. [Google Scholar] [CrossRef]
- Coecke, S.; Eskes, C.; Gartlon, J.; Kinsner, A.; Price, A.; Van Vliet, E.; Prieto, P.; Boveri, M.; Bremer, S.; Adler, S.; et al. The Value of Alternative Testing for Neurotoxicity in the Context of Regulatory Needs. Environ. Toxicol. Pharmacol. 2006, 21, 153–167. [Google Scholar] [CrossRef]
- Tate, K.; Kirk, B.; Tseng, A.; Ulffers, A.; Litwa, K. Effects of the Selective Serotonin Reuptake Inhibitor Fluoxetine on Developing Neural Circuits in a Model of the Human Fetal Cortex. Int. J. Mol. Sci. 2021, 22, 10457. [Google Scholar] [CrossRef]
- Tukker, A.M.; Bouwman, L.M.S.; van Kleef, R.G.D.M.; Hendriks, H.S.; Legler, J.; Westerink, R.H.S. Perfluorooctane Sulfonate (PFOS) and Perfluorooctanoate (PFOA) Acutely Affect Human A1β2γ2L GABAA Receptor and Spontaneous Neuronal Network Function in Vitro. Sci. Rep. 2020, 10, 5311. [Google Scholar] [CrossRef]
- Sirenko, O.; Parham, F.; Dea, S.; Sodhi, N.; Biesmans, S.; Mora-Castilla, S.; Ryan, K.; Behl, M.; Chandy, G.; Crittenden, C.; et al. Functional and Mechanistic Neurotoxicity Profiling Using Human IPSC-Derived Neural 3D Cultures. Toxicol. Sci. 2019, 167, 58–76. [Google Scholar] [CrossRef] [PubMed]
- van Melis, L.V.J.; Heusinkveld, H.J.; Langendoen, C.; Peters, A.; Westerink, R.H.S. Organophosphate Insecticides Disturb Neuronal Network Development and Function via Non-AChE Mediated Mechanisms. Neurotoxicology 2022, 94, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Loser, D.; Grillberger, K.; Hinojosa, M.G.; Blum, J.; Haufe, Y.; Danker, T.; Johansson, Y.; Möller, C.; Nicke, A.; Bennekou, S.H.; et al. Acute Effects of the Imidacloprid Metabolite Desnitro-Imidacloprid on Human NACh Receptors Relevant for Neuronal Signaling. Arch. Toxicol. 2021, 95, 3695–3716. [Google Scholar] [CrossRef]
- Kosodo, Y.; Röper, K.; Haubensak, W.; Marzesco, A.M.; Corbeil, D.; Huttner, W.B. Asymmetric Distribution of the Apical Plasma Membrane during Neurogenic Divisions of Mamalian Neuroepithelial Cells. EMBO J. 2004, 23, 2314–2324. [Google Scholar] [CrossRef] [PubMed]
- Bayatti, N.; Sarma, S.; Shaw, C.; Eyre, J.A.; Vouyiouklis, D.A.; Lindsay, S.; Clowry, G.J. Progressive Loss of PAX6, TBR2, NEUROD and TBR1 MRNA Gradients Correlates with Translocation of EMX2 to the Cortical Plate during Human Cortical Development. Eur. J. Neurosci. 2008, 28, 1449–1456. [Google Scholar] [CrossRef]
- Estivill-Torrus, G.; Pearson, H.; van Heyningen, V.; Price, D.J.; Rashbass, P. Pax6 Is Required to Regulate the Cell Cycle and the Rate of Progression from Symmetrical to Asymmetrical Division in Mammalian Cortical Progenitors. Development 2002, 129, 455–466. [Google Scholar] [CrossRef]
- Togo, K.; Fukusumi, H.; Shofuda, T.; Ohnishi, H.; Yamazaki, H.; Hayashi, M.K.; Kawasaki, N.; Takei, N.; Nakazawa, T.; Saito, Y.; et al. Postsynaptic Structure Formation of Human IPS Cell-Derived Neurons Takes Longer than Presynaptic Formation during Neural Differentiation in Vitro. Mol. Brain 2021, 14, 149. [Google Scholar] [CrossRef]
- Reemst, K.; Noctor, S.C.; Lucassen, P.J.; Hol, E.M. The Indispensable Roles of Microglia and Astrocytes during Brain Development. Front. Hum. Neurosci. 2016, 10, 566. [Google Scholar] [CrossRef]
- Mahmoud, S.; Gharagozloo, M.; Simard, C.; Gris, D. Astrocytes Maintain Glutamate Homeostasis in the CNS by Controlling the Balance between Glutamate Uptake and Release. Cells 2019, 8, 184. [Google Scholar] [CrossRef]
- Oyanagi, K.; Tashiro, T.; Negishi, T. Cell-Type-Specific and Differentiation-Status-Dependent Variations in Cytotoxicity of Tributyltin in Cultured Rat Cerebral Neurons and Astrocytes. J. Toxicol. Sci. 2015, 40, 459–468. [Google Scholar] [CrossRef]
- Laurenza, I.; Pallocca, G.; Mennecozzi, M.; Scelfo, B.; Pamies, D.; Bal-Price, A. A Human Pluripotent Carcinoma Stem Cell-Based Model for in Vitro Developmental Neurotoxicity Testing: Effects of Methylmercury, Lead and Aluminum Evaluated by Gene Expression Studies. Int. J. Dev. Neurosci. 2013, 31, 679–691. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Peng, J.; Behl, M.; Sipes, N.S.; Shockley, K.R.; Rao, M.S.; Tice, R.R.; Zeng, X. Comparative Neurotoxicity Screening in Human IPSC-Derived Neural Stem Cells, Neurons and Astrocytes. Brain Res. 2016, 1638, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, F.; Liao, Y.; Jin, Y.; Sun, G. Effects of Arsenite in Astrocytes on Neuronal Signaling Transduction. Toxicology 2013, 303, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Guttenplan, K.A.; Weigel, M.K.; Prakash, P.; Wijewardhane, P.R.; Hasel, P.; Rufen-Blanchette, U.; Münch, A.E.; Blum, J.A.; Fine, J.; Neal, M.C.; et al. Neurotoxic Reactive Astrocytes Induce Cell Death via Saturated Lipids. Nature 2021, 599, 102–107. [Google Scholar] [CrossRef]
- Stary, C.M.; Sun, X.; Giffard, R.G. Astrocytes Protect against Isoflurane Neurotoxicity by Buffering Pro-Brain–Derived Neurotrophic Factor. Anesthesiology 2015, 123, 810–819. [Google Scholar] [CrossRef]
- Brüll, M.; Spreng, A.S.; Gutbier, S.; Loser, D.; Krebs, A.; Reich, M.; Kraushaar, U.; Britschgi, M.; Patsch, C.; Leist, M. Incorporation of Stem Cell-Derived Astrocytes into Neuronal Organoids to Allow Neuro-Glial Interactions in Toxicological Studies. ALTEX 2020, 37, 409–428. [Google Scholar] [CrossRef]
- Crofton, K.M.; Bassan, A.; Behl, M.; Chushak, Y.G.; Fritsche, E.; Gearhart, J.M.; Marty, M.S.; Mumtaz, M.; Pavan, M.; Ruiz, P.; et al. Current Status and Future Directions for a Neurotoxicity Hazard Assessment Framework That Integrates in Silico Approaches. Comput. Toxicol. 2022, 22, 100223. [Google Scholar] [CrossRef]
- Betarbet, R.; Sherer, T.B.; Mackenzie, G.; Garcia-osuna, M.; Panov, A.V.; Greenamyre, J.T. Chronic Systemic Pesticide Exposure Produces Pd Symptoms Betarbet. Nat. Neurosci. 2000, 26, 1301–1306. [Google Scholar] [CrossRef]
- Jokanović, M. Neurotoxic Effects of Organophosphorus Pesticides and Possible Association with Neurodegenerative Diseases in Man: A Review. Toxicology 2018, 410, 125–131. [Google Scholar] [CrossRef]
- Pauly, M.G.; Krajka, V.; Stengel, F.; Seibler, P.; Klein, C.; Capetian, P. Adherent vs. Free-Floating Neural Induction by Dual SMAD Inhibition for Neurosphere Cultures Derived from Human Induced Pluripotent Stem Cells. Front. Cell Dev. Biol. 2018, 6, 3. [Google Scholar] [CrossRef]
- Nadadhur, A.G.; Leferink, P.S.; Holmes, D.; Hinz, L.; Cornelissen-Steijger, P.; Gasparotto, L.; Heine, V.M. Patterning Factors during Neural Progenitor Induction Determine Regional Identity and Differentiation Potential in Vitro. Stem Cell Res. 2018, 32, 25–34. [Google Scholar] [CrossRef] [PubMed]
- McCready, F.P.; Gordillo-Sampedro, S.; Pradeepan, K.; Martinez-Trujillo, J.; Ellis, J. Multielectrode Arrays for Functional Phenotyping of Neurons from Induced Pluripotent Stem Cell Models of Neurodevelopmental Disorders. Biology 2022, 11, 316. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, Y.; Masu, M.; Ishii, T.; Shigemoto, R.; Nakanishi, S. A Family of Metabotropic Glutamate Receptors. Neuron 1992, 8, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Bodzęta, A.; Scheefhals, N.; MacGillavry, H.D. Membrane Trafficking and Positioning of MGluRs at Presynaptic and Postsynaptic Sites of Excitatory Synapses. Neuropharmacology 2021, 200, 108799. [Google Scholar] [CrossRef] [PubMed]
- Panatier, A.; Poulain, D.A.; Oliet, S.H.R. Regulation of Transmitter Release by High-Affinity Group III MGluRs in the Supraoptic Nucleus of the Rat Hypothalamus. Neuropharmacology 2004, 47, 333–341. [Google Scholar] [CrossRef]
- Mateo, Z.; Porter, J.T. Group II Metabotropic Glutamate Receptors Inhibit Glutamate Release at Thalamocortical Synapses in the Developing Somatosensory Cortex. Neuroscience 2007, 146, 1062–1072. [Google Scholar] [CrossRef]
- Bocchio, M.; Lukacs, I.P.; Stacey, R.; Plaha, P.; Apostolopoulos, V.; Livermore, L.; Sen, A.; Ansorge, O.; Gillies, M.J.; Somogyi, P.; et al. Group II Metabotropic Glutamate Receptors Mediate Presynaptic Inhibition of Excitatory Transmission in Pyramidal Neurons of the Human Cerebral Cortex. Front. Cell. Neurosci. 2019, 12, 508. [Google Scholar] [CrossRef]
- Van Den Pol, A.N.; Gao, X.B.; Patrylo, P.R.; Ghosh, P.K.; Obrietan, K. Glutamate Inhibits GABA Excitatory Activity in Developing Neurons. J. Neurosci. 1998, 18, 10749–10761. [Google Scholar] [CrossRef]
- Görtz, P.; Henning, U.; Theiss, S.; Lange-Asschenfeldt, C. Effect Fingerprints of Antipsychotic Drugs on Neural Networks in Vitro. J. Neural Transm. 2019, 126, 1363–1371. [Google Scholar] [CrossRef]
- Gemperle, A.Y.; Enz, A.; Pozza, M.F.; Lüthi, A.; Olpe, H.R. Effects of Clozapine, Haloperidol and Iloperidone on Neurotransmission and Synaptic Plasticity in Prefrontal Cortex and Their Accumulation in Brain Tissue: An in Vitro Study. Neuroscience 2003, 117, 681–695. [Google Scholar] [CrossRef]
- Chen, W.; Zhu, F.; Guo, J.; Sheng, J.; Li, W.; Zhao, X.; Wang, G.; Li, K. Chronic Haloperidol Increases Voltage-Gated Na+ Currents in Mouse Cortical Neurons. Biochem. Biophys. Res. Commun. 2014, 450, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Dzyubenko, E.; Juckel, G.; Faissner, A. The Antipsychotic Drugs Olanzapine and Haloperidol Modify Network Connectivity and Spontaneous Activity of Neural Networks in Vitro. Sci. Rep. 2017, 7, 11609. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, P.; Martin, M.; LeFew, W.R.; Ross, J.; Houck, K.A.; Shafer, T.J. Multi-Well Microelectrode Array Recordings Detect Neuroactivity of ToxCast Compounds. Neurotoxicology 2014, 44, 204–217. [Google Scholar] [CrossRef] [PubMed]
- Smulders, C.J.G.M.; Van Kleef, R.G.D.M.; de Groot, A.; Gotti, C.; Vijverberg, H.P.M. A Noncompetitive, Sequential Mechanism for Inhibition of Rat α 4β2 Neuronal Nicotinic Acetylcholine Receptors by Carbamate Pesticides. Toxicol. Sci. 2004, 82, 219–227. [Google Scholar] [CrossRef]
- McConnell, E.R.; McClain, M.A.; Ross, J.; LeFew, W.R.; Shafer, T.J. Evaluation of Multi-Well Microelectrode Arrays for Neurotoxicity Screening Using a Chemical Training Set. Neurotoxicology 2012, 33, 1048–1057. [Google Scholar] [CrossRef]
- Defranchi, E.; Novellino, A.; Whelan, M.; Vogel, S.; Ramirez, T.; van Ravenzwaay, B.; Landsiedel, R. Feasibility Assessment of Micro-Electrode Chip Assay as a Method of Detecting Neurotoxicity in Vitro. Front. Neuroeng. 2011, 4, 6. [Google Scholar] [CrossRef]
- Alloisio, S.; Nobile, M.; Novellino, A. Multiparametric Characterisation of Neuronal Network Activity for in Vitro Agrochemical Neurotoxicity Assessment. Neurotoxicology 2015, 48, 152–165. [Google Scholar] [CrossRef]
- Dingemans, M.M.L.; Schütte, M.G.; Wiersma, D.M.M.; de Groot, A.; van Kleef, R.G.D.M.; Wijnolts, F.M.J.; Westerink, R.H.S. Chronic 14-Day Exposure to Insecticides or Methylmercury Modulates Neuronal Activity in Primary Rat Cortical Cultures. Neurotoxicology 2016, 57, 194–202. [Google Scholar] [CrossRef]
- Kocaturk, S.; Guven, E.B.; Shah, F.; Tepper, J.M.; Assous, M. Cholinergic Control of Striatal GABAergic Microcircuits. Cell Rep. 2022, 41, 111531. [Google Scholar] [CrossRef]
- Schvartz, D.; González-Ruiz, V.; Walter, N.; Antinori, P.; Jeanneret, F.; Tonoli, D.; Boccard, J.; Zurich, M.G.; Rudaz, S.; Monnet-Tschudi, F.; et al. Protein Pathway Analysis to Study Development-Dependent Effects of Acute and Repeated Trimethyltin (TMT) Treatments in 3D Rat Brain Cell Cultures. Toxicol. Vitr. 2019, 60, 281–292. [Google Scholar] [CrossRef]
- Brock, T.O.; O’Callaghan, J.P. Quantitative Changes in the Synaptic Vesicle Proteins Synapsin I and P38 and the Astrocyte-Specific Protein Glial Fibrillary Acidic Protein Are Associated with Chemical-Induced Injury to the Rat Central Nervous System. J. Neurosci. 1987, 7, 931–942. [Google Scholar] [CrossRef]
- Amin, H.; Marinaro, F.; Tonelli, D.D.P.; Berdondini, L. Developmental Excitatory-to-Inhibitory GABA-Polarity Switch Is Disrupted in 22q11.2 Deletion Syndrome: A Potential Target for Clinical Therapeutics. Sci. Rep. 2017, 7, 15752. [Google Scholar] [CrossRef] [PubMed]
- Deidda, G.; Parrini, M.; Naskar, S.; Bozarth, I.F.; Contestabile, A.; Cancedda, L. Reversing Excitatory GABA A R Signaling Restores Synaptic Plasticity and Memory in a Mouse Model of Down Syndrome. Nat. Med. 2015, 21, 318–326. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Nomura, T.; Xu, J.; Contractor, A. The Developmental Switch in GABA Polarity Is Delayed in Fragile X Mice. J. Neurosci. 2014, 34, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Braat, S.; Kooy, R.F. The GABAA Receptor as a Therapeutic Target for Neurodevelopmental Disorders. Neuron 2015, 86, 1119–1130. [Google Scholar] [CrossRef]
- Modafferi, S.; Zhong, X.; Kleensang, A.; Murata, Y.; Fagiani, F.; Pamies, D.; Hogberg, H.T.; Calabrese, V.; Lachman, H.; Hartung, T.; et al. Gene–Environment Interactions in Developmental Neurotoxicity: A Case Study of Synergy between Chlorpyrifos and Chd8 Knockout in Human Brainspheres. Environ. Health Perspect. 2021, 129, 077001. [Google Scholar] [CrossRef]
- Zhong, X.; Harris, G.; Smirnova, L.; Zufferey, V.; Baldino Russo, F.; Baleeiro Beltrao Braga, P.C.; Chesnut, M.; Zurich, M.G.; Hogberg, H.T.; Hartung, T.; et al. Antidepressant Paroxetine Exerts Developmental Neurotoxicity in an IPSC-Derived 3D Human Brain Model. Front. Cell. Neurosci. 2020, 14, 25. [Google Scholar] [CrossRef]
- Rockley, K.L.; Roberts, R.A.; Morton, M.J. Innovative Models for In Vitro Detection of Seizure. Toxicol. Res. 2019, 8, 784–788. [Google Scholar] [CrossRef]
- Tukker, A.M.; Westerink, R.H.S. Novel Test Strategies for in Vitro Seizure Liability Assessment. Expert Opin. Drug Metab. Toxicol. 2021, 17, 923–936. [Google Scholar] [CrossRef]
- Blum, J.; Masjosthusmann, S.; Bartmann, K.; Bendt, F.; Dolde, X.; Dönmez, A.; Förster, N.; Holzer, A.-K.; Hübenthal, U.; Keßel, H.E.; et al. Establishment of a Human Cell-Based in Vitro Battery to Assess Developmental Neurotoxicity Hazard of Chemicals. Chemosphere 2023, 311, 137035. [Google Scholar] [CrossRef]
- Crofton, K.M.; Mundy, W.R. External Scientific Report on the Interpretation of Data from the Developmental Neurotoxicity In Vitro Testing Assays for Use in Integrated Approaches for Testing and Assessment. EFSA Support. Publ. 2021, 18, 6924E. [Google Scholar] [CrossRef]



| Mode | Parameter | 2D-NIM | GNEIB | Stemdiff | |||
|---|---|---|---|---|---|---|---|
| CINDA+ | Electro | CINDA+ | Electro | CINDA+ | Electro | ||
| General electrical activity after 7 weeks on MEA | Active Wells [%] | 83 | 0 | 83 | 25 | 54 | 25 |
| Active electrodes [%] | 58 | 0 | 31 | 5 | 16 | 11 | |
| wMFR [Hz] (±SEM) | 6.86 (±0.82) | - | 6.29 (±1.25) | 1.94 (±0.65) | 0.84 (±0.29) | 4.3 (±0.63) | |
| mean burst frequency [Hz] (±SEM) | 0.31 (±0.04) | - | 0.36 (±0.06) | 0.009 (±0) | 0.06 (±0.02) | 0.25 (±0.03) | |
| mean network burst frequency [Hz] (±SEM) | 0.08 (±0.02) | - | 0.15 (±0.03) | - | 0.04 (±0.02) | 0.11 (±0.02) | |
| mean network burst percentage [%] (±SEM) | 42.86 (±5.04) | - | 42.28 (±6.32) | - | 41.68 (±14.39) | 48.02 (±3.13) | |
| Glutamatergic response | units responding to 50 µM glu with an increase (as % of unsorted units) | 20 | 27 | 30 | 43 | 15 | 26 |
| mean fold change in responding units to 50 µM glu with an increase (±SEM) | 3.98 (±1.12) | 4.73 (±2.28) | 5.43 (±2.18) | 7.71 (±2.23) | 2.12 (±0.42) | 1.80 (±0.20) | |
| units responding to 50 µM glu with a decrease (as % of unsorted units) | 57 | 54 | 37 | 27 | 77 | 52 | |
| mean fold change in responding units to 50 µM glu with a decrease (±SEM) | 0.16 (±0.03) | 0.13 (±0.06) | 0.32 (±0.05) | 0.13 (±0.05) | 0.10 (±0.03) | 0.50 (±0.06) | |
| units responding to 50 µM glu and 50 µM AP5/NBQX (as % of unsorted units) | 9 | 12 | 16 | 40 | 15 | 26 | |
| mean fold change in responding units to 50 µM glu and 50 µM AP5/NBQX (±SEM) | 0.23 (±0.08) | 0.33 (±0.18) | 0.23 (±0.05) | 0.03 (±0.01) | 0.04 (±0.02) | 0.17 (±0.07) | |
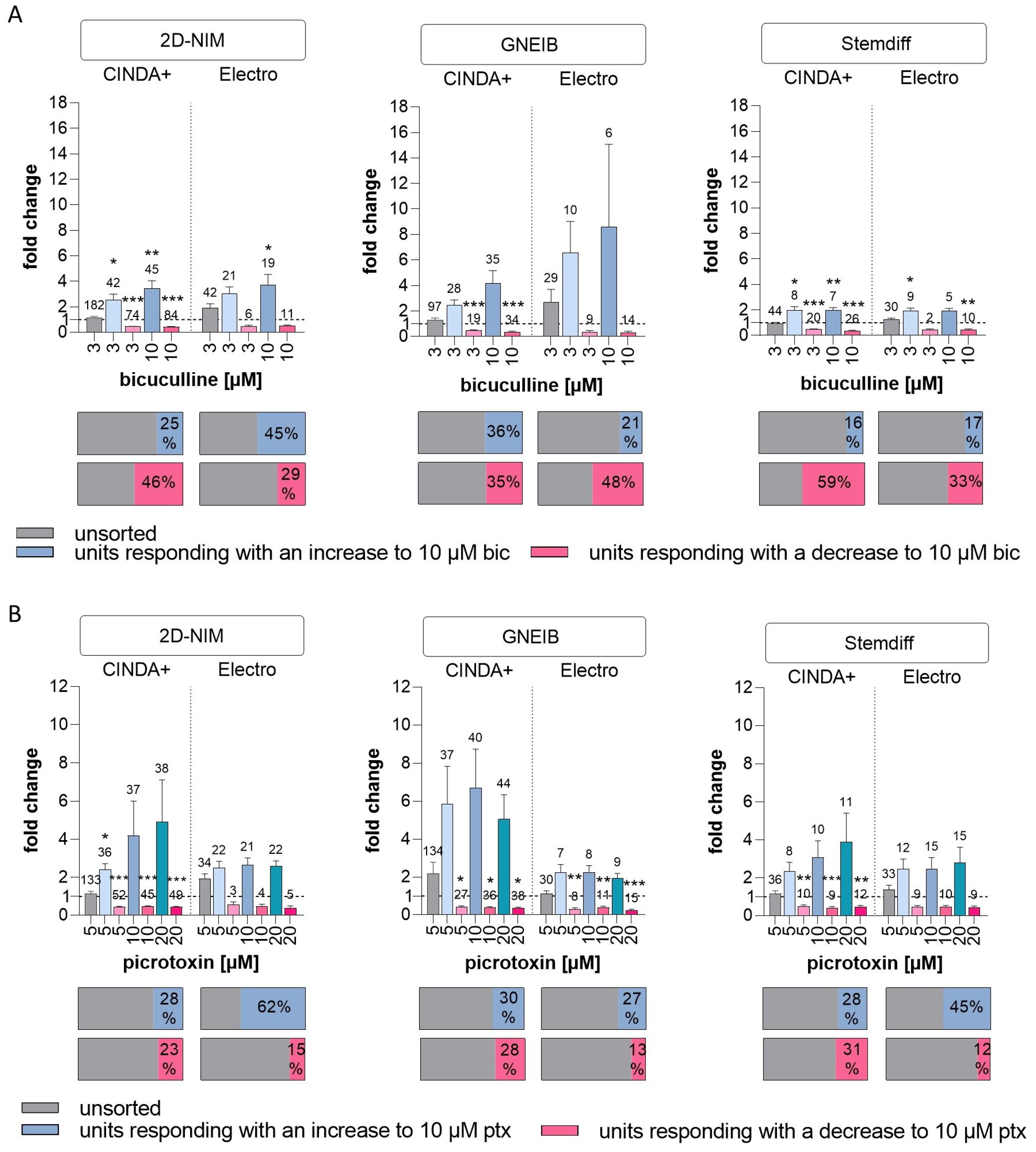
| GABAergic response | units responding with an increase to 10 µM bic (as % of unsorted units) | 25 | 45 | 36 | 21 | 16 | 17 |
| mean fold change in responding units with an increase to 10 µM bic (±SEM) | 3.45 (±0.60) | 3.7 (±0.85) | 4.17 (±0.99) | 8.58 (±6.48) | 1.99 (±0.20) | 1.92 (±0.2) | |
| units responding with a decrease to 10 µM bic (as % of unsorted units) | 46 | 29 | 35 | 48 | 59 | 33 | |
| mean fold change in responding units with a decrease to 10 µM bic (±SEM) | 0.43 (±0.02) | 0.50 (±0.07) | 0.37 (±0.04) | 0.33 (±0.08) | 0.37 (±0.04) | 0.42 (±0.09) | |
| units responding with an increase to 10 µM ptx (as % of unsorted units) | 28 | 62 | 30 | 27 | 28 | 45 | |
| mean fold change in responding units with an increase to 10 µM ptx (±SEM) | 4.19 (±1.81) | 2.66 (±0.36) | 6.69 (±2.05) | 2.25 (±0.35) | 3.06 (±0.89) | 2.47 (±0.59) | |
| units responding with a decrease to 10 µM ptx (as % of unsorted units) | 23 | 15 | 28 | 13 | 31 | 12 | |
| mean fold change in responding units with a decrease to 10 µM ptx (±SEM) | 0.47 (±0.04) | 0.48 (±0.11) | 0.39 (±0.04) | 0.39 (±0.07) | 0.40 (±0.08) | 0.46 (±0.07) | |
| Dopaminergic response | units responding with an increase to 1 µM halo (as % of unsorted units) | 20 | 0 | 16 | 6 | 15 | 9 |
| mean fold change in responding units with an increase to 1 µM halo (±SEM) | 3.23 (±0.53) | - | 7.82 (±1.88) | 10.25 (±0) | 1.71 (±0.26) | 1.99 (±0.44) | |
| units responding with a decrease to 1 µM halo (as % of unsorted units) | 57 | 85 | 65 | 89 | 73 | 47 | |
| mean fold change in responding units with a decrease to 1 µM halo (±SEM) | 0.28 (±0.02) | 0.15 (±0.05) | 0.21 (±0.02) | 0.13 (±0.04) | 0.29 (±0.04) | 0.32 (±0.06) | |
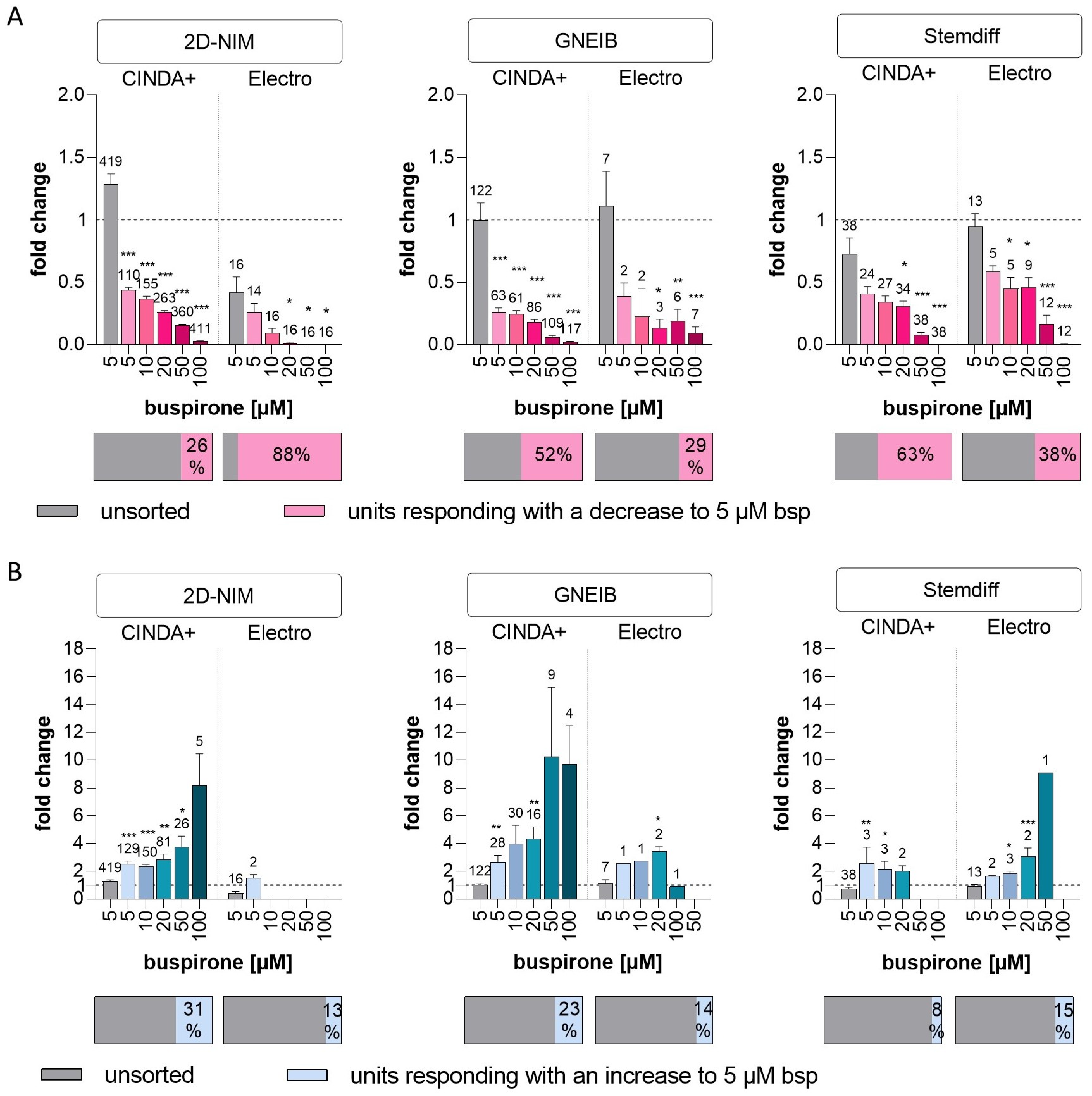
| Serotonergic response | units responding with a decrease to 5 µM bsp (as % of unsorted units) | 26 | 88 | 52 | 29 | 63 | 38 |
| mean fold change in responding units to 5 µM bsp (±SEM) | 0.44 (±0.02) | 0.26 (±0.07) | 0.26 (±0.03) | 0.39 (±0.10) | 0.41 (±0.06) | 0.59 (±0.05) | |
| units responding with an increase to 5 µM bsp (as % of unsorted units) | 31 | 13 | 23 | 14 | 8 | 15 | |
| mean fold change in responding units with an increase to 5 µM bsp (±SEM) | 2.49 (±0.24) | 1.51 (±0.26) | 2.67 (±0.47) | 2.56 (±0) | 2.56 (±1.17) | 1.63 (±0.06) | |
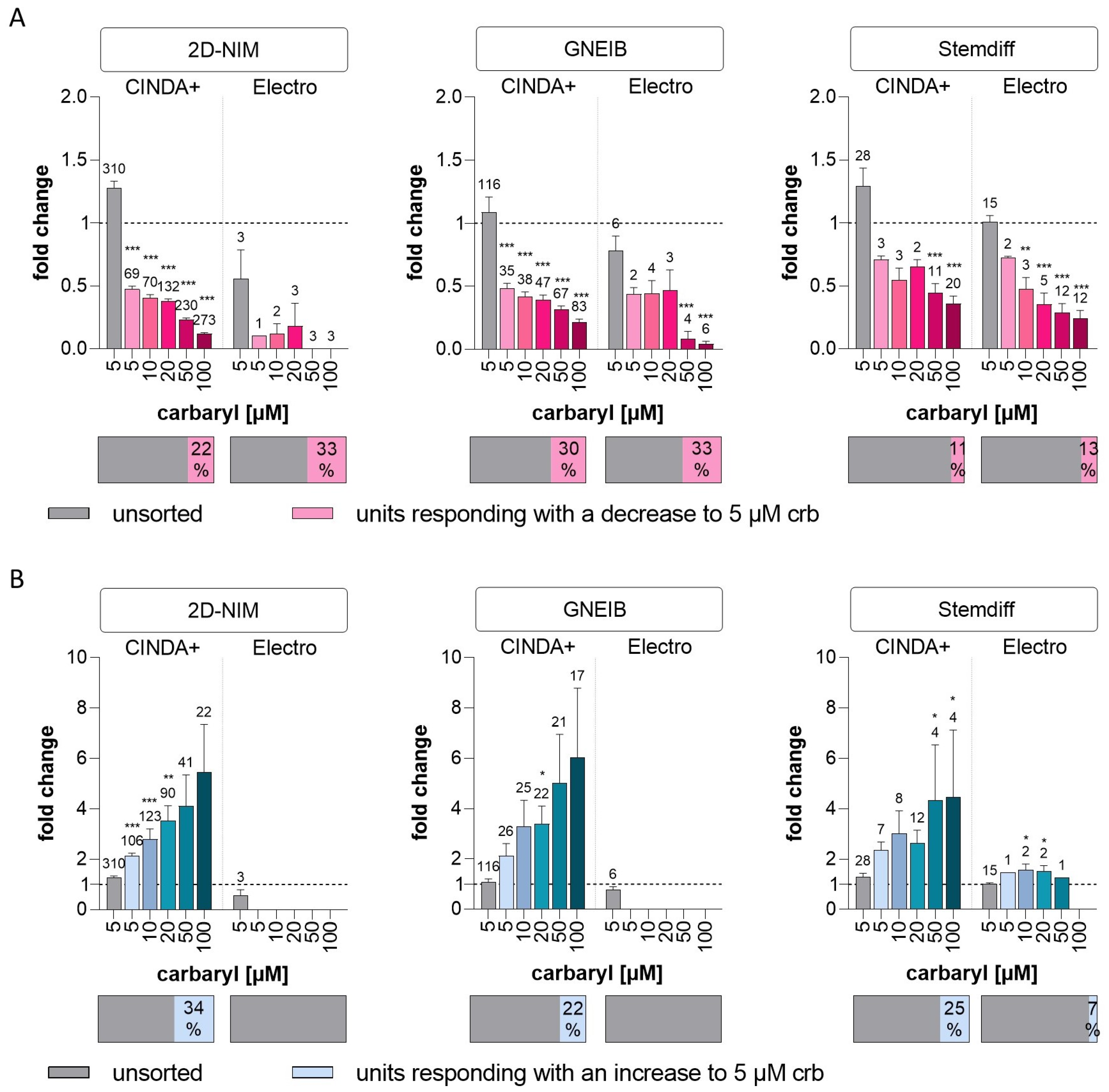
| Cholinergic response | units responding with a decrease to 5 µM crb (as % of unsorted units) | 22 | 33 | 30 | 33 | 11 | 13 |
| mean fold change in responding units with decreased wMFR to 5 µM crb (±SEM) | 0.47 (±0.03) | 0.10 (±0) | 0.48 (±0.04) | 0.44 (±0.05) | 0.71 (±0.03) | 0.72 (±0.01) | |
| units responding with an increase to 5 µM crb (as % of unsorted units) | 34 | 0 | 22 | 0 | 25 | 7 | |
| mean fold change in responding units with increased wMFR to 5 µM crb (±SEM) | 2.14 (±0.10) | - | 2.11 (±0.49) | - | 2.35 (±0.33) | 1.47 (±0) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hartmann, J.; Henschel, N.; Bartmann, K.; Dönmez, A.; Brockerhoff, G.; Koch, K.; Fritsche, E. Molecular and Functional Characterization of Different BrainSphere Models for Use in Neurotoxicity Testing on Microelectrode Arrays. Cells 2023, 12, 1270. https://doi.org/10.3390/cells12091270
Hartmann J, Henschel N, Bartmann K, Dönmez A, Brockerhoff G, Koch K, Fritsche E. Molecular and Functional Characterization of Different BrainSphere Models for Use in Neurotoxicity Testing on Microelectrode Arrays. Cells. 2023; 12(9):1270. https://doi.org/10.3390/cells12091270
Chicago/Turabian StyleHartmann, Julia, Noah Henschel, Kristina Bartmann, Arif Dönmez, Gabriele Brockerhoff, Katharina Koch, and Ellen Fritsche. 2023. "Molecular and Functional Characterization of Different BrainSphere Models for Use in Neurotoxicity Testing on Microelectrode Arrays" Cells 12, no. 9: 1270. https://doi.org/10.3390/cells12091270
APA StyleHartmann, J., Henschel, N., Bartmann, K., Dönmez, A., Brockerhoff, G., Koch, K., & Fritsche, E. (2023). Molecular and Functional Characterization of Different BrainSphere Models for Use in Neurotoxicity Testing on Microelectrode Arrays. Cells, 12(9), 1270. https://doi.org/10.3390/cells12091270