The miR-183 Cluster: Biogenesis, Functions, and Cell Communication via Exosomes in Cancer
Abstract
1. Introduction
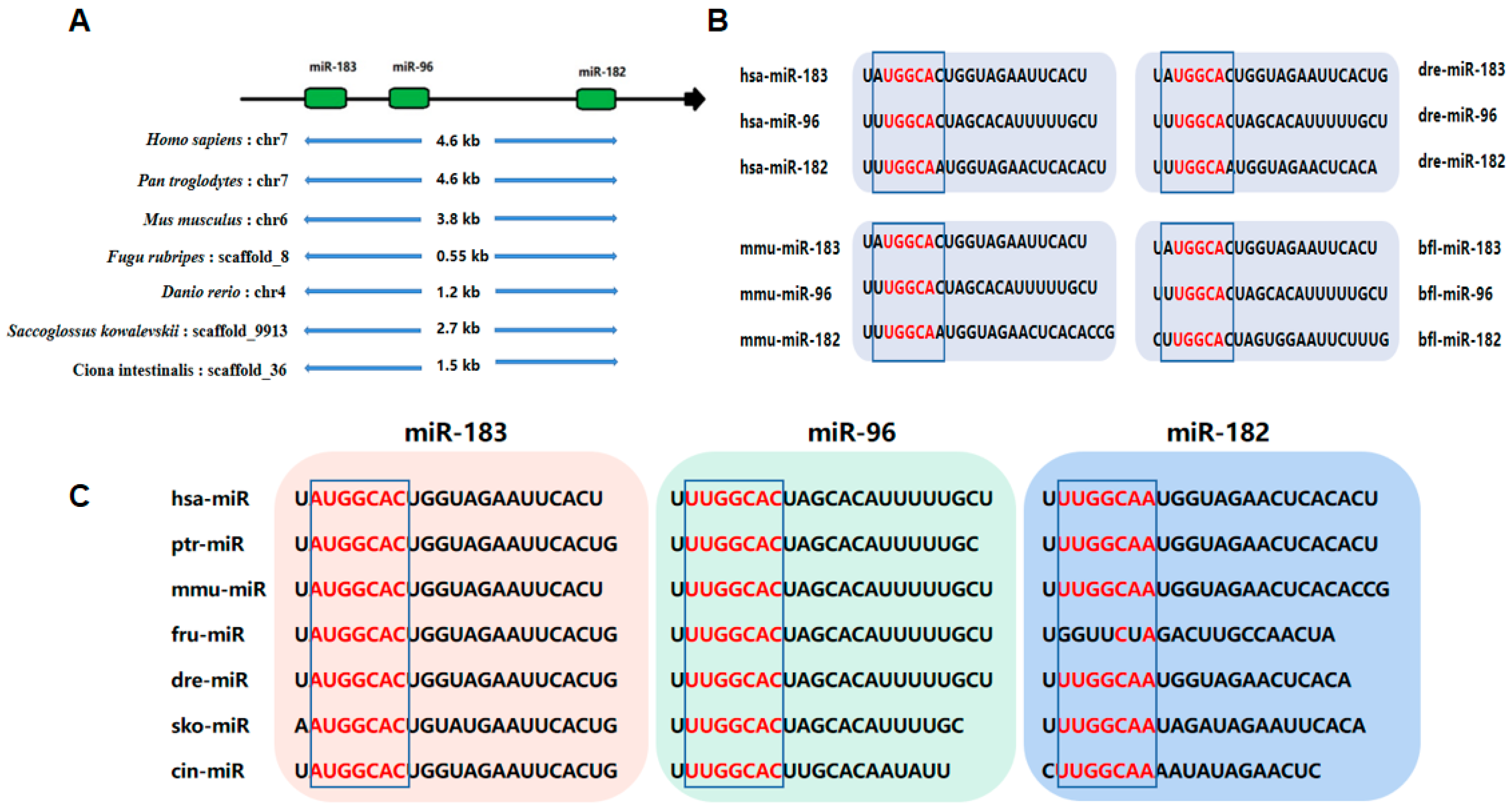
2. Biogenesis of miR-183 Cluster
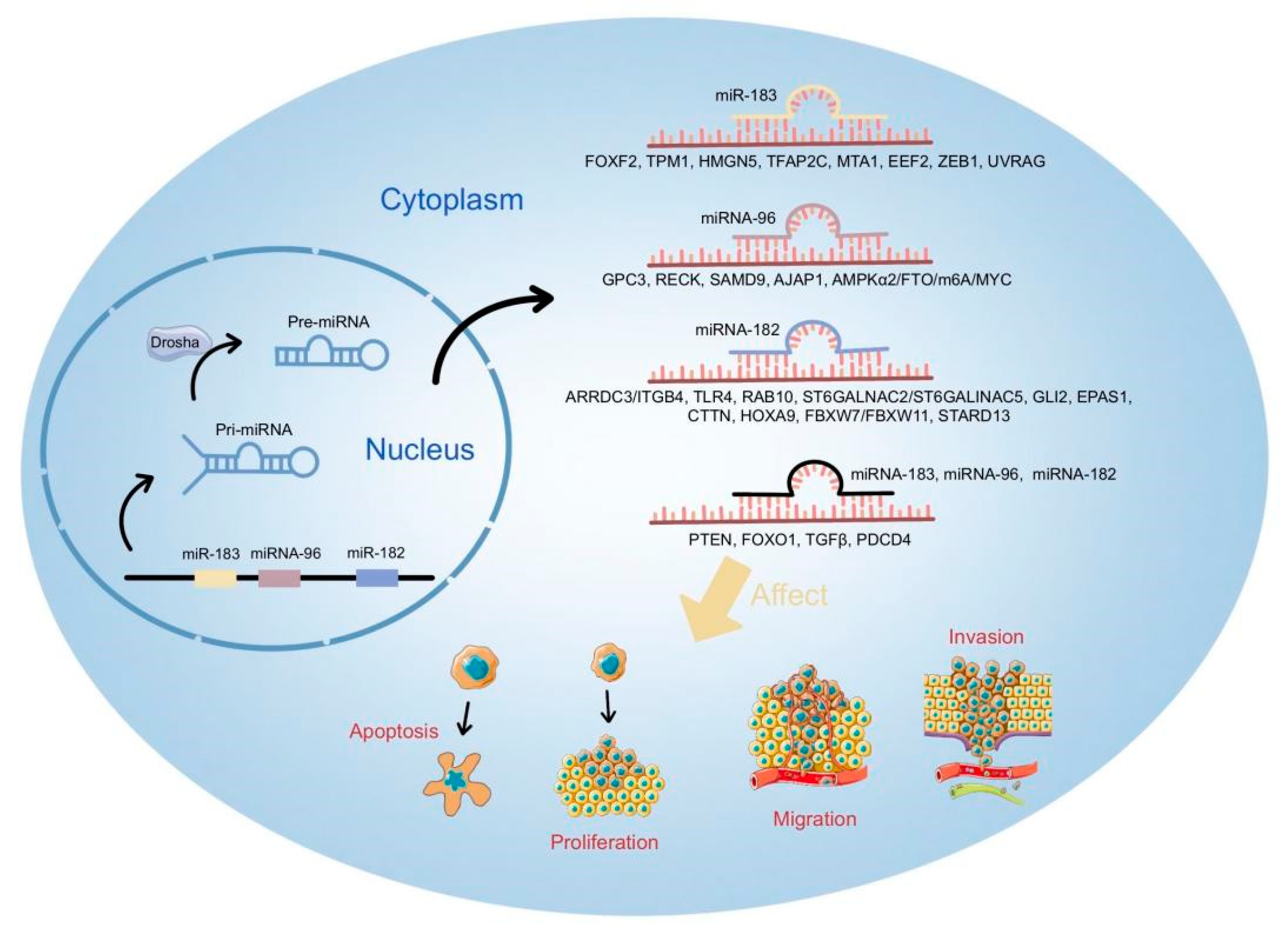
3. Biological Roles and Mechanisms of miR-183 Cluster in Cancer
3.1. miR-183 in Cancer
3.2. miR-182 in Cancer
3.3. miR-96 in Cancer
3.4. Key Factors Targeted by miR-183 Cluster
4. miR-183 Cluster and Other Noncoding RNAs
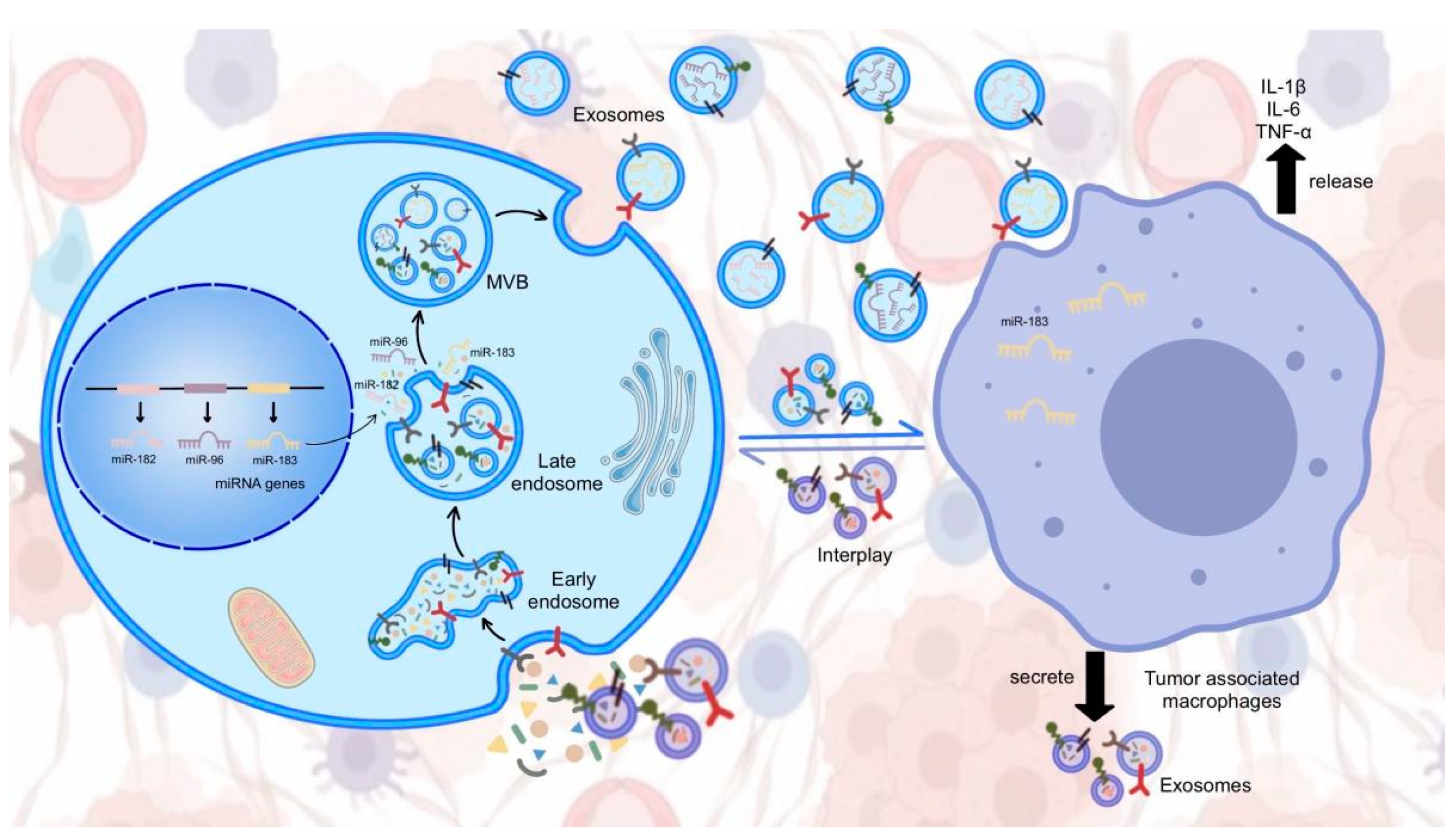
5. Exosomal miR-183 Cluster in Cancer Cell Communication
6. Discussion and Perspectives
7. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Zhang, S.; Zeng, H.; Wang, S.; Sun, K.; Chen, R.; Li, L.; Wei, W.; He, J. Cancer Incidence and Mortality in China, 2016. J. Natl. Cancer Cent. 2022, 2, 1–9. [Google Scholar] [CrossRef]
- Ruiz-Cordero, R.; Devine, W.P. Targeted Therapy and Checkpoint Immunotherapy in Lung Cancer. Surg. Pathol. Clin. 2020, 13, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Nan, A.; Chen, L.; Li, X.; Jia, Y.; Qiu, M.; Dai, X.; Zhou, H.; Zhu, J.; Zhang, H.; et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol. Cancer 2020, 19, 101. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, Y.; Zhao, L.; Pan, Y.; Shan, Y.; Li, Y.; Jia, L. Upregulation of microRNA-135b and microRNA-182 pro-motes chemoresistance of colorectal cancer by targeting ST6GALNAC2 via PI3K/AKT pathway. Mol. Carcinog. 2017, 56, 2669–2680. [Google Scholar] [CrossRef] [PubMed]
- Vietri, M.T.; D’Elia, G.; Caliendo, G.; Resse, M.; Casamassimi, A.; Passariello, L.; Albanese, L.; Cioffi, M.; Molinari, A.M. He-reditary Prostate Cancer: Genes Related, Target Therapy and Prevention. Int. J. Mol. Sci. 2021, 22, 3753. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Wu, Y.; Yang, J.; Yang, D.; Fang, X. Progress in the treatment of advanced gastric cancer. Tumor Biol. 2017, 39, 1010428317714626. [Google Scholar] [CrossRef]
- Davey, M.; Davies, M.; Lowery, A.; Miller, N.; Kerin, M. The Role of MicroRNA as Clinical Biomarkers for Breast Cancer Surgery and Treatment. Int. J. Mol. Sci. 2021, 22, 8290. [Google Scholar] [CrossRef]
- Visone, R.; Croce, C.M. Mirnas and Cancer. Am. J. Pathol. 2009, 174, 1131–1138. [Google Scholar] [CrossRef]
- Biller, L.H.; Schrag, D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA 2021, 325, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Yu, I.S.; Kopetz, S. The Emergence of Targetable Pathways in Colorectal Cancer. Clin. Adv. Hematol. Oncol. 2021, 19, 774–783. [Google Scholar] [PubMed]
- Jung, E.; Choi, J.; Kim, J.S.; Han, T.S. Microrna-Based Therapeutics for Drug-Resistant Colorectal Cancer. Pharmaceuticals 2021, 14, 136. [Google Scholar] [CrossRef] [PubMed]
- Machlowska, J.; Baj, J.; Sitarz, M.; Maciejewski, R.; Sitarz, R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int. J. Mol. Sci. 2020, 21, 4012. [Google Scholar] [CrossRef] [PubMed]
- Jokar, N.; Velikyan, I.; Ahmadzadehfar, H.; Rekabpour, S.J.; Jafari, E.; Ting, H.H.; Biersack, H.J.; Assadi, M. Theranostic Approach in Breast Cancer: A Treasured Tailor for Future Oncology. Clin. Nucl. Med. 2021, 46, e410–e420. [Google Scholar] [CrossRef] [PubMed]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 2019, 76, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Mollaei, H.; Safaralizadeh, R.; Rostami, Z. MicroRNA replacement therapy in cancer. J. Cell. Physiol. 2019, 234, 12369–12384. [Google Scholar] [CrossRef]
- Kataria, P.; Surela, N.; Chaudhary, A.; Das, J. MiRNA: Biological Regulator in Host-Parasite Interaction during Malaria Infection. Int. J. Environ. Res. Public Health 2022, 19, 2395. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Ko, N.-Y.; Chen, L.-R.; Chen, K.-H. The Role of Micro RNA and Long-Non-Coding RNA in Osteoporosis. Int. J. Mol. Sci. 2020, 21, 4886. [Google Scholar] [CrossRef]
- Hill, M.; Tran, N. miRNA interplay: Mechanisms and consequences in cancer. Dis. Model. Mech. 2021, 14, dmm047662. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, X.-M.; Ding, L.; Zhang, X.-J.; Ma, Z.-L. mTOR signaling-related MicroRNAs and Cancer involvement. J. Cancer 2018, 9, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Ichiyama, K.; Dong, C. The role of miR-183 cluster in immunity. Cancer Lett. 2019, 443, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed]
- Mihelich, B.L.; Dambal, S.; Lin, S.; Nonn, L. miR-182, of the miR-183 cluster family, is packaged in exosomes and is detected in human exosomes from serum, breast cells and prostate cells. Oncol. Lett. 2016, 12, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wang, X.; Yang, Y.; Chen, W.; Zhang, K.; Teng, B.; Huang, C.; Zhao, Q.; Qiu, Z. Hypoxic Tumor-Derived Exosomal Long Noncoding RNA UCA1 Promotes Angiogenesis via miR-96-5p/AMOTL2 in Pancreatic Cancer. Mol. Ther. Nucleic Acids 2020, 22, 179–195. [Google Scholar] [CrossRef]
- Mao, S.; Zhao, J.; Zhang, Z.-J.; Zhao, Q. MiR-183-5p overexpression in bone mesenchymal stem cell-derived exosomes protects against myocardial ischemia/reperfusion injury by targeting FOXO1. Immunobiology 2022, 227, 152204. [Google Scholar] [CrossRef]
- Akbar, R.; Ullah, K.; Rahman, T.U.; Cheng, Y.; Pang, H.-Y.; Jin, L.Y.; Wang, Q.-J.; Huang, H.-F.; Sheng, J.-Z. miR-183-5p regulates uterine receptivity and enhances embryo implantation. J. Mol. Endocrinol. 2020, 64, 43–52. [Google Scholar] [CrossRef]
- Dambal, S.; Shah, M.; Mihelich, B.; Nonn, L. The microRNA-183 cluster: The family that plays together stays together. Nucleic Acids Res. 2015, 43, 7173–7188. [Google Scholar] [CrossRef]
- Zhou, T.; Zhang, G.-J.; Zhou, H.; Xiao, H.-X.; Li, Y. Overexpression of microRNA-183 in human colorectal cancer and its clinical significance. Eur. J. Gastroenterol. Hepatol. 2014, 26, 229–233. [Google Scholar] [CrossRef]
- Kundu, S.T.; Byers, L.A.; Peng, D.H.; Roybal, J.D.; Diao, L.; Wang, J.; Tong, P.; Creighton, C.J.; Gibbons, D.L. The Mir-200 Cluster and the Mir-183~96~182 Cluster Target Foxf2 to Inhibit Invasion and Metastasis in Lung Cancers. Oncogene 2016, 35, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Di, M.; Liang, J.; Shi, S.; Tan, Q.; Wang, Z. MicroRNA-183 in Cancer Progression. J. Cancer 2020, 11, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Waseem, M.; Ahmad, M.K.; Serajuddin, M.; Bhaskar, V.; Sankhwar, S.N.; Mahdi, A.A. MicroRNA-183-5p: A New Potential Marker for Prostate Cancer. Indian J. Clin. Biochem. 2019, 34, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Golbakhsh, M.; Boddouhi, B.; Hatami, N.; Goudarzi, P.K.; Shakeri, M.; Yahaghi, E.; Taheriazam, A. Down-regulation of microRNA-182 and microRNA-183 predicts progression of osteosarcoma. Arch. Med. Sci. 2017, 6, 1352–1356. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ma, Z.; Liu, X.; Zhang, C.; Hu, Y.; Ding, L.; Qi, P.; Wang, J.; Lu, S.; Li, Y. MiR-183-5p is required for non-small cell lung cancer progression by repressing PTEN. Biomed. Pharmacother. 2019, 111, 1103–1111. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, M.; Li, Y.; Luo, J.; Wang, Y.; Geng, W.; Yang, Z.; Ma, H.; Bai, Y. Mir-183 Promotes Radioresistance of Lung Adenocarcinoma H1299 Cells Via Epitheli-al-Mesenchymal Transition. Braz. J. Med. Biol. Res. 2021, 54, E9700. [Google Scholar] [CrossRef]
- Huangfu, L.; Liang, H.; Wang, G.; Su, X.; Li, L.; Du, Z.; Hu, M.; Dong, Y.; Bai, X.; Liu, T.; et al. miR-183 regulates autophagy and apoptosis in colorectal cancer through targeting of UVRAG. Oncotarget 2016, 7, 4735–4745. [Google Scholar] [CrossRef]
- Dai, Y.; Gao, X. Inhibition of cancer cell-derived exosomal microRNA-183 suppresses cell growth and metastasis in prostate cancer by upregulating TPM1. Cancer Cell Int. 2021, 21, 145. [Google Scholar] [CrossRef]
- Li, Y.; He, S.; Zhan, Y.; He, A.; Gong, Y.; Ji, G.; Huang, C.; Peng, D.; Guan, B.; Li, X.; et al. microRNA-183-3p Inhibits Progression of Human Prostate Cancer by Downregulating High-Mobility Group Nucleosome Binding Domain 5. DNA Cell Biol. 2019, 38, 840–848. [Google Scholar] [CrossRef]
- Li, W.; Cui, X.; Qi, A.; Yan, L.; Wang, T.; Li, B. Mir-183-5p Acts as a Potential Prognostic Biomarker in Gastric Cancer and Regulates Cell Func-tions by Modulating Eef2. Pathol. Res. Pract. 2019, 215, 152636. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, Y.; Han, L.; Sun, S.; Shu, Y. miR-183 inhibits autophagy and apoptosis in gastric cancer cells by targeting ultraviolet radiation resistance-associated gene. Int. J. Mol. Med. 2018, 42, 3562–3570. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Shen, J.; Yue, H.; Cao, Z. miRNA-183-5p.1 promotes the migration and invasion of gastric cancer AGS cells by targeting TPM1. Oncol. Rep. 2019, 42, 2371–2381. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Michels, B.E.; Tosun, O.E.; Jung, J.; Kappes, J.; Ibing, S.; Nataraj, N.B.; Sahay, S.; Schneider, M.; Wörner, A.; et al. 5′isomiR-183-5p|+2 elicits tumor suppressor activity in a negative feedback loop with E2F1. J. Exp. Clin. Cancer Res. 2022, 41, 190. [Google Scholar] [CrossRef]
- Yang, C.-L.; Zheng, X.-L.; Ye, K.; Ge, H.; Sun, Y.-N.; Lu, Y.-F.; Fan, Q.-X. MicroRNA-183 Acts as a Tumor Suppressor in Human Non-Small Cell Lung Cancer by Down-Regulating MTA1. Cell. Physiol. Biochem. 2018, 46, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Bian, W.; Zhang, H.; Tang, M.; Zhang, S.; Wang, L.; Liu, L.; Wang, W. Potential Role of microRNA-183 as a Tumor Suppressor in Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2018, 51, 2065–2072. [Google Scholar] [CrossRef]
- Sun, X.; Xu, Y.; Zhang, S.; Li, X.; Wang, Y.; Zhang, Y.; Zhao, X.; Li, Y.; Wang, Y. Microrna-183 Suppresses The Vitality, Invasion and Migration of Human Osteosarcoma Cells by Targeting Metastasis-Associated Protein 1. Exp. Ther. Med. 2018, 15, 5058–5064. [Google Scholar] [CrossRef]
- Kang, J.; Kim, W.; Lee, S.; Kwon, D.; Chun, J.; Son, B.; Kim, E.; Lee, J.-M.; Youn, H.; Youn, B. TFAP2C promotes lung tumorigenesis and aggressiveness through miR-183- and miR-33a-mediated cell cycle regulation. Oncogene 2017, 36, 1585–1596. [Google Scholar] [CrossRef]
- Zhang, T.; Li, W.; Gu, M.; Wang, Z.; Zhou, S.; Hao, X.; Li, W.; Xu, S. Clinical Significance of miR-183-3p and miR-182-5p in NSCLC and Their Correlation. Cancer Manag. Res. 2021, 13, 3539–3550. [Google Scholar] [CrossRef]
- Chen, G.; Yu, L.; Dong, H.; Liu, Z.; Sun, Y. MiR-182 enhances radioresistance in non-small cell lung cancer cells by regulating FOXO3. Clin. Exp. Pharmacol. Physiol. 2019, 46, 137–143. [Google Scholar] [CrossRef]
- Yang, W.; Yin, Y.; Bi, L.; Wang, Y.; Yao, J.; Xu, L.; Jiao, L. MiR-182-5p promotes the Metastasis and Epithelial-mesenchymal Transition in Non-small Cell Lung Cancer by Targeting EPAS1. J. Cancer 2021, 12, 7120–7129. [Google Scholar] [CrossRef]
- Gao, L.; Yan, S.B.; Yang, J.; Kong, J.L.; Shi, K.; Ma, F.C.; Huang, L.Z.; Luo, J.; Yin, S.Y.; He, R.Q.; et al. Mir-182-5p and Its Target Hoxa9 in Non-Small Cell Lung Cancer: A Clinical and In-Silico Exploration with the Combination of Rt-Qpcr, Mirna-Seq and Mirna-Chip. BMC Med. Genom. 2020, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, W.; Wu, G.; Peng, C.; Liu, J. miR-182-5p Serves as an Oncogene in Lung Adenocarcinoma through Binding to STARD13. Comput. Math. Methods Med. 2021, 2021, 7074343. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Liu, Y.-H.; Wang, L.-L.; Wang, J.; Zhao, Z.-H.; Qu, J.-F.; Wang, S.-F. MiR-182 promotes cell proliferation by suppressing FBXW7 and FBXW11 in non-small cell lung cancer. Am. J. Transl. Res. 2018, 10, 1131–1142. [Google Scholar] [PubMed]
- Ning, F.-L.; Wang, F.; Li, M.-L.; Yu, Z.-S.; Hao, Y.-Z.; Chen, S.-S. MicroRNA-182 modulates chemosensitivity of human non-small cell lung cancer to cisplatin by targeting PDCD4. Diagn. Pathol. 2014, 9, 143. [Google Scholar] [CrossRef] [PubMed]
- Seidl, C.; Panzitt, K.; Bertsch, A.; Brcic, L.; Schein, S.; Mack, M.; Leithner, K.; Prinz, F.; Olschewski, H.; Kornmueller, K.; et al. MicroRNA-182-5p regulates hedgehog signaling pathway and chemosensitivity of cisplatin-resistant lung adenocarcinoma cells via targeting GLI2. Cancer Lett. 2020, 469, 266–276. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Gong, H.; Yuan, Y.; Li, Y.; Wang, C.; Li, W.; Zhang, Z.; Liu, M.; Liu, H.; et al. miR-182 suppresses invadopodia formation and metastasis in non-small cell lung cancer by targeting cortactin gene. J. Exp. Clin. Cancer Res. 2018, 37, 145. [Google Scholar] [CrossRef]
- Jia, L.; Luo, S.; Ren, X.; Li, Y.; Hu, J.; Liu, B.; Zhao, L.; Shan, Y.; Zhou, H. miR-182 and miR-135b Mediate the Tumorigenesis and Invasiveness of Colorectal Cancer Cells via Targeting ST6GALNAC2 and PI3K/AKT Pathway. Dig. Dis. Sci. 2017, 62, 3447–3459. [Google Scholar] [CrossRef]
- Lin, M.; Li, Y.; Xian, J.; Chen, J.; Feng, Y.; Mao, C.; Pan, Y.; Li, Z.; Zeng, Y.; Yang, L.; et al. Long non-coding RNA AGER-1 inhibits colorectal cancer progression through sponging miR-182. Int. J. Biol. Markers 2020, 35, 10–18. [Google Scholar] [CrossRef]
- Bai, L.; Luo, L.; Gao, W.; Bu, C.; Huang, J. miR-182 modulates cell proliferation and invasion in prostate cancer via targeting ST6GALNAC5. Braz. J. Med. Biol. Res. 2021, 54, e9695. [Google Scholar] [CrossRef]
- Shiina, M.; Hashimoto, Y.; Kulkarni, P.; Dasgupta, P.; Shahryari, V.; Yamamura, S.; Tanaka, Y.; Dahiya, R. Role of Mir-182/Pdcd4 Axis in Aggressive Behavior of Prostate Cancer in The African Americans. BMC Cancer 2021, 21, 1028. [Google Scholar] [CrossRef]
- Wallis, C.J.; Gordanpour, A.; Bendavid, J.S.; Sugar, L.; Nam, R.K.; Seth, A. MiR-182 Is Associated with Growth, Migration and Invasion in Prostate Cancer via Suppression of FOXO1. J. Cancer 2015, 6, 1295–1305. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Xu, C.; Fang, Z.; Li, Y.; Liu, H.; Wang, Y.; Xu, C.; Sun, Y. Androgen Receptor Regulated Microrna Mir-182-5p Promotes Prostate Cancer Progression by Targeting the Arrdc3/Itgb4 Pathway. Biochem. Biophys. Res. Commun. 2016, 474, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lu, G.; Shao, Y.; Xu, D. MiR-182 promotes prostate cancer progression through activating Wnt/β-catenin signal pathway. Biomed. Pharmacother. 2018, 99, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zuo, X.; Meng, X. Circ_002059 Suppresses Cell Proliferation and Migration of Gastric Cancer Via Mir-182/Mtss1 Axis. Acta Biochim. Biophys. Sin. 2021, 53, 454–462. [Google Scholar] [CrossRef]
- Duan, X.; Yu, X.; Li, Z. Circular Rna Hsa_Circ_0001658 Regulates Apoptosis and Autophagy in Gastric Cancer through Microrna-182/Ras-Related Protein Rab-10 Signaling Axis. Bioengineered 2022, 13, 2387–2397. [Google Scholar] [CrossRef]
- Soheilifar, M.H.; Vaseghi, H.; Seif, F.; Ariana, M.; Ghorbanifar, S.; Habibi, N.; Papari Barjasteh, F.; Pornour, M. Concomitant Overexpression of Mir-182-5p and Mir-182-3p Raises the Possibility of Il-17-Producing Treg Formation in Breast Cancer by Targeting Cd3d, Itk, Foxo1, and Nfats: A Meta-Analysis and Exper-imental Study. Cancer Sci. 2021, 112, 589–603. [Google Scholar] [CrossRef]
- Ma, C.; He, D.; Tian, P.; Wang, Y.; He, Y.; Wu, Q.; Jia, Z.; Zhang, X.; Zhang, P.; Ying, H.; et al. Mir-182 Targeting Reprograms Tumor-Associated Macrophages and Limits Breast Cancer Pro-gression. Proc. Natl. Acad. Sci. USA 2022, 119, e2114006119. [Google Scholar] [CrossRef]
- Wang, F.; Wu, D.; Xu, Z.; Chen, J.; Zhang, J.; Li, X.; Chen, S.; He, F.; Xu, J.; Su, L.; et al. miR-182-5p affects human bladder cancer cell proliferation, migration and invasion through regulating Cofilin 1. Cancer Cell Int. 2019, 19, 42. [Google Scholar] [CrossRef]
- Soliman, S.E.; Elabd, N.S.; El-Kousy, S.M.; Awad, M.F. Down regulation of miR-30a-5p and miR-182–5p in gastric cancer: Clinical impact and survival analysis. Biochem. Biophys. Rep. 2021, 27, 101079. [Google Scholar] [CrossRef]
- Zheng, Q.; Ding, H.; Wang, L.; Yan, Y.; Wan, Y.; Yi, Y.; Tao, L.; Zhu, C. Circulating Exosomal miR-96 as a Novel Biomarker for Radioresistant Non-Small-Cell Lung Cancer. J. Oncol. 2021, 2021, 5893981. [Google Scholar] [CrossRef]
- Fei, X.; Zhang, J.; Zhao, Y.; Sun, M.; Zhao, H.; Li, S. miR-96 promotes invasion and metastasis by targeting GPC3 in non-small cell lung cancer cells. Oncol. Lett. 2018, 15, 9081–9086. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Pu, X.; Wang, Q.; Cao, J.; Xu, F.; Xu, L.I.; Li, K. Mir-96 Induces Cisplatin Chemoresistance in Non-Small Cell Lung Cancer Cells by Downreg-ulating Samd9. Oncol. Lett. 2016, 11, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Chen, J.; Li, Z.; Li, L.; Chen, J.; Guo, Y. microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKα2-FTO-m6A/MYC axis. J. Exp. Clin. Cancer Res. 2020, 39, 240. [Google Scholar] [CrossRef] [PubMed]
- Iseki, Y.; Shibutani, M.; Maeda, K.; Nagahara, H.; Fukuoka, T.; Matsutani, S.; Hirakawa, K.; Ohira, M. MicroRNA-96 Promotes Tumor Invasion in Colorectal Cancer via RECK. Anticancer Res. 2018, 38, 2031–2035. [Google Scholar]
- Yu, J.-J.; Wu, Y.-X.; Zhao, F.-J.; Xia, S.-J. miR-96 promotes cell proliferation and clonogenicity by down-regulating of FOXO1 in prostate cancer cells. Med. Oncol. 2014, 31, 910. [Google Scholar] [CrossRef]
- He, X.; Zou, K. MiRNA-96-5p contributed to the proliferation of gastric cancer cells by targeting FOXO3. J. Biochem. 2020, 167, 101–108. [Google Scholar] [CrossRef]
- Lang, C.; Xu, M.; Zhao, Z.; Chen, J.; Zhang, L. Microrna-96 Expression Induced by Low-Dose Cisplatin Or Doxorubicin Regulates Chemo-sensitivity, Cell Death and Proliferation in Gastric Cancer Sgc7901 Cells by Targeting Foxo1. Oncol. Lett. 2018, 16, 4020–4026. [Google Scholar]
- Yang, N.; Zhou, J.; Li, Q.; Han, F.; Yu, Z. miR-96 exerts carcinogenic effect by activating AKT/GSK-3β/β-catenin signaling pathway through targeting inhibition of FOXO1 in hepatocellular carcinoma. Cancer Cell Int. 2019, 19, 38. [Google Scholar] [CrossRef]
- Bao, Y.-H.; Wang, Y.; Liu, Y.; Wang, S.; Wu, B. MiR-96 expression in prostate cancer and its effect on the target gene regulation. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4548–4556. [Google Scholar]
- Xie, W.; Sun, F.; Chen, L.; Cao, X. miR-96 promotes breast cancer metastasis by suppressing MTSS1. Oncol. Lett. 2018, 15, 3464–3471. [Google Scholar] [CrossRef]
- He, C.; Zhang, Q.; Gu, R.; Lou, Y.; Liu, W. Mir-96 Regulates Migration and Invasion of Bladder Cancer through Epithelial-Mesenchymal Transition in Response to Transforming Growth Factor-Β1. J. Cell. Biochem. 2018, 119, 7807–7817. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Chen, Y.; Wu, J.; Cui, X.; Zheng, S.; Yan, H.; Wu, Y.; Wang, F. Lncrna Fgf14-As2 Represses Growth of Prostate Carcinoma Cells Via Modulating Mir-96-5p/Ajap1 Axis. J. Clin. Lab. Anal. 2021, 35, e24012. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Sun, L.; Liang, L. LncRNA HOXC-AS3 Suppresses the Formation of Mature miR-96 in Ovarian Cancer Cells to Promote Cell Proliferation. Reprod. Sci. 2021, 28, 2342–2349. [Google Scholar] [CrossRef]
- Yadav, R.K.; Chauhan, A.S.; Zhuang, L.; Gan, B. FoxO transcription factors in cancer metabolism. Semin. Cancer Biol. 2018, 50, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Jiramongkol, Y.; Lam, E.W.-F. FOXO transcription factor family in cancer and metastasis. Cancer Metastasis Rev. 2020, 39, 681–709. [Google Scholar] [CrossRef]
- McLoughlin, H.S.; Wan, J.; Spengler, R.M.; Xing, Y.; Davidson, B.L. Human-specific microRNA regulation of FOXO1: Implications for microRNA recognition element evolution. Hum. Mol. Genet. 2014, 23, 2593–2603. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Chen, J.; He, L.; Stiles, B.L. PTEN: Tumor Suppressor and Metabolic Regulator. Front. Endocrinol. 2018, 9, 338. [Google Scholar] [CrossRef]
- Zhao, Y.-S.; Yang, W.-C.; Xin, H.-W.; Han, J.-X.; Ma, S.-G. MiR-182-5p Knockdown Targeting PTEN Inhibits Cell Proliferation and Invasion of Breast Cancer Cells. Yonsei Med. J. 2019, 60, 148–157. [Google Scholar] [CrossRef]
- Vahabi, M.; Pulito, C.; Sacconi, A.; Donzelli, S.; D’Andrea, M.; Manciocco, V.; Pellini, R.; Paci, P.; Sanguineti, G.; Strigari, L.; et al. miR-96-5p targets PTEN expression affecting radio-chemosensitivity of HNSCC cells. J. Exp. Clin. Cancer Res. 2019, 38, 141. [Google Scholar] [CrossRef]
- Haddadi, N.; Lin, Y.; Travis, G.; Simpson, A.M.; Nassif, N.T.; McGowan, E.M. Pten/Ptenp1: ‘Regulating The Regulator of Rtk-Dependent Pi3k/Akt Signalling’, New Targets for Cancer Therapy. Mol. Cancer 2018, 17, 37. [Google Scholar] [CrossRef]
- Vidarsdottir, L.; Azimi, A.; Das, I.; Sigvaldadottir, I.; Rahmanto, A.S.; Petri, A.; Kauppinen, S.; Ingvar, C.; Jönsson, G.; Olsson, H.; et al. PTENP1-AS contributes to BRAF inhibitor resistance and is associated with adverse clinical outcome in stage III melanoma. Sci. Rep. 2021, 11, 11023. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Qin, T.; Mao, J.; Zhang, J.; Fan, S.; Lu, Y.; Sun, Z.; Zhang, Q.; Song, B.; Li, L. Ptenp1/Mir-20a/Pten Axis Contributes to Breast Cancer Progression by Regulating Pten Via Pi3k/Akt Pathway. J. Exp. Clin. Cancer Res. 2019, 38, 256. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.Q.; Yang, X.W.; Chen, Y.B.; Zhang, D.W.; Jiang, X.F.; Xue, P. Exosomal Mir-21 Regulates The Tets/Ptenp1/Pten Pathway to Promote Hepatocellular Carcinoma Growth. Mol. Cancer 2019, 18, 148. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Sheng, W.; Meng, Y.; Cao, Y.; Li, R. LncRNA PTENP1 inhibits cervical cancer progression by suppressing miR-106b. Artif. Cells Nanomed. Biotechnol. 2020, 48, 393–407. [Google Scholar] [CrossRef]
- Ou, L.; Xiang, T.-Y.; Hao, X.-Y.; Wang, D.-Z.; Zeng, Q. Reduced long non-coding RNA PTENP1 contributed to proliferation and invasion via miR-19b/MTUS1 axis in patients with cervical cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4132–4144. [Google Scholar]
- Batlle, E.; Massagué, J. Transforming Growth Factor-Β Signaling in Immunity and Cancer. Immunity 2019, 50, 924–940. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, C.; Zhou, B.; Jiang, D. miR-183 modulated cell proliferation and apoptosis in ovarian cancer through the TGF-β/Smad4 signaling pathway. Int. J. Mol. Med. 2019, 43, 1734–1746. [Google Scholar] [CrossRef]
- Qi, H.; Cao, Q.; Liu, Q. MicroRNA-183 exerts a protective role in lupus nephritis through blunting the activation of TGF-β/Smad/TLR3 pathway via reducing Tgfbr1. Exp. Cell Res. 2020, 394, 112138. [Google Scholar] [CrossRef]
- Shang, R.; Baek, S.C.; Kim, K.; Kim, B.; Kim, V.N.; Lai, E.C. Genomic Clustering Facilitates Nuclear Processing of Suboptimal Pri-Mirna Loci. Mol. Cell. 2020, 78, 303–316.e4. [Google Scholar] [CrossRef]
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef]
- Han, T.-S.; Hur, K.; Cho, H.-S.; Ban, H. Epigenetic Associations between lncRNA/circRNA and miRNA in Hepatocellular Carcinoma. Cancers 2020, 12, 2622. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhou, Y.; Li, H. LncRNA, miRNA and lncRNA-miRNA interaction in viral infection. Virus Res. 2018, 257, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ding, Y.; Wu, Y.; Wang, X. Identification of The Complex Regulatory Relationships Related to Gastric Cancer from Lncrna-Mirna-Mrna Network. J. Cell. Biochem. 2020, 121, 876–887. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lou, W.; Ding, B.; Yang, B.; Lu, H.; Kong, Q.; Fan, W. A Novel Mrna-Mirna-Lncrna Competing Endogenous Rna Triple Sub-Network Associated with Prognosis of Pancreatic Cancer. Aging 2019, 11, 2610–2627. [Google Scholar] [CrossRef]
- Yang, S.; Wang, D.-D.; Zhou, S.-Y.; Zhang, Q.; Wang, J.-Y.; Zhong, S.-L.; Zhang, H.-D.; Wang, X.-Y.; Xia, X.; Chen, W.; et al. Identification of circRNA–miRNA networks for exploring an underlying prognosis strategy for breast cancer. Epigenomics 2020, 12, 101–125. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Liu, Y.; Wu, W.; Wu, K.; Zhang, W. Reconstruction and analysis of circRNA-miRNA-mRNA network in the pathology of cervical cancer. Oncol. Rep. 2019, 41, 2209–2225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Piao, H.-Y.; Guo, S.; Wang, Y.; Zhang, T.; Zheng, Z.-C.; Zhao, Y. LINC00163 inhibits the invasion and metastasis of gastric cancer cells as a ceRNA by sponging miR-183 to regulate the expression of AKAP12. Int. J. Clin. Oncol. 2020, 25, 570–583. [Google Scholar] [CrossRef]
- Cao, C.; Han, S.; Yuan, Y.; Wu, Y.; Lian, W.; Zhang, X.; Pan, L.; Li, M. Downregulated Circular Rna Hsa_Circ_0000291 Suppresses Migration and Proliferation of Gastric Cancer via Targeting the Mir-183/Itgb1 Axis. Cancer Manag. Res. 2019, 11, 9675–9683. [Google Scholar] [CrossRef]
- Gao, W.; Lin, S.; Cheng, C.; Zhu, A.; Hu, Y.; Shi, Z.; Zhang, X.; Hong, Z. Long Non-Coding Rna Casc2 Regulates Sprouty2 Via Functioning as a Competing Endog-enous Rna for Mir-183 to Modulate The Sensitivity of Prostate Cancer Cells to Docetaxel. Arch. Biochem. Biophys. 2019, 665, 69–78. [Google Scholar] [CrossRef]
- Shen, T.; Cheng, X.; Liu, X.; Xia, C.; Zhang, H.; Pan, D.; Zhang, X.; Li, Y. Circ_0026344 restrains metastasis of human colorectal cancer cells via miR-183. Artif. Cells Nanomed. Biotechnol. 2019, 47, 4038–4045. [Google Scholar] [CrossRef]
- Zhang, Q.; Zheng, J.; Liu, L. The Long Noncoding Rna Pcgem1 Promotes Cell Proliferation, Migration and Invasion Via Tar-geting The Mir-182/Fbxw11 Axis in Cervical Cancer. Cancer Cell Int. 2019, 19, 304. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Liu, M.; Wang, S. CircRNA hsa_circRNA_0001776 inhibits proliferation and promotes apoptosis in endometrial cancer via downregulating LRIG2 by sponging miR-182. Cancer Cell Int. 2020, 20, 412. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, H.; Du, Y.; Liu, P.; Zhang, J.; Li, Y.; Shen, H.; Xing, L.; Xue, X.; Chen, J.; et al. Long noncoding RNA TP53TG1 promotes pancreatic ductal adenocarcinoma development by acting as a molecular sponge of microRNA-96. Cancer Sci. 2019, 110, 2760–2772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Li, Y.; Wang, M.; Gu, J.; Xu, W.; Cai, H.; Fang, X.; Zhang, X. Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer 2022, 21, 56. [Google Scholar] [CrossRef]
- Kimiz-Gebologlu, I.; Oncel, S.S. Exosomes: Large-scale production, isolation, drug loading efficiency, and biodistribution and uptake. J. Control. Release 2022, 347, 533–543. [Google Scholar] [CrossRef]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef]
- Sun, Z.; Shi, K.; Yang, S.; Liu, J.; Zhou, Q.; Wang, G.; Song, J.; Li, Z.; Zhang, Z.; Yuan, W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol. Cancer 2018, 17, 147. [Google Scholar] [CrossRef]
- Li, B.; Cao, Y.; Sun, M.; Feng, H. Expression, Regulation, and Function of Exosome-Derived Mirnas in Cancer Progression and Therapy. FASEB J. 2021, 35, E21916. [Google Scholar] [CrossRef]
- Guo, J.; Duan, Z.; Zhang, C.; Wang, W.; He, H.; Liu, Y.; Wu, P.; Wang, S.; Song, M.; Chen, H.; et al. Mouse 4t1 Breast Cancer Cell-Derived Exosomes Induce Proinflammatory Cytokine Pro-duction in Macrophages Via Mir-183. J. Immunol. 2020, 205, 2916–2925. [Google Scholar] [CrossRef]
- Zhang, S.; Li, D.; Zhao, M.; Yang, F.; Sang, C.; Yan, C.; Wang, Z.; Li, Y. Exosomal Mir-183-5p Shuttled by M2 Polarized Tumor-Associated Macrophage Promotes The Development of Colon Cancer Via Targeting Them4 Mediated Pi3k/Akt and Nf-Κb Pathways. Front. Oncol. 2021, 11, 672684. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Wang, J.-J.; Zhao, L.-J.; Yang, X.-R.; Yu, Y.-L. Exosomal miR-182 regulates the effect of RECK on gallbladder cancer. World J. Gastroenterol. 2020, 26, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yuan, H.; Xu, H.; Zhao, H.; Xiong, N. Hypoxic Cancer-Secreted Exosomal miR-182-5p Promotes Glioblastoma Angiogenesis by Targeting Kruppel-like Factor 2 and 4. Mol. Cancer Res. 2020, 18, 1218–1231. [Google Scholar] [CrossRef] [PubMed]
- Shang, A.; Wang, X.; Gu, C.; Liu, W.; Sun, J.; Zeng, B.; Chen, C.; Ji, P.; Wu, J.; Quan, W.; et al. Exosomal miR-183-5p promotes angiogenesis in colorectal cancer by regulation of FOXO1. Aging 2020, 12, 8352–8371. [Google Scholar] [CrossRef]
- Simonson, B.; Das, S. Microrna Therapeutics: The Next Magic Bullet? Mini Rev. Med. Chem. 2015, 15, 467–474. [Google Scholar]
- Lee, S.W.L.; Paoletti, C.; Campisi, M.; Osaki, T.; Adriani, G.; Kamm, R.D.; Mattu, C.; Chiono, V. MicroRNA delivery through nanoparticles. J. Control. Release 2019, 313, 80–95. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 Study of Mrx34, A Liposomal Mir-34a Mimic, in Patients with Advanced Solid Tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Sharma, S.; Pukale, S.; Sahel, D.K.; Singh, P.; Mittal, A.; Chitkara, D. Folate Targeted Hybrid Lipo-Polymeric Nanoplexes Containing Docetaxel and Mirna-34a for Breast Cancer Treatment. Mater. Sci. Eng. 2021, 128, 112305. [Google Scholar] [CrossRef]
- Lolli, A.; Sivasubramaniyan, K.; Vainieri, M.L.; Oieni, J.; Kops, N.; Yayon, A.; van Osch, G.J. Hydrogel-Based Delivery of Antimir-221 Enhances Cartilage Regeneration by Endogenous Cells. J. Control. Release 2019, 309, 220–230. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, J.; Wu, J.; Fan, Q.; Zhou, J.; Wu, J.; Liu, S.; Zang, J.; Ye, J.; Xiao, M.; et al. Exosome-Mediated Targeted Delivery of Mir-210 for Angiogenic Therapy After Cerebral Ischemia in Mice. J. Nanobiotechnol. 2019, 17, 29. [Google Scholar] [CrossRef] [PubMed]
- Gulfidan, G.; Soylu, M.; Demirel, D.; Erdonmez, H.B.C.; Beklen, H.; Sarica, P.O.; Arga, K.Y.; Turanli, B. Systems Biomarkers for Papillary Thyroid Cancer Prognosis and Treatment through Multi-Omics Networks. Arch Biochem. Biophys. 2022, 715, 109085. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, C.; Zhou, T.; Liu, X.; Liu, X.; Li, X.; Chen, D. Role of Exosomal Proteins in Cancer Diagnosis. Mol. Cancer 2017, 16, 145. [Google Scholar] [CrossRef] [PubMed]
- Sandfeld-Paulsen, B.; Jakobsen, K.R.; Bæk, R.; Folkersen, B.H.; Rasmussen, T.R.; Meldgaard, P.; Varming, K.; Jørgensen, M.M.; Sorensen, B.S. Exosomal Proteins as Diagnostic Biomarkers in Lung Cancer. J. Thorac. Oncol. 2016, 11, 1701–1710. [Google Scholar] [CrossRef]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes Facilitate Therapeutic Targeting of Oncogenic Kras in Pancreatic Cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef]
- Tang, X.H.; Guo, T.; Gao, X.Y.; Wu, X.L.; Xing, X.F.; Ji, J.F.; Li, Z.Y. Exosome-Derived Noncoding Rnas in Gastric Cancer: Functions and Clinical Applications. Mol. Cancer 2021, 20, 99. [Google Scholar] [CrossRef]

| Cancer | Regulation | Tissue No. | Cell Lines | Functions | Target Gene | Source |
|---|---|---|---|---|---|---|
| NSCLC | × | × | H157, 344SQ (mouse) | Inhibit EMT, migration, and invasion | Foxf2 | [31] |
| NSCLC | Up | n = 8 | NCL-H292, NCL-H838, MRC-5, WI-26 VA4 | Promote occurrence and invasion | TFAP2C | [47] |
| NSCLC | Up | n = 76 | A549, H226, BEAS-2B, H1395 | Promote proliferation | × | [48] |
| NSCLC | Up | n = 55 | A549, SPC-A-1, H1299 | Promote migration and proliferation | PTEN | [35] |
| Lung cancer | Up | × | H1299 | Promote EMT and radioresistance | ZEB1 | [36] |
| NSCLC | Up | n = 194 | SPC-A-1 | Inhibit EMT, migration, and proliferation | MTA1 | [44] |
| CRC | Up | n = 94 | × | Invasion | × | [30] |
| CRC | Down | × | HCT116, HT29 | Promote apoptosis | UVRAG | [37] |
| PCa | Up | n = 50 | × | × | × | [33] |
| PCa | Up | × | LNCaP, PC-3 | Promote migration, proliferation, and invasion | TPM1 | [38] |
| PCa | Down | n = 12 | Vcap, C4-2 | Inhibit proliferation and migration. Promote apoptosis | HMGN5 | [39] |
| GC | Down | n = 102 | BGC823, MKN45 | Inhibit proliferation and migration | EEF2 | [40] |
| GC | Up | n = 24 | GES-1, MKN-7, AGS, HGC-27 | Promote migration and invasion | TPM1 | [42] |
| GC | Down | × | MKN28 | Inhibit invasion | UVRAG | [41] |
| HCC | Down | n = 10 | HepG2 | Inhibit proliferation and migration | MTA1 | [45] |
| OS | Down | n = 40 | × | Associated with progression and metastasis of osteosarcoma | × | [34] |
| OS | Down | n = 25 | MG63 | Inhibit the vitality, invasion and migration | MTA1 | [46] |
| Cancer | Regulation | Tissue | Cell Lines | Functions | Target Gene | Source |
|---|---|---|---|---|---|---|
| NSCLC | Up | × | NCI-H460, H1299 | Enhance radioresistance | FOXO3 | [49] |
| NSCLC | Up | × | NCI-H1975, NCI-H460, A549, 95-D | Promote EMT and migration | EPAS1 | [50] |
| NSCLC | Up | n = 124 | × | Oncogenic role | HOXA9 | [51] |
| Lung cancer | Up | × | A549, PC-9 | Promote migration, proliferation, and invasion | STARD13 | [52] |
| NSCLC | Up | n = 11 | H460 | Promote proliferation | FBXW7, FBXW11 | [53] |
| NSCLC | Up | × | A549 | Chemoresistance | PDCD4 | [54] |
| Lung cancer | Down | n = 27 | NCI-H460, A549, CisR | Cisplatin resistance, tumorigenesis | GLI2 | [55] |
| NSCLC | Down | n = 55 | A549, H1299 | Inhibit migration and invasion | CTTN | [56] |
| CRC | Up | n = 31 | HCT8, LoVo | Promote chemoresistance | ST6GALNAC2 | [6] |
| CRC | Up | n = 33 | SW480, SW620 | Tumorigenesis and invasion | ST6GALNAC2 | [57] |
| CRC | Up | n = 159 | DLD1, HCT15 | Inhibit migration and proliferation | lncAGER | [58] |
| PCa | Up | n = 25 | PC-3, Du145 | Promote proliferation and invasion | ST6GALNAC5 | [59] |
| Pca | Up | n = 82 | MDA-Pca-2b, DU145, LNCaP | Invasion | PDCD4 | [60] |
| Pca | Up | n = 147 | DU145, LNCaP | Promote proliferation, migration, and invasion | FOXO1 | [61] |
| PCa | Up | n = 65 | HTB81, LNCaP | Promote proliferation, migration, and invasion and inhibit apoptosis | ARRDC3/ITGB4 | [62] |
| PCa | Up | n = 25 | PC-3, LNCaP | Promote proliferation, colony formation, migration, and invasion and inhibit apoptosis | Wnt/β-catenin | [63] |
| GC | Down | n = 148 | × | × | × | [69] |
| GC | Up | × | GC-823, HGC-27 | Inhibit migration and proliferation | Circ002059, MTSS1 | [64] |
| GC | Down | × | AGS, HGC27 | Regulate viability, autophagy, and apoptosis | Circ0001658, RAB10 | [65] |
| Breast cancer | Up | n = 42 | Jurkat | Raise the possibility of IL-17–producing Treg formation | CD3d, ITK, FOXO1, NFATs | [66] |
| Breast cancer | Up | × | Py8119, AT3, SCP28, Jurkat, CTLL2, PBMC | Promote breast cancer progression | TGFβ, TLR4 | [67] |
| Bladder cancer | Down | n = 8 | RT4, T24 | Promote proliferation, migration, and invasion | Cofilin 1 | [68] |
| Cancer | Regulation | Tissue | Cell Lines | Functions | Target Gene | Source |
|---|---|---|---|---|---|---|
| NSCLC | Up | n = 52 | × | Non-invasive diagnostic and prognostic marker of radioresistant NSCLC | × | [70] |
| NSCLC | Up | n = 57 | A549, H460 | Promote migration and invasion | GPC3 | [71] |
| NSCLC | Up | n = 5 | H358, H23 | Induce cisplatin chemoresistance | SAMD9 | [72] |
| CRC | Up | n = 60 | SW480, SW620, HCT-8 | Promote proliferation and inhibit apoptosis | AMPKα2/FTO/m6A/MYC | [73] |
| CRC | × | × | HCT-116 | Promote invasion | RECK | [74] |
| PCa | Up | × | PC-3 | Promote migration | FOXO1, FOXO3a | [79] |
| PCa | Up | n = 13 | PcaP, PC3 | Promote proliferation | FOXO1 | [75] |
| PCa | Up | × | DU145, LNCaP | Inhibit proliferation | LncRNA FGF14-AS2, AJAP1 | [82] |
| GC | Up | n = 70 | MKN45, SGC7901 | Promote proliferation | FOXO3 | [76] |
| GC | Up | × | SGC7901 | Promote proliferation, migration, and invasion | FOXO1 | [77] |
| brest cancer | Up | n = 44 | MCF-7, MDA-MB-231 | Promote migration | MTSS1 | [80] |
| HCC | Up | n = 60 | HepG2 | Inhibit proliferation, migration, and invasion | FOXO1, AKT/GSK-3β/β-catenin | [78] |
| bladder cancer | × | × | HT1376 | Regulate migration and invasion | TGF-β1, FOXO1 | [81] |
| OC | Up | n = 62 | Caov-3, OVCAR3 | Promote migration | LncRNA HOXC-AS3 | [83] |
| Cancer | miRNA | ceRNA | Functions | Source |
|---|---|---|---|---|
| GC | miR-183 | LINCC00163 | Inhibit GC via miR-183/AKAP12 axis | [107] |
| GC | miR-183 | Circ0000291 | Promote GC via miR-183/ITGB1 axis | [108] |
| PCa | miR-183 | LncCASC2 | Compete with miR-183 to rescue the expression of SPRY2 inhibit cancer | [109] |
| CRC | miR-183 | Circ0026344 | Inhibit CRC via miR-183 | [110] |
| CC | miR-182 | LncPCGEM1 | Activate the NF-κB and β-catenin/TCF signaling pathways | [111] |
| EC | miR-182 | Circ0001776 | Inhibit EC via miR-182/LRIG2 axis | [112] |
| PDAC | miR-96 | LncTP53TG1 | Promote tumor via miR-96/KRAS axis | [113] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Meng, W.; Guo, Z.; Liu, M.; He, Y.; Li, Y.; Ma, Z. The miR-183 Cluster: Biogenesis, Functions, and Cell Communication via Exosomes in Cancer. Cells 2023, 12, 1315. https://doi.org/10.3390/cells12091315
Li S, Meng W, Guo Z, Liu M, He Y, Li Y, Ma Z. The miR-183 Cluster: Biogenesis, Functions, and Cell Communication via Exosomes in Cancer. Cells. 2023; 12(9):1315. https://doi.org/10.3390/cells12091315
Chicago/Turabian StyleLi, Shuhui, Wei Meng, Ziyi Guo, Min Liu, Yanyun He, Yanli Li, and Zhongliang Ma. 2023. "The miR-183 Cluster: Biogenesis, Functions, and Cell Communication via Exosomes in Cancer" Cells 12, no. 9: 1315. https://doi.org/10.3390/cells12091315
APA StyleLi, S., Meng, W., Guo, Z., Liu, M., He, Y., Li, Y., & Ma, Z. (2023). The miR-183 Cluster: Biogenesis, Functions, and Cell Communication via Exosomes in Cancer. Cells, 12(9), 1315. https://doi.org/10.3390/cells12091315





