Novel Filamin C Myofibrillar Myopathy Variants Cause Different Pathomechanisms and Alterations in Protein Quality Systems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Methods
Standard Protocol Approvals, Registrations, and Patient Consents
2.3. FLNC Mutation Detection and Analysis of Mutant Allele Expression by RT-PCR
2.4. Histochemistry and Immunohistochemistry
2.5. Cloning of the Truncated p.Y2704X FLNc Construct, and Expression and Purification of Recombinant Proteins
2.6. Cross-Linking of FLNc Polypeptides
2.7. Proteolytic Susceptibility Studies
2.8. Transcript Studies on cDNA of Patient Skeletal Muscle Biopsies
2.9. Western Blotting of Control and Patient Samples
3. Results
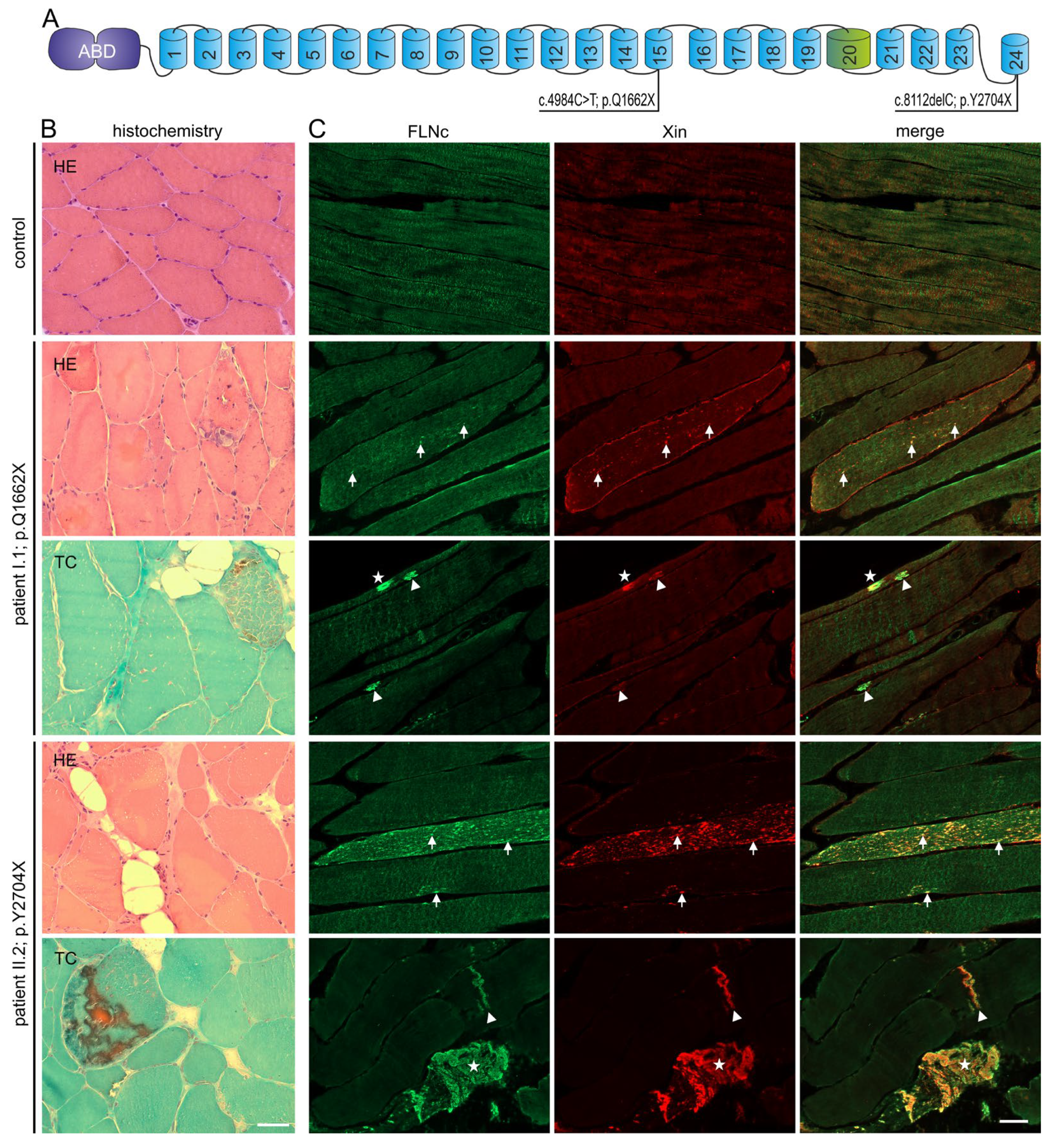
3.1. Identification of Two Novel FLNC Variants
3.2. Clinical Features
3.3. Histopathological and Ultrastructural Studies
3.4. Magnetic Resonance Imaging (MRI) Studies
3.5. Expression Analysis of Mutant and Normal FLNC RNA and FLNc Protein
3.6. Analysis of Stability and Dimerization Capability of the p.Y2704X Variant Protein
3.7. Transcript Studies on RNA Extracted from Human Muscle Biopsies
3.8. Analysis of Protein Expression of PQS Markers
3.9. Intracellular Distribution of PQS Markers
4. Discussion
4.1. New FLNC Variants Lead to Selective Skeletal Muscle Involvement in p.Y2704X and an Overlap of Skeletal and Cardiac Alterations in p.Q1662X
4.2. Not Only the Expression of Toxic FLNc Protein but Also FLNc Haploinsufficiency May Lead to Intracellular Protein Aggregation
4.3. PQS Is Altered Differently in MFM-Filaminopathy Muscle Tissue
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goldfarb, L.G.; Park, K.Y.; Cervenáková, L.; Gorokhova, S.; Lee, H.S.; Vasconcelos, O.; Nagle, J.W.; Semino-Mora, C.; Sivakumar, K.; Dalakas, M.C. Missense Mutations in Desmin Associated with Familial Cardiac and Skeletal Myopathy. Nat. Genet. 1998, 19, 402–403. [Google Scholar] [CrossRef] [PubMed]
- Vicart, P.; Caron, A.; Guicheney, P.; Li, Z.; Prévost, M.C.; Faure, A.; Chateau, D.; Chapon, F.; Tomé, F.; Dupret, J.M.; et al. A Missense Mutation in the AlphaB-Crystallin Chaperone Gene Causes a Desmin-Related Myopathy. Nat. Genet. 1998, 20, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Selcen, D.; Engel, A.G. Mutations in Myotilin Cause Myofibrillar Myopathy. Neurology 2004, 62, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Selcen, D.; Engel, A.G. Mutations in ZASP Define a Novel Form of Muscular Dystrophy in Humans. Ann. Neurol. 2005, 57, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Vorgerd, M.; van der Ven, P.F.M.; Bruchertseifer, V.; Löwe, T.; Kley, R.A.; Schröder, R.; Lochmüller, H.; Himmel, M.; Koehler, K.; Fürst, D.O.; et al. A Mutation in the Dimerization Domain of Filamin C Causes a Novel Type of Autosomal Dominant Myofibrillar Myopathy. Am. J. Hum. Genet. 2005, 77, 297–304. [Google Scholar] [CrossRef]
- Selcen, D.; Muntoni, F.; Burton, B.K.; Pegoraro, E.; Sewry, C.; Bite, A.V.; Engel, A.G. Mutation in BAG3 Causes Severe Dominant Childhood Muscular Dystrophy. Ann. Neurol. 2009, 65, 83–89. [Google Scholar] [CrossRef]
- Selcen, D.; Bromberg, M.B.; Chin, S.S.; Engel, A.G. Reducing Bodies and Myofibrillar Myopathy Features in FHL1 Muscular Dystrophy. Neurology 2011, 77, 1951–1959. [Google Scholar] [CrossRef]
- Pfeffer, G.; Barresi, R.; Wilson, I.J.; Hardy, S.A.; Griffin, H.; Hudson, J.; Elliott, H.R.; Ramesh, A.V.; Radunovic, A.; Winer, J.B.; et al. Titin Founder Mutation Is a Common Cause of Myofibrillar Myopathy with Early Respiratory Failure. J. Neurol. Neurosurg. Psychiatry 2014, 85, 331–338. [Google Scholar] [CrossRef]
- Sarparanta, J.; Jonson, P.H.; Golzio, C.; Sandell, S.; Luque, H.; Screen, M.; McDonald, K.; Stajich, J.M.; Mahjneh, I.; Vihola, A.; et al. Mutations Affecting the Cytoplasmic Functions of the Co-Chaperone DNAJB6 Cause Limb-Girdle Muscular Dystrophy. Nat. Genet. 2012, 44, 450–455. [Google Scholar] [CrossRef]
- Ghaoui, R.; Cooper, S.T.; Lek, M.; Jones, K.; Corbett, A.; Reddel, S.W.; Needham, M.; Liang, C.; Waddell, L.B.; Nicholson, G.; et al. Use of Whole-Exome Sequencing for Diagnosis of Limb-Girdle Muscular Dystrophy. JAMA Neurol. 2015, 72, 1424. [Google Scholar] [CrossRef]
- Reimann, L.; Wiese, H.; Leber, Y.; Schwäble, A.N.; Fricke, A.L.; Rohland, A.; Knapp, B.; Peikert, C.D.; Drepper, F.; van der Ven, P.F.M.; et al. Myofibrillar Z-Discs Are a Protein Phosphorylation Hot Spot with Protein Kinase C (PKCα) Modulating Protein Dynamics. Mol. Cell. Proteom. 2017, 16, 346–367. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.; Kuhn, C.; Katus, H.A.; Frey, N. The Sarcomeric Z-Disc: A Nodal Point in Signalling and Disease. J. Mol. Med. Berl. Ger. 2006, 84, 446–468. [Google Scholar] [CrossRef]
- Maerkens, A.; Kley, R.A.; Olivé, M.; Theis, V.; van der Ven, P.F.M.; Reimann, J.; Milting, H.; Schreiner, A.; Uszkoreit, J.; Eisenacher, M.; et al. Differential Proteomic Analysis of Abnormal Intramyoplasmic Aggregates in Desminopathy. J. Proteom. 2013, 90, 14–27. [Google Scholar] [CrossRef] [PubMed]
- van den Bogaart, F.J.A.; Claeys, K.G.; Kley, R.A.; Kusters, B.; Schrading, S.; Kamsteeg, E.J.; Voermans, N.C. Widening the Spectrum of Filamin-C Myopathy: Predominantly Proximal Myopathy Due to the p.A193T Mutation in the Actin-Binding Domain of FLNC. Neuromuscul. Disord. 2017, 27, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Juo, L.-Y.; Liao, W.-C.; Shih, Y.-L.; Yang, B.-Y.; Liu, A.-B.; Yan, Y.-T. HSPB7 Interacts with Dimerized FLNC and Its Absence Results in Progressive Myopathy in Skeletal Muscles. J. Cell Sci. 2016, 129, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Kley, R.A.; van der Ven, P.F.M.; Olivé, M.; Höhfeld, J.; Goldfarb, L.G.; Fürst, D.O.; Vorgerd, M. Impairment of Protein Degradation in Myofibrillar Myopathy Caused by FLNC/Filamin C Mutations. Autophagy 2013, 9, 422–423. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.; Kley, R.A.; Strach, K.; Meyer, C.; Sommer, T.; Eger, K.; Rolfs, A.; Meyer, W.; Pou, A.; Pradas, J.; et al. Distinct Muscle Imaging Patterns in Myofibrillar Myopathies. Neurology 2008, 71, 758–765. [Google Scholar] [CrossRef]
- Fürst, D.O.; Goldfarb, L.G.; Kley, R.A.; Vorgerd, M.; Olivé, M.; van der Ven, P.F.M. Filamin C-Related Myopathies: Pathology and Mechanisms. Acta Neuropathol. 2013, 125, 33–46. [Google Scholar] [CrossRef]
- Wattjes, M.P.; Kley, R.A.; Fischer, D. Neuromuscular Imaging in Inherited Muscle Diseases. Eur. Radiol. 2010, 20, 2447–2460. [Google Scholar] [CrossRef]
- Linnemann, A.; van der Ven, P.F.M.; Vakeel, P.; Albinus, B.; Simonis, D.; Bendas, G.; Schenk, J.A.; Micheel, B.; Kley, R.A.; Fürst, D.O. The Sarcomeric Z-Disc Component Myopodin Is a Multiadapter Protein That Interacts with Filamin and Alpha-Actinin. Eur. J. Cell Biol. 2010, 89, 681–692. [Google Scholar] [CrossRef]
- Faulkner, G.; Pallavicini, A.; Comelli, A.; Salamon, M.; Bortoletto, G.; Ievolella, C.; Trevisan, S.; Kojic’, S.; Dalla Vecchia, F.; Laveder, P.; et al. FATZ, a Filamin-, Actinin-, and Telethonin-Binding Protein of the Z-Disc of Skeletal Muscle. J. Biol. Chem. 2000, 275, 41234–41242. [Google Scholar] [CrossRef] [PubMed]
- Frey, N.; Olson, E.N. Calsarcin-3, a Novel Skeletal Muscle-Specific Member of the Calsarcin Family, Interacts with Multiple Z-Disc Proteins. J. Biol. Chem. 2002, 277, 13998–14004. [Google Scholar] [CrossRef] [PubMed]
- Takada, F.; Vander Woude, D.L.; Tong, H.Q.; Thompson, T.G.; Watkins, S.C.; Kunkel, L.M.; Beggs, A.H. Myozenin: An Alpha-Actinin- and Gamma-Filamin-Binding Protein of Skeletal Muscle Z Lines. Proc. Natl. Acad. Sci. USA 2001, 98, 1595–1600. [Google Scholar] [CrossRef] [PubMed]
- Begay, R.L.; Graw, S.L.; Sinagra, G.; Asimaki, A.; Rowland, T.J.; Slavov, D.B.; Gowan, K.; Jones, K.L.; Brun, F.; Merlo, M.; et al. Filamin C Truncation Mutations Are Associated with Arrhythmogenic Dilated Cardiomyopathy and Changes in the Cell-Cell Adhesion Structures. JACC Clin. Electrophysiol. 2018, 4, 504–514. [Google Scholar] [CrossRef]
- Brun, F.; Gigli, M.; Graw, S.L.; Judge, D.P.; Merlo, M.; Murray, B.; Calkins, H.; Sinagra, G.; Taylor, M.R.; Mestroni, L.; et al. FLNC Truncations Cause Arrhythmogenic Right Ventricular Cardiomyopathy. J. Med. Genet. 2020, 57, 254–257. [Google Scholar] [CrossRef]
- Himmel, M.; Van Der Ven, P.F.M.; Stöcklein, W.; Fürst, D.O. The Limits of Promiscuity: Isoform-Specific Dimerization of Filamins. Biochemistry 2003, 42, 430–439. [Google Scholar] [CrossRef]
- Pudas, R.; Kiema, T.-R.; Butler, P.J.G.; Stewart, M.; Ylänne, J. Structural Basis for Vertebrate Filamin Dimerization. Structure 2005, 13, 111–119. [Google Scholar] [CrossRef]
- Wu, T.; Xu, Y.; Zhang, L.; Liang, Z.; Zhou, X.; Evans, S.M.; Chen, J. Filamin C Is Essential for Mammalian Myocardial Integrity. PLoS Genet. 2023, 19, e1010630. [Google Scholar] [CrossRef]
- Molt, S.; Bührdel, J.B.; Yakovlev, S.; Schein, P.; Orfanos, Z.; Kirfel, G.; Winter, L.; Wiche, G.; van der Ven, P.F.M.; Rottbauer, W.; et al. Aciculin Interacts with Filamin C and Xin and Is Essential for Myofibril Assembly, Remodeling and Maintenance. J. Cell Sci. 2014, 127, 3578–3592. [Google Scholar] [CrossRef]
- Gontier, Y.; Taivainen, A.; Fontao, L.; Sonnenberg, A.; van der Flier, A.; Carpen, O.; Faulkner, G.; Borradori, L. The Z-Disc Proteins Myotilin and FATZ-1 Interact with Each Other and Are Connected to the Sarcolemma via Muscle-Specific Filamins. J. Cell Sci. 2005, 118, 3739–3749. [Google Scholar] [CrossRef]
- van der Ven, P.F.; Wiesner, S.; Salmikangas, P.; Auerbach, D.; Himmel, M.; Kempa, S.; Hayess, K.; Pacholsky, D.; Taivainen, A.; Schröder, R.; et al. Indications for a Novel Muscular Dystrophy Pathway. Gamma-Filamin, the Muscle-Specific Filamin Isoform, Interacts with Myotilin. J. Cell Biol. 2000, 151, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Thompson, T.G.; Chan, Y.M.; Hack, A.A.; Brosius, M.; Rajala, M.; Lidov, H.G.; McNally, E.M.; Watkins, S.; Kunkel, L.M. Filamin 2 (FLN2): A Muscle-Specific Sarcoglycan Interacting Protein. J. Cell Biol. 2000, 148, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Paranavitane, V.; Stephens, L.R.; Hawkins, P.T. Structural Determinants of LL5β Subcellular Localisation and Association with Filamin C. Cell. Signal. 2007, 19, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Höhfeld, J.; Benzing, T.; Bloch, W.; Fürst, D.O.; Gehlert, S.; Hesse, M.; Hoffmann, B.; Hoppe, T.; Huesgen, P.F.; Köhn, M.; et al. Maintaining Proteostasis under Mechanical Stress. EMBO Rep. 2021, 22, e52507. [Google Scholar] [CrossRef]
- Odgerel, Z.; van der Ven, P.F.M.; Fürst, D.O.; Goldfarb, L.G. DNA Sequencing Errors in Molecular Diagnostics of Filamin Myopathy. Clin. Chem. Lab. Med. 2010, 48, 1409–1414. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Kley, R.A.; Hellenbroich, Y.; van der Ven, P.F.M.; Furst, D.O.; Huebner, A.; Bruchertseifer, V.; Peters, S.A.; Heyer, C.M.; Kirschner, J.; Schroder, R.; et al. Clinical and Morphological Phenotype of the Filamin Myopathy: A Study of 31 German Patients. Brain 2007, 130, 3250–3264. [Google Scholar] [CrossRef]
- Obermann, W.M.; Gautel, M.; Weber, K.; Fürst, D.O. Molecular Structure of the Sarcomeric M Band: Mapping of Titin and Myosin Binding Domains in Myomesin and the Identification of a Potential Regulatory Phosphorylation Site in Myomesin. EMBO J. 1997, 16, 211–220. [Google Scholar] [CrossRef]
- Löwe, T.; Kley, R.A.; van der Ven, P.F.M.; Himmel, M.; Huebner, A.; Vorgerd, M.; Fürst, D.O. The Pathomechanism of Filaminopathy: Altered Biochemical Properties Explain the Cellular Phenotype of a Protein Aggregation Myopathy. Hum. Mol. Genet. 2007, 16, 1351–1358. [Google Scholar] [CrossRef]
- Schuld, J.; Orfanos, Z.; Chevessier, F.; Eggers, B.; Heil, L.; Uszkoreit, J.; Unger, A.; Kirfel, G.; van der Ven, P.F.M.; Marcus, K.; et al. Homozygous Expression of the Myofibrillar Myopathy-Associated p.W2710X Filamin C Variant Reveals Major Pathomechanisms of Sarcomeric Lesion Formation. Acta Neuropathol. Commun. 2020, 8, 154. [Google Scholar] [CrossRef]
- Nieto-Torres, J.L.; Shanahan, S.-L.; Chassefeyre, R.; Chaiamarit, T.; Zaretski, S.; Landeras-Bueno, S.; Verhelle, A.; Encalada, S.E.; Hansen, M. LC3B Phosphorylation Regulates FYCO1 Binding and Directional Transport of Autophagosomes. Curr. Biol. 2021, 31, 3440–3449.e7. [Google Scholar] [CrossRef] [PubMed]
- Kölbel, H.; Roos, A.; van der Ven, P.F.M.; Evangelista, T.; Nolte, K.; Johnson, K.; Töpf, A.; Wilson, M.; Kress, W.; Sickmann, A.; et al. First Clinical and Myopathological Description of a Myofibrillar Myopathy with Congenital Onset and Homozygous Mutation in FLNC. Hum. Mutat. 2020, 41, 1600–1614. [Google Scholar] [CrossRef] [PubMed]
- Duff, R.M.; Tay, V.; Hackman, P.; Ravenscroft, G.; McLean, C.; Kennedy, P.; Steinbach, A.; Schöffler, W.; van der Ven, P.F.M.; Fürst, D.O.; et al. Mutations in the N-Terminal Actin-Binding Domain of Filamin C Cause a Distal Myopathy. Am. J. Hum. Genet. 2011, 88, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Gemelli, C.; Prada, V.; Fiorillo, C.; Fabbri, S.; Maggi, L.; Geroldi, A.; Gibertini, S.; Mandich, P.; Trevisan, L.; Fossa, P.; et al. A Novel Mutation in the N-Terminal Acting-Binding Domain of Filamin C Protein Causing a Distal Myofibrillar Myopathy. J. Neurol. Sci. 2019, 398, 75–78. [Google Scholar] [CrossRef]
- Zhang, Y.-T.; Pu, C.-Q.; Ban, R.; Liu, H.-X.; Shi, Q.; Lu, X.-H. Clinical, Pathological, and Genetic Features of Two Chinese Cases with Filamin C Myopathy. Chin. Med. J. 2018, 131, 2986–2988. [Google Scholar] [CrossRef]
- Shatunov, A.; Olivé, M.; Odgerel, Z.; Stadelmann-Nessler, C.; Irlbacher, K.; van Landeghem, F.; Bayarsaikhan, M.; Lee, H.-S.; Goudeau, B.; Chinnery, P.F.; et al. In-Frame Deletion in the Seventh Immunoglobulin-like Repeat of Filamin C in a Family with Myofibrillar Myopathy. Eur. J. Hum. Genet. 2009, 17, 656–663. [Google Scholar] [CrossRef]
- Luan, X.; Hong, D.; Zhang, W.; Wang, Z.; Yuan, Y. A Novel Heterozygous Deletion–Insertion Mutation (2695–2712 Del/GTTTGT Ins) in Exon 18 of the Filamin C Gene Causes Filaminopathy in a Large Chinese Family. Neuromuscul. Disord. 2010, 20, 390–396. [Google Scholar] [CrossRef]
- Miao, J.; Su, F.; Liu, X.; Wei, X.; Yuan, Y.; Yu, X. A Case Report: A Heterozygous Deletion (2791_2805 Del) in Exon 18 of the Filamin C Gene Causing Filamin C-Related Myofibrillar Myopathies in a Chinese Family. BMC Neurol. 2018, 18, 79. [Google Scholar] [CrossRef]
- Avila-Smirnow, D.; Gueneau, L.; Batonnet-Pichon, S.; Delort, F.; Bécane, H.-M.; Claeys, K.; Beuvin, M.; Goudeau, B.; Jais, J.-P.; Nelson, I.; et al. Cardiac Arrhythmia and Late-Onset Muscle Weakness Caused by a Myofibrillar Myopathy with Unusual Histopathological Features Due to a Novel Missense Mutation in FLNC. Rev. Neurol. 2016, 172, 594–606. [Google Scholar] [CrossRef]
- Kiselev, A.; Vaz, R.; Knyazeva, A.; Khudiakov, A.; Tarnovskaya, S.; Liu, J.; Sergushichev, A.; Kazakov, S.; Frishman, D.; Smolina, N.; et al. De Novo Mutations in FLNC Leading to Early-Onset Restrictive Cardiomyopathy and Congenital Myopathy. Hum. Mutat. 2018, 39, 1161–1172. [Google Scholar] [CrossRef]
- Matsumura, T.; Inoue, K.; Toyooka, K.; Inoue, M.; Iida, A.; Saito, Y.; Nishikawa, T.; Moriuchi, K.; Beck, G.; Nishino, I.; et al. Clinical Trajectory of a Patient with Filaminopathy Who Developed Arrhythmogenic Cardiomyopathy, Myofibrillar Myopathy, and Multiorgan Tumors. Neuromuscul. Disord. 2021, 31, 1282–1286. [Google Scholar] [CrossRef] [PubMed]
- Muravyev, A.; Vershinina, T.; Tesner, P.; Sjoberg, G.; Fomicheva, Y.; Čajbiková, N.N.; Kozyreva, A.; Zhuk, S.; Mamaeva, E.; Tarnovskaya, S.; et al. Rare Clinical Phenotype of Filaminopathy Presenting as Restrictive Cardiomyopathy and Myopathy in Childhood. Orphanet J. Rare Dis. 2022, 17, 358. [Google Scholar] [CrossRef] [PubMed]
- Guergueltcheva, V.; Peeters, K.; Baets, J.; Ceuterick-de Groote, C.; Martin, J.J.; Suls, A.; De Vriendt, E.; Mihaylova, V.; Chamova, T.; Almeida-Souza, L.; et al. Distal Myopathy with Upper Limb Predominance Caused by Filamin C Haploinsufficiency. Neurology 2011, 77, 2105–2114. [Google Scholar] [CrossRef]
- Rossi, D.; Palmio, J.; Evilä, A.; Galli, L.; Barone, V.; Caldwell, T.A.; Policke, R.A.; Aldkheil, E.; Berndsen, C.E.; Wright, N.T.; et al. A Novel FLNC Frameshift and an OBSCN Variant in a Family with Distal Muscular Dystrophy. PLoS ONE 2017, 12, e0186642. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, J.; Han, C.; Li, Y.; Guo, Y.; Tong, X. A Mutation in the Filamin c Gene Causes Myofibrillar Myopathy with Lower Motor Neuron Syndrome: A Case Report. BMC Neurol. 2019, 19, 198. [Google Scholar] [CrossRef] [PubMed]
- Tasca, G.; Odgerel, Z.; Monforte, M.; Aurino, S.; Clarke, N.F.; Waddell, L.B.; Udd, B.; Ricci, E.; Goldfarb, L.G. Novel FLNC Mutation in a Patient with Myofibrillar Myopathy in Combination with Late-Onset Cerebellar Ataxia. Muscle Nerve 2012, 46, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Verdonschot, J.A.J.; Vanhoutte, E.K.; Claes, G.R.F.; Helderman-van den Enden, A.T.J.M.; Hoeijmakers, J.G.J.; Hellebrekers, D.M.E.I.; Haan, A.; Christiaans, I.; Lekanne Deprez, R.H.; Boen, H.M.; et al. A Mutation Update for the FLNC Gene in Myopathies and Cardiomyopathies. Hum. Mutat. 2020, 41, 1091–1111. [Google Scholar] [CrossRef]
- Lee, H.-C.H.; Wong, S.; Sheng, B.; Pan, N.-Y.K.; Leung, Y.-K.F.; Lau, K.-K.D.; Cheng, Y.S.; Ho, L.-C.; Li, R.; Lee, C.-N.; et al. Clinical and Pathological Characterization of FLNC-Related Myofibrillar Myopathy Caused by Founder Variant c.8129G>A in Hong Kong Chinese. Clin. Genet. 2020, 97, 747–757. [Google Scholar] [CrossRef]
- Park, Y.-E.; Kim, D.-S.; Shin, J.-H. A Novel Nonsense Mutation in the Dimerization Domain of FLNC Causing Mild Myofibrillar Myopathy. Clin. Neurol. Neurosurg. 2022, 221, 107386. [Google Scholar] [CrossRef]
- Kley, R.A.; Serdaroglu-Oflazer, P.; Leber, Y.; Odgerel, Z.; van der Ven, P.F.M.; Olivé, M.; Ferrer, I.; Onipe, A.; Mihaylov, M.; Bilbao, J.M.; et al. Pathophysiology of Protein Aggregation and Extended Phenotyping in Filaminopathy. Brain J. Neurol. 2012, 135, 2642–2660. [Google Scholar] [CrossRef]
- Kley, R.A.; Leber, Y.; Schrank, B.; Zhuge, H.; Orfanos, Z.; Kostan, J.; Onipe, A.; Sellung, D.; Güttsches, A.K.; Eggers, B.; et al. FLNC-Associated Myofibrillar Myopathy. Neurol. Genet. 2021, 7, e590. [Google Scholar] [CrossRef]
- Chevessier, F.; Schuld, J.; Orfanos, Z.; Plank, A.-C.; Wolf, L.; Maerkens, A.; Unger, A.; Schlötzer-Schrehardt, U.; Kley, R.A.; Von Hörsten, S.; et al. Myofibrillar Instability Exacerbated by Acute Exercise in Filaminopathy. Hum. Mol. Genet. 2015, 24, 7207–7220. [Google Scholar] [CrossRef] [PubMed]
- Gigli, M.; Stolfo, D.; Graw, S.L.; Merlo, M.; Gregorio, C.; Nee Chen, S.; Dal Ferro, M.; PaldinoMD, A.; De Angelis, G.; Brun, F.; et al. Phenotypic Expression, Natural History, and Risk Stratification of Cardiomyopathy Caused by Filamin C Truncating Variants. Circulation 2021, 144, 1600–1611. [Google Scholar] [CrossRef]
- Agarwal, R.; Paulo, J.A.; Toepfer, C.N.; Ewoldt, J.K.; Sundaram, S.; Chopra, A.; Zhang, Q.; Gorham, J.; DePalma, S.R.; Chen, C.S.; et al. Filamin C Cardiomyopathy Variants Cause Protein and Lysosome Accumulation. Circ. Res. 2021, 129, 751–766. [Google Scholar] [CrossRef] [PubMed]
- Ulbricht, A.; Eppler, F.J.; Tapia, V.E.; van der Ven, P.F.M.; Hampe, N.; Hersch, N.; Vakeel, P.; Stadel, D.; Haas, A.; Saftig, P.; et al. Cellular Mechanotransduction Relies on Tension-Induced and Chaperone-Assisted Autophagy. Curr. Biol. 2013, 23, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Hiniker, A.; Daniels, B.H.; Lee, H.S.; Margeta, M. Comparative Utility of LC3, P62 and TDP-43 Immunohistochemistry in Differentiation of Inclusion Body Myositis from Polymyositis and Related Inflammatory Myopathies. Acta Neuropathol. Commun. 2013, 1, 29. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Ye, L.; Huang, W.F.; Guo, L.J.; Xu, Z.G.; Wu, H.L.; Yang, C.; Liu, H.F. P62 Links the Autophagy Pathway and the Ubiqutin–Proteasome System upon Ubiquitinated Protein Degradation. Cell. Mol. Biol. Lett. 2016, 21, 29. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sellung, D.; Heil, L.; Daya, N.; Jacobsen, F.; Mertens-Rill, J.; Zhuge, H.; Döring, K.; Piran, M.; Milting, H.; Unger, A.; et al. Novel Filamin C Myofibrillar Myopathy Variants Cause Different Pathomechanisms and Alterations in Protein Quality Systems. Cells 2023, 12, 1321. https://doi.org/10.3390/cells12091321
Sellung D, Heil L, Daya N, Jacobsen F, Mertens-Rill J, Zhuge H, Döring K, Piran M, Milting H, Unger A, et al. Novel Filamin C Myofibrillar Myopathy Variants Cause Different Pathomechanisms and Alterations in Protein Quality Systems. Cells. 2023; 12(9):1321. https://doi.org/10.3390/cells12091321
Chicago/Turabian StyleSellung, Dominik, Lorena Heil, Nassam Daya, Frank Jacobsen, Janine Mertens-Rill, Heidi Zhuge, Kristina Döring, Misagh Piran, Hendrik Milting, Andreas Unger, and et al. 2023. "Novel Filamin C Myofibrillar Myopathy Variants Cause Different Pathomechanisms and Alterations in Protein Quality Systems" Cells 12, no. 9: 1321. https://doi.org/10.3390/cells12091321







