Distinct Traits of Structural and Regulatory Evolutional Conservation of Human Genes with Specific Focus on Major Cancer Molecular Pathways
Abstract
1. Introduction
2. Materials and Methods
2.1. dN/dS Data
2.2. Retroelement Enrichment Data
2.3. Functional Classification of Histone Modifications
2.4. Analysis of Gene Ontologies
2.5. Correlation Analysis
2.6. Molecular Pathways Databank
2.7. Pathway Visualizations
3. Results
3.1. Study Design
3.2. Aggregation of NGRE Scores
- -
- We first grouped the histone tag metrics according to their relation to active chromatin (H3K4me3, H3K9ac, H3K27ac, H3K4me1) or heterochromatin (H3K27me3, H3K9me3). Active chromatin NGREac = (NGREH3K9ac + NGRE H3K27ac + NGRE H3K4me3 + NGREH3K4me1)/4; heterochromatin NGREhc = (NGREH3K27me3 + NGREH3K9me3)/2.
- -
- Finally, using three functional tag components, we calculated aggregated NGRE metrics for the first time, which can be regarded as joint indexes of regulatory evolution for every gene under analysis (NGREAGG):
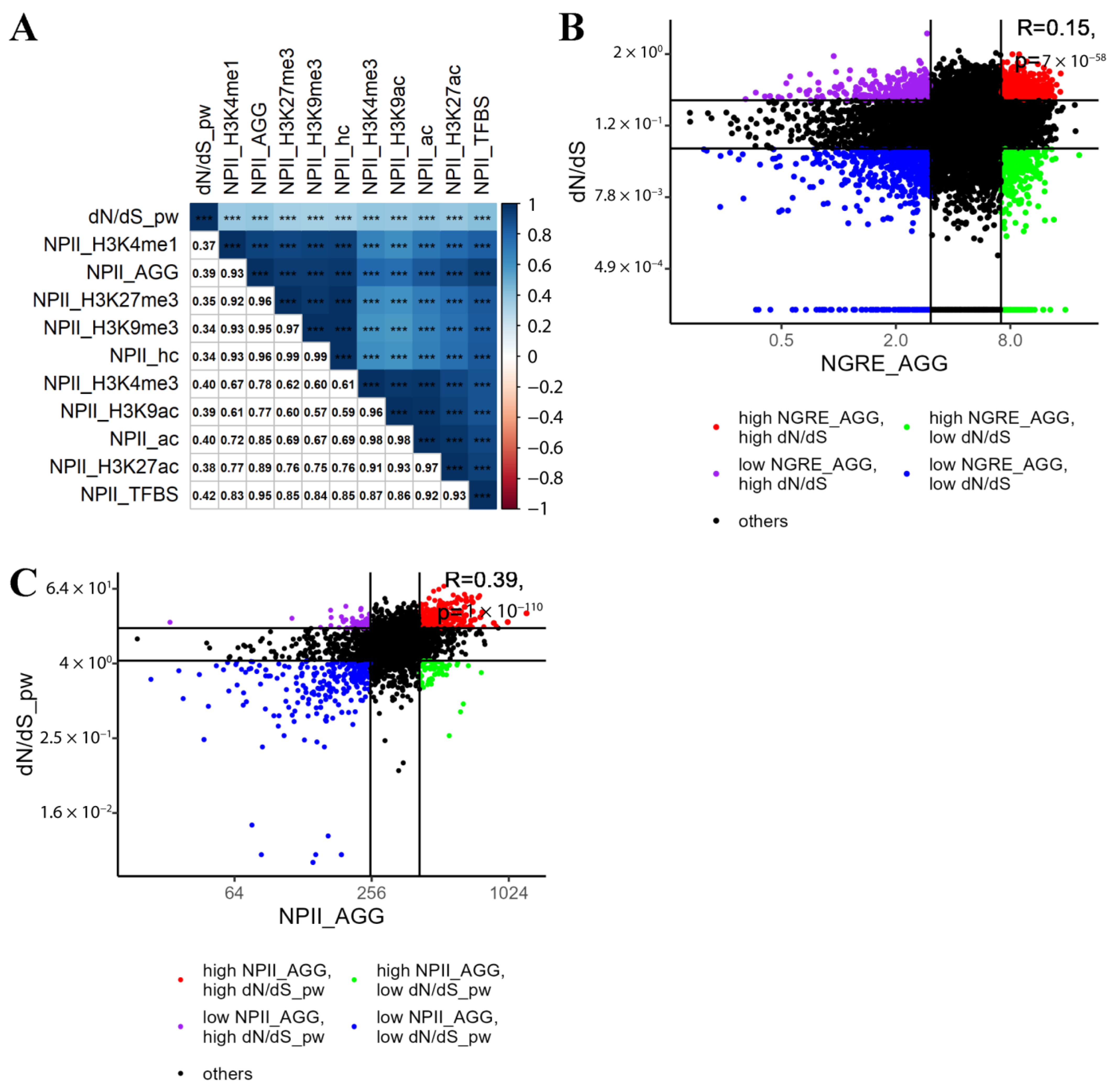
3.3. Correlation between Evolutionary Rate Metrics for Individual Genes
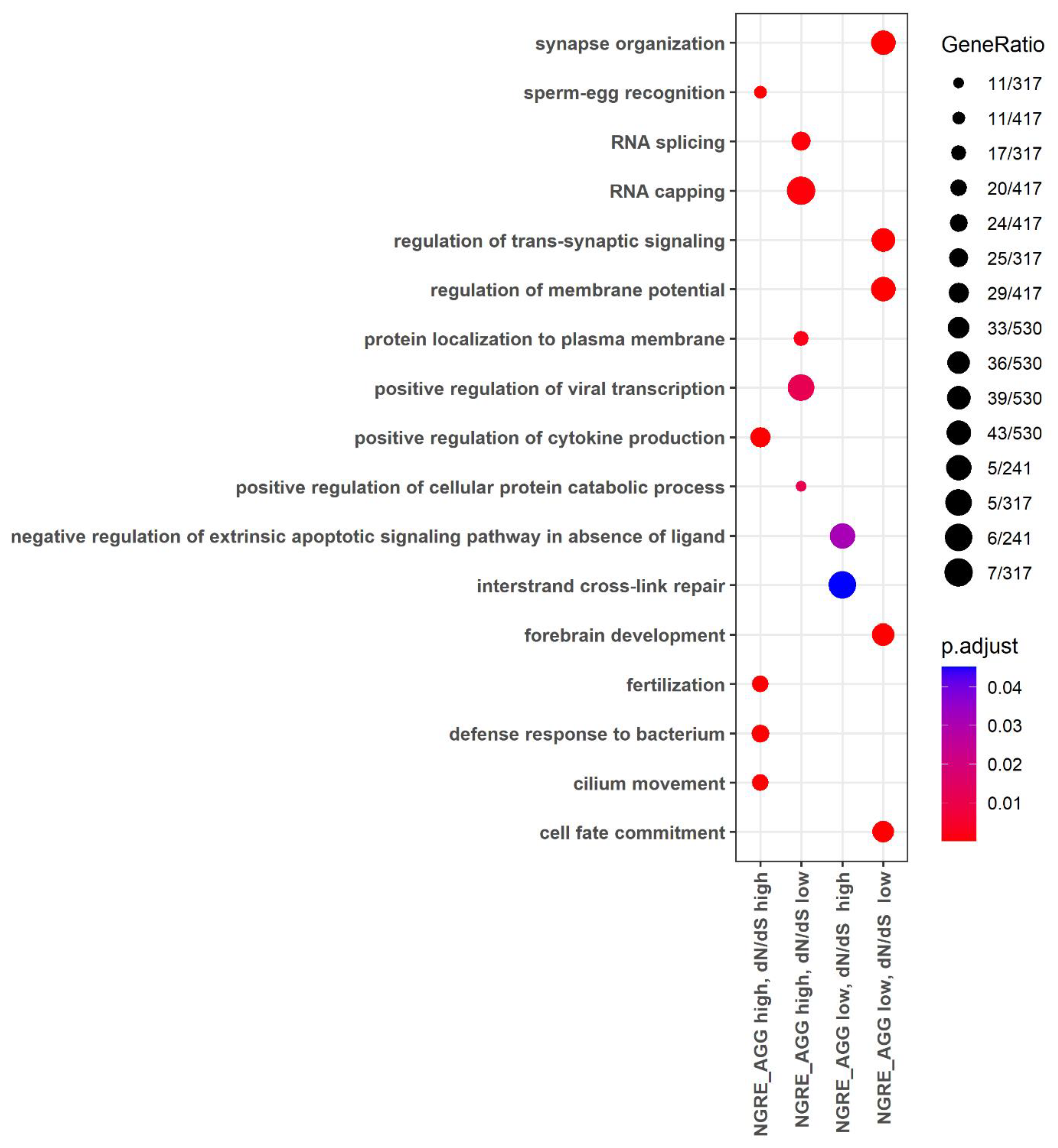
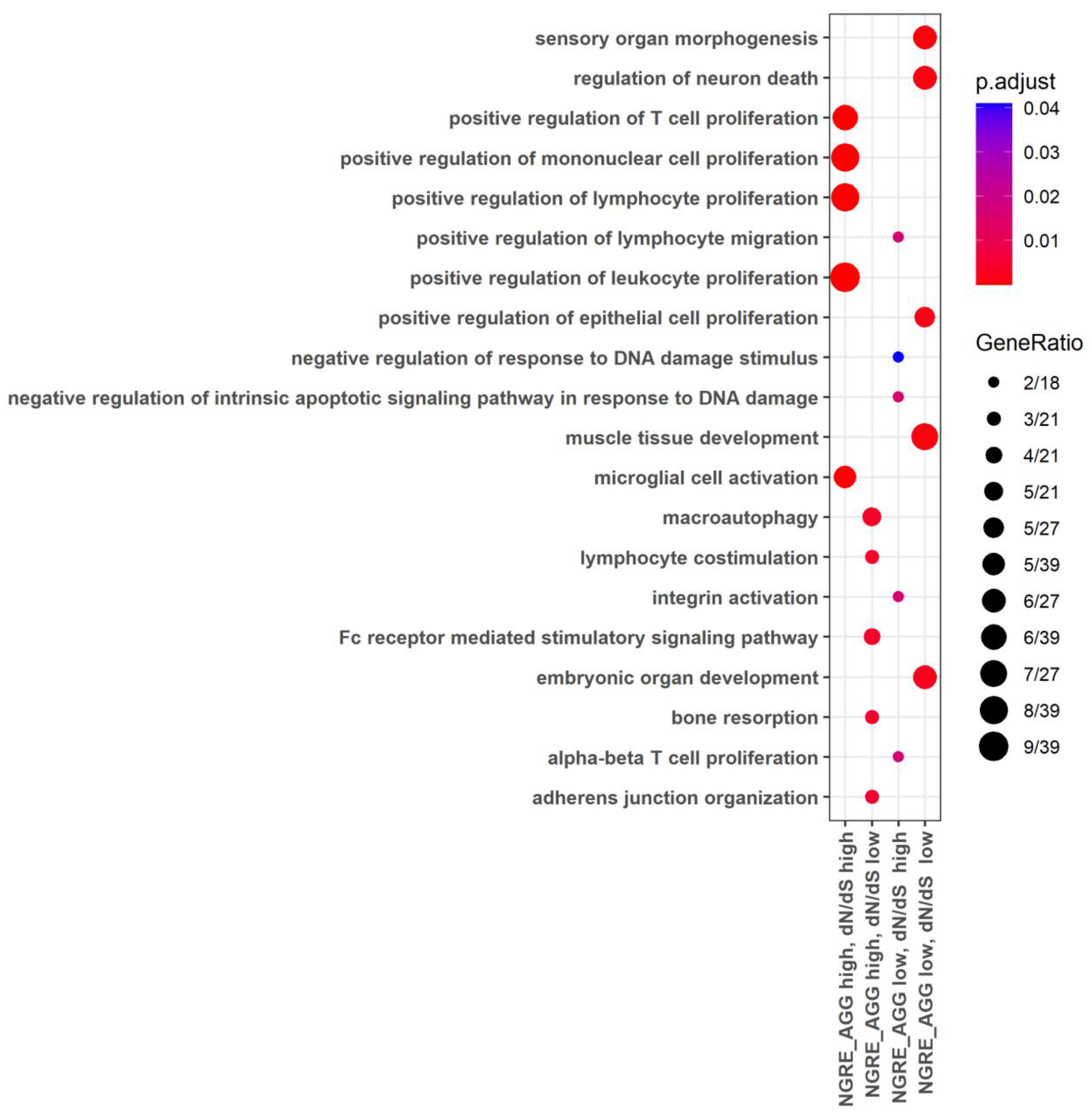
3.4. Functional Groups of Genes with the Lowest and with the Highest Evolutionary Rate Ranks
3.5. Molecular Pathways with the Lowest and with the Highest Evolutionary Rate Ranks
3.6. Database of Human Genes Structural and Regulatory Evolutionary Rate
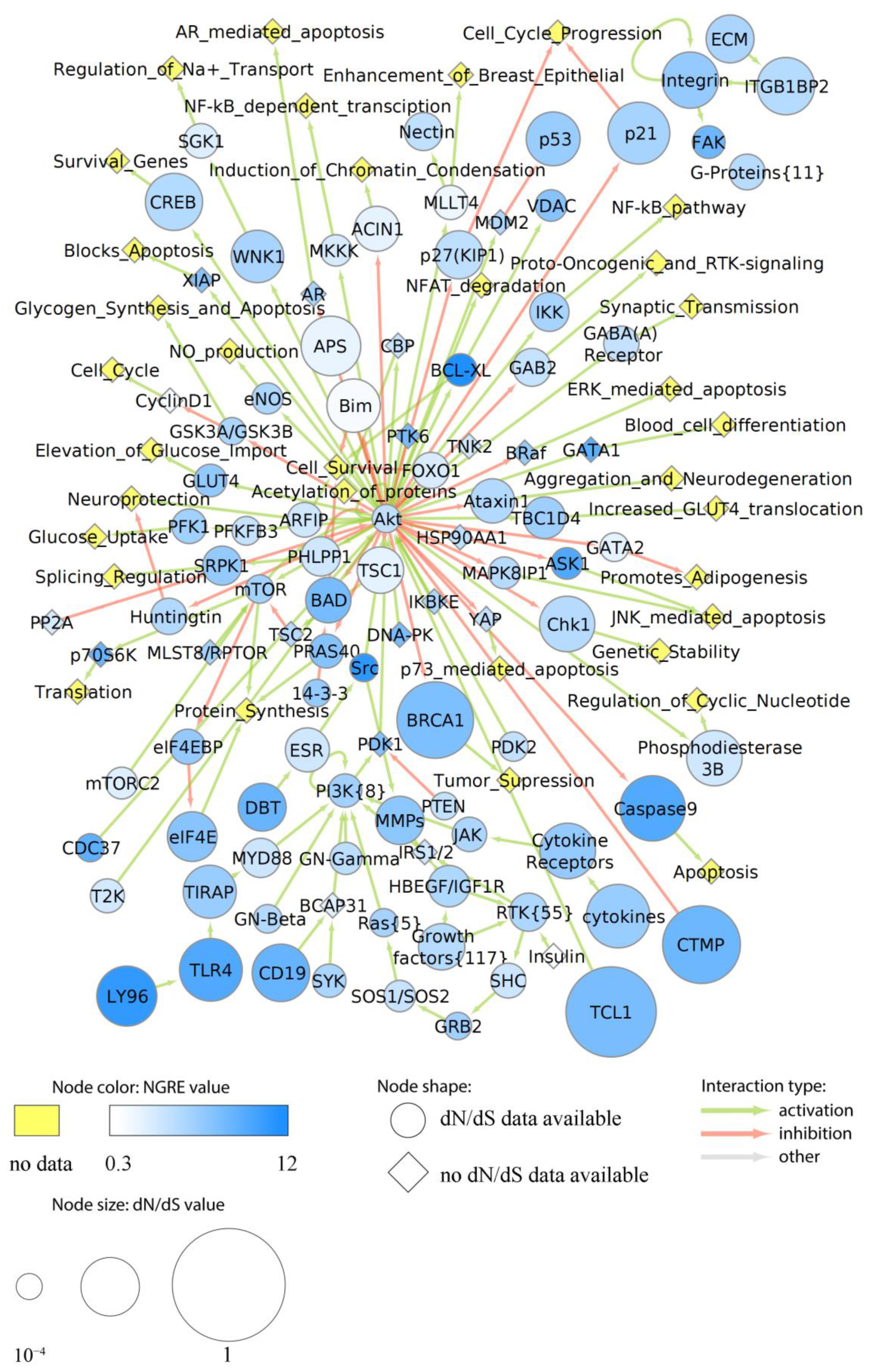
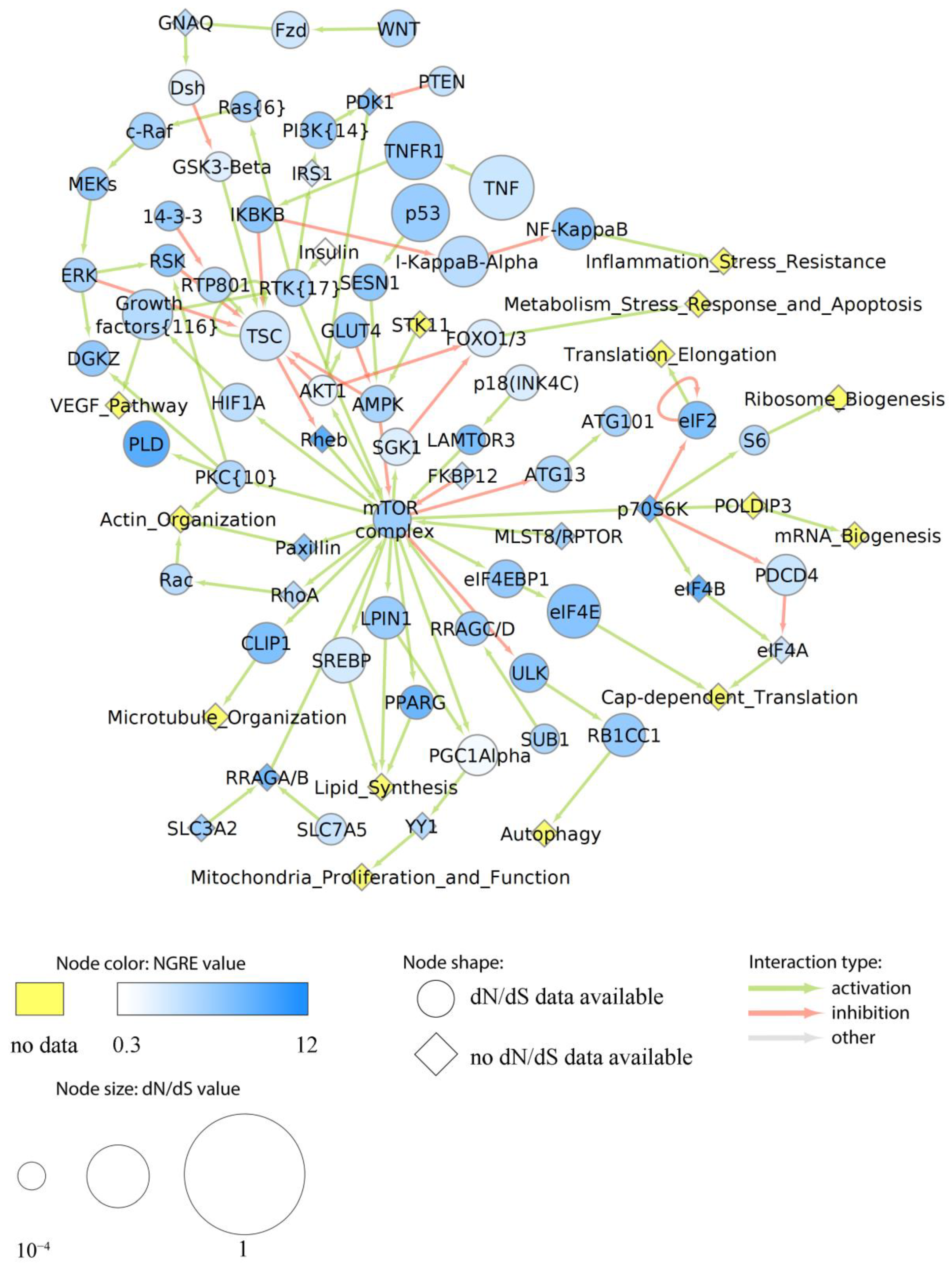
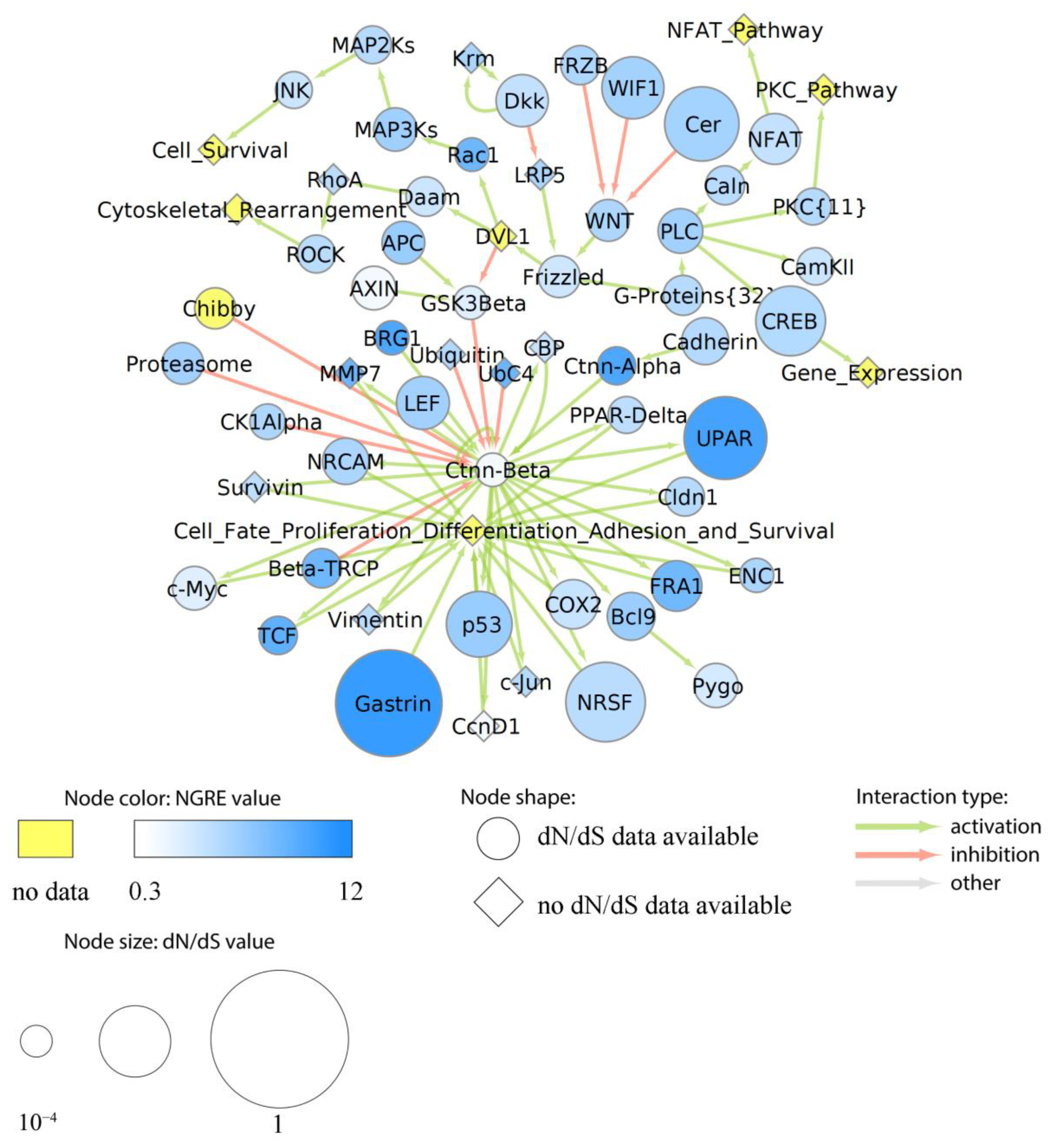
3.7. Major Cancer-Related Pathways in the Context of Structural and Regulatory Evolution of Human Genes
3.7.1. Rank of Cancer Pathways by Evolutional Metrics
3.7.2. Evolution of Functional Nodes of Cancer Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McGrath, C. Human Genetics: A Look in the Mirror. Genome Biol. Evol. 2020, 12, 1256–1257. [Google Scholar] [CrossRef]
- Hill, M.S.; Vande Zande, P.; Wittkopp, P.J. Molecular and evolutionary processes generating variation in gene expression. Nat. Rev. Genet. 2021, 22, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Peter, I.S.; Davidson, E.H. Evolution of gene regulatory networks controlling body plan development. Cell 2011, 144, 970–985. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, A.S.; Meerson, M.B.; Gerashchenko, M.V.; Kulakovskiy, I.V.; Dmitriev, S.E.; Gladyshev, V.N. Multifaceted deregulation of gene expression and protein synthesis with age. Proc. Natl. Acad. Sci. USA 2020, 117, 15581–15590. [Google Scholar] [CrossRef] [PubMed]
- Kinzina, E.D.; Podolskiy, D.I.; Dmitriev, S.E.; Gladyshev, V.N. Patterns of Aging Biomarkers, Mortality, and Damaging Mutations Illuminate the Beginning of Aging and Causes of Early-Life Mortality. Cell Rep. 2019, 29, 4276–4284.e3. [Google Scholar] [CrossRef]
- Suntsova, M.V.; Buzdin, A.A. Differences between human and chimpanzee genomes and their implications in gene expression, protein functions and biochemical properties of the two species. BMC Genom. 2020, 21, 535. [Google Scholar] [CrossRef]
- Kryazhimskiy, S.; Plotkin, J.B. The Population Genetics of dN/dS. PLoS Genet. 2008, 4, 1000304. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, A.; Crawford, D.L. Neutral and adaptive variation in gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 5425–5430. [Google Scholar] [CrossRef]
- Nikitin, D.; Penzar, D.; Garazha, A.; Sorokin, M.; Tkachev, V.; Borisov, N.; Poltorak, A.; Prassolov, V.; Buzdin, A.A. Profiling of human molecular pathways affected by retrotransposons at the level of regulation by transcription factor proteins. Front. Immunol. 2018, 9, 30. [Google Scholar] [CrossRef] [PubMed]
- Schumann, G.G.; Gogvadze, E.V.; Osanai-Futahashi, M.; Kuroki, A.; Münk, C.; Fujiwara, H.; Ivics, Z.; Buzdin, A.A. Unique Functions of Repetitive Transcriptomes. Int. Rev. Cell Mol. Biol. 2010, 285, 115–188. [Google Scholar]
- Gogvadze, E.; Buzdin, A. Retroelements and their impact on genome evolution and functioning. Cell. Mol. Life Sci. 2009, 66, 3727–3742. [Google Scholar] [CrossRef] [PubMed]
- Nefedova, L.N.; Kim, A. The role of retroelements in the evolution of animal genomes. Zhurnal Obs. Biol. 2021, 82, 13–25. [Google Scholar] [CrossRef]
- Nikitin, D.; Kolosov, N.; Murzina, A.; Pats, K.; Zamyatin, A.; Tkachev, V.; Sorokin, M.; Kopylov, P.; Buzdin, A. Retroelement-Linked H3K4me1 Histone Tags Uncover Regulatory Evolution Trends of Gene Enhancers and Feature Quickly Evolving Molecular Processes in Human Physiology. Cells 2019, 8, 1219. [Google Scholar] [CrossRef]
- Yi, S.V.; Goodisman, M.A.D. The impact of epigenetic information on genome evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20200114. [Google Scholar] [CrossRef]
- Simon, A.J.; Morrow, B.R.; Ellington, A.D. {Retroelement-Based} Genome Editing and Evolution. ACS Synth. Biol. 2018, 7, 2600–2611. [Google Scholar] [CrossRef]
- Yan, F.; Yu, X.; Duan, Z.; Lu, J.; Jia, B.; Qiao, Y.; Sun, C.; Wei, C. Discovery and characterization of the evolution, variation and functions of diversity-generating retroelements using thousands of genomes and metagenomes. BMC Genom. 2019, 20, 595. [Google Scholar] [CrossRef]
- O’Connor, T.; Bodén, M.; Bailey, T.L. {CisMapper}: Predicting regulatory interactions from transcription factor {ChIP-seq} data. Nucleic Acids Res. 2017, 45, e19. [Google Scholar] [CrossRef]
- Sadakierska-Chudy, A.; Filip, M. A comprehensive view of the epigenetic landscape. Part {II}: Histone post-translational modification, nucleosome level, and chromatin regulation by {ncRNAs}. Neurotox. Res. 2015, 27, 172–197. [Google Scholar] [CrossRef] [PubMed]
- Igolkina, A.A.; Zinkevich, A.; Karandasheva, K.O.; Popov, A.A.; Selifanova, M.V.; Nikolaeva, D.; Tkachev, V.; Penzar, D.; Nikitin, D.M.; Buzdin, A. H3K4me3, H3K9ac, H3K27ac, H3K27me3 and H3K9me3 Histone Tags Suggest Distinct Regulatory Evolution of Open and Condensed Chromatin Landmarks. Cells 2019, 8, 1034. [Google Scholar] [CrossRef]
- Nikitin, D.; Garazha, A.; Sorokin, M.; Penzar, D.; Tkachev, V.; Markov, A.; Gaifullin, N.; Borger, P.; Poltorak, A.; Buzdin, A. Retroelement—Linked Transcription Factor Binding Patterns Point to Quickly Developing Molecular Pathways in Human Evolution. Cells 2019, 8, 130. [Google Scholar] [CrossRef] [PubMed]
- Scally, A.; Dutheil, J.Y.; Hillier, L.W.; Jordan, G.E.; Goodhead, I.; Herrero, J.; Hobolth, A.; Lappalainen, T.; Mailund, T.; Marques-Bonet, T.; et al. Insights into hominid evolution from the gorilla genome sequence. Nature 2012, 483, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, J.; Wang, T.; Xie, H. Construction and Analyses of Human Large-Scale Tissue Specific Networks. PLoS ONE 2014, 9, e115074. [Google Scholar] [CrossRef]
- Danino, Y.M.; Even, D.; Ideses, D.; Juven-Gershon, T. The core promoter: At the heart of gene expression. Biochim. Biophys. Acta 2015, 1849, 1116–1131. [Google Scholar] [CrossRef]
- Zolotovskaia, M.A.; Tkachev, V.S.; Guryanova, A.A.; Simonov, A.M.; Raevskiy, M.M.; Efimov, V.V.; Wang, Y.; Sekacheva, M.I.; Garazha, A.V.; Borisov, N.M.; et al. OncoboxPD: Human 51 672 molecular pathways database with tools for activity calculating and visualization. Comput. Struct. Biotechnol. J. 2022, 20, 2280–2291. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.r-project.org/ (accessed on 23 July 2019).
- Wei, T.; Simko, V. R Package “Corrplot”: Visualization of a Correlation Matrix. Available online: https://github.com/taiyun/corrplot (accessed on 7 January 2022).
- Schaefer, C.F.; Anthony, K.; Krupa, S.; Buchoff, J.; Day, M.; Hannay, T.; Buetow, K.H. PID: The Pathway Interaction Database. Nucleic Acids Res. 2009, 37, D674–D679. [Google Scholar] [CrossRef]
- QIAGEN—Pathway-Central. Available online: https://www.qiagen.com/us/shop/genes-and-pathways/pathway-central/ (accessed on 19 September 2018).
- Fabregat, A.; Sidiropoulos, K.; Garapati, P.; Gillespie, M.; Hausmann, K.; Haw, R.; Jassal, B.; Jupe, S.; Korninger, F.; McKay, S.; et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016, 44, D481–D487. [Google Scholar] [CrossRef]
- Nishimura, D. BioCarta. Biotech Softw. Internet Rep. 2001, 2, 117–120. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Romero, P.; Wagg, J.; Green, M.L.; Kaiser, D.; Krummenacker, M.; Karp, P.D. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2004, 6, R2. [Google Scholar] [CrossRef] [PubMed]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, C.; Chen, N.; Gu, H.; Yen, A.; Cao, L.; Wang, E.; Wang, L. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 2016, 7, 33440. [Google Scholar] [CrossRef]
- Treda, C.; Popeda, M.; Ksiazkiewicz, M.; Grzela, D.P.; Walczak, M.P.; Banaszczyk, M.; Peciak, J.; Stoczynska-Fidelus, E.; Rieske, P. EGFR Activation Leads to Cell Death Independent of PI3K/AKT/mTOR in an AD293 Cell Line. PLoS ONE 2016, 11, e0155230. [Google Scholar] [CrossRef]
- Edeling, M.; Ragi, G.; Huang, S.; Pavenstädt, H.; Susztak, K. Developmental signalling pathways in renal fibrosis: The roles of Notch, Wnt and Hedgehog. Nat. Rev. Nephrol. 2016, 12, 426–439. [Google Scholar]
- Kumar, V.; Vashishta, M.; Kong, L.; Wu, X.; Lu, J.J.; Guha, C.; Dwarakanath, B.S. The Role of Notch, Hedgehog, and Wnt Signaling Pathways in the Resistance of Tumors to Anticancer Therapies. Front. Cell Dev. Biol. 2021, 9, 857. [Google Scholar] [CrossRef] [PubMed]
- Stefani, C.; Miricescu, D.; Stanescu-Spinu, I.I.; Nica, R.I.; Greabu, M.; Totan, A.R.; Jinga, M. Growth Factors, PI3K/AKT/mTOR and MAPK Signaling Pathways in Colorectal Cancer Pathogenesis: Where Are We Now? Int. J. Mol. Sci. 2021, 22, 10260. [Google Scholar]
- Kamdje, A.H.N.; Kamga, P.T.; Simo, R.T.; Vecchio, L.; Etet, P.F.S.; Muller, J.M.; Bassi, G.; Lukong, E.; Goel, R.K.; Amvene, J.M.; et al. Developmental pathways associated with cancer metastasis: Notch, Wnt, and Hedgehog. Cancer Biol. Med. 2017, 14, 109–120. [Google Scholar]
- DuBridge, R.B.; Tang, P.; Hsia, H.C.; Leong, P.M.; Miller, J.H.; Calos, M.P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 1987, 7, 379. [Google Scholar]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Hollingsworth, M.A.; Swanson, B.J. Mucins in cancer: Protection and control of the cell surface. Nat. Rev. Cancer 2004, 4, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Roy, L.D.; Sahraei, M.; Subramani, D.B.; Besmer, D.; Nath, S.; Tinder, T.L.; Bajaj, E.; Shanmugam, K.; Lee, Y.Y.; Hwang, S.I.L.; et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene 2010, 30, 1449–1459. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.J.; Zapata, L.; Werner, B.; Barnes, C.P.; Sottoriva, A.; Graham, T.A. Measuring the distribution of fitness effects in somatic evolution by combining clonal dynamics with dN/dS ratios. eLife 2020, 9, e48714. [Google Scholar] [CrossRef] [PubMed]
- Zapata, L.; Pich, O.; Serrano, L.; Kondrashov, F.A.; Ossowski, S.; Schaefer, M.H. Negative selection in tumor genome evolution acts on essential cellular functions and the immunopeptidome. Genome Biol. 2018, 19, 67. [Google Scholar] [CrossRef] [PubMed]



| Functional Tag | Cell Line |
|---|---|
| Active Chromatin: H3K4me1, H3K4me3, H3K9ac, H3K27ac; Heterochromatin: H3K27me3, H3K9me3 | GM12878—lymphoblastoid cells, Hela-S3—cervical carcinoma cells, HepG2—hepatocellular carcinoma cells, K562—leukemia cells, MCF-7—breast cancer cells |
| Transcriptional Factor Binding Sites (TFBS) for 563 transcriptional factor proteins | GM12878—lymphoblastoid cells, Hela-S3—cervical carcinoma cells, HepG2—hepatocellular carcinoma cells, K562—leukemia cells, MCF-7—breast cancer cells, HEK293—immortalized embryonal kidney cells, HEK293T—daughter cell line that was derived from HEK293 by transfecting with a plasmid expressing a temperature-sensitive version of the SV40 large T antigen [42] A549—alveolar adenocarcinoma cells, SK-N-SH—neuroblastoma cells, HCT116—colon carcinoma cells, Ishikawa—endometrial adenocarcinoma cells, MCF-10A—non-tumorigenic epithelial cells, GM12891—lymphoblastoid cells |
| Threshold for Top or Bottom | Low dN/dS, Low NPIIAGG | High dN/dS, High NPIIAGG | High dN/dS, Low NPIIAGG | Low dN/dS, High NPIIAGG |
|---|---|---|---|---|
| 20% | 242 | 249 | 47 | 66 |
| 10% | 154 | 158 | 12 | 34 |
| 5% | 85 | 83 | 2 | 17 |
| 1% | 20 | 19 | 1 (Scavenging by Class B Receptors pathway) | 1 (Regulation of nuclear SMAD2 3 signaling pathway: negative regulation of cell growth) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zakharova, G.; Modestov, A.; Pugacheva, P.; Mekic, R.; Savina, E.; Guryanova, A.; Rachkova, A.; Yakushov, S.; Alimov, A.; Kulaeva, E.; et al. Distinct Traits of Structural and Regulatory Evolutional Conservation of Human Genes with Specific Focus on Major Cancer Molecular Pathways. Cells 2023, 12, 1299. https://doi.org/10.3390/cells12091299
Zakharova G, Modestov A, Pugacheva P, Mekic R, Savina E, Guryanova A, Rachkova A, Yakushov S, Alimov A, Kulaeva E, et al. Distinct Traits of Structural and Regulatory Evolutional Conservation of Human Genes with Specific Focus on Major Cancer Molecular Pathways. Cells. 2023; 12(9):1299. https://doi.org/10.3390/cells12091299
Chicago/Turabian StyleZakharova, Galina, Alexander Modestov, Polina Pugacheva, Rijalda Mekic, Ekaterina Savina, Anastasia Guryanova, Anastasia Rachkova, Semyon Yakushov, Andrei Alimov, Elizaveta Kulaeva, and et al. 2023. "Distinct Traits of Structural and Regulatory Evolutional Conservation of Human Genes with Specific Focus on Major Cancer Molecular Pathways" Cells 12, no. 9: 1299. https://doi.org/10.3390/cells12091299
APA StyleZakharova, G., Modestov, A., Pugacheva, P., Mekic, R., Savina, E., Guryanova, A., Rachkova, A., Yakushov, S., Alimov, A., Kulaeva, E., Fedoseeva, E., Kleyman, A., Vasin, K., Tkachev, V., Garazha, A., Sekacheva, M., Suntsova, M., Sorokin, M., Buzdin, A., & Zolotovskaia, M. A. (2023). Distinct Traits of Structural and Regulatory Evolutional Conservation of Human Genes with Specific Focus on Major Cancer Molecular Pathways. Cells, 12(9), 1299. https://doi.org/10.3390/cells12091299









