Amyloidogenic and Neuroinflammatory Molecular Pathways Are Contrasted Using Menaquinone 4 (MK4) and Reduced Menaquinone 7 (MK7R) in Association with Increased DNA Methylation in SK-N-BE Neuroblastoma Cell Line
Abstract
:1. Introduction
2. Materials and Methods
2.1. Vitamin K Derivatives
2.2. Cell Culture
2.3. UPLC-PDA Chromatography for Vitamin K Determination
2.4. Quantitative Real-Time PCR Analysis
2.5. Protein Analysis
2.6. Enzyme-Linked Immunosorbent Assay (ELISA)
2.7. DNA Methylation Assay to Profile CpG and Non-CpG Methylation
2.8. Statistical Analysis
3. Results
3.1. Solubility and Uptake
3.2. Neurodegeneration-Associated Pathway
3.3. Neuroinflammation-Associated Pathway
3.4. DNA Methylation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dam, B.Y.H.; Schønheyder, F. The occurrence and chemical nature of vitamin k. Biochem. J. 1936, 30, 897–901. [Google Scholar] [CrossRef] [PubMed]
- Yagami, T.; Ueda, K.; Asakura, K.; Sakaeda, T.; Nakazato, H.; Kuroda, T.; Hata, S.; Sakaguchi, G.; Itoh, N.; Nakano, T.; et al. Gas6 rescues cortical neurons from amyloid beta protein-induced apoptosis. Neuropharmacology 2002, 43, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Ohsaki, Y.; Shirakawa, H.; Hiwatashi, K.; Furukawa, Y.; Mizutani, T.; Komai, M. Vitamin K suppresses lipopolysaccharide-induced inflammation in the rat. Biosci. Biotechnol. Biochem. 2006, 70, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Ohsaki, Y.; Shirakawa, H.; Miura, A.; Giriwono, P.E.; Sato, S.; Ohashi, A.; Iribe, M.; Goto, T.; Komai, M. Vitamin K suppresses the lipopolysaccharide-induced expression of inflammatory cytokines in cultured macrophage-like cells via the inhibition of the activation of nuclear factor κB through the repression of IKKα/β phosphorylation. J. Nutr. Biochem. 2010, 21, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- Mukai, K.; Itoh, S.; Morimoto, H. Stopped-flow kinetic study of vitamin E regeneration reaction with biological hydroquinones (reduced forms of ubiquinone, vitamin K and tocopherolquinone) in solution. J. Biol. Chem. 1992, 267, 22277–22281. [Google Scholar] [CrossRef] [PubMed]
- Vervoort, L.M.; Ronden, J.E.; Thijssen, H.H. The potent antioxidant activity of the vitamin K cycle in microsomal lipid peroxidation. Biochem. Pharmacol. 1997, 54, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Ambrożewicz, E.; Muszyńska, M.; Tokajuk, G.; Grynkiewicz, G.; Žarković, N.; Skrzydlewska, E. Beneficial effects of vitamins K and D3 on redox balance of human osteoblasts cultured with hydroxypatite-based biomaterials. Cells 2019, 8, 325. [Google Scholar] [CrossRef]
- Xv, F.; Chen, J.; Duan, L.; Li, S. Research progress on the anticancer effects of vitamin K2. Oncol. Lett. 2018, 15, 8926–8934. [Google Scholar] [CrossRef]
- Wen, L.; Chen, J.; Duan, L.; Li, S. Vitamin K-dependent proteins involved in bone and cardiovascular health. Mol. Med. Rep. 2018, 18, 3–15. [Google Scholar] [CrossRef]
- Alisi, L.; Cao, R.; De Angelis, C.; Cafolla, A.; Caramia, F.; Cartocci, G.; Librando, A.; Fiorelli, M. The relationships between vitamin K and cognition: A Review of Current Evidence. Front. Neurol. 2019, 10, 239. [Google Scholar] [CrossRef]
- Simes, D.C.; Viegas, C.S.B.; Araújo, N.; Marreiros, C. Vitamin K as a powerful micronutrient in aging and age-related diseases: Pros and cons from clinical studies. Int. J. Mol. Sci. 2019, 20, 4150. [Google Scholar] [CrossRef] [PubMed]
- Ferland, G. Vitamin K, an emerging nutrient in brain function. BioFactors 2012, 38, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Tamboli, I.Y.; Hampel, H.; Tien, N.T.; Tolksdorf, K.; Breiden, B.; Mathews, P.M.; Saftig, P.; Sandhoff, K.; Walter, J. Sphingolipid storage affects autophagic metabolism of the amyloid precursor protein and promotes Abeta generation. J. Neurosci. 2011, 31, 1837–1849. [Google Scholar] [CrossRef] [PubMed]
- Allison, A.C. The possible role of vitamin K deficiency in the pathogenesis of Alzheimer’s disease and in augmenting brain damage associated with cardiovascular disease. Med. Hypotheses 2001, 57, 151–155. [Google Scholar] [CrossRef]
- Presse, N.; Shatenstein, B.; Kergoat, M.J.; Ferland, G. Low vitamin K intakes in community-dwelling elders at an early stage of Alzheimer’s disease. J. Am. Diet Assoc. 2008, 108, 2095–2099. [Google Scholar] [CrossRef] [PubMed]
- Presse, N.; Belleville, S.; Gaudreau, P.; Greenwood, C.E.; Kergoat, M.J.; Morais, J.A.; Payette, H.; Shatenstein, B.; Ferland, G. Vitamin K status and cognitive function in healthy older adults. Neurobiol. Aging 2013, 34, 2777–2783. [Google Scholar] [CrossRef] [PubMed]
- Emekli-Alturfan, E.; Alturfan, A.A. The emerging relationship between vitamin K and neurodegenerative diseases: A review of current evidence. Mol. Biol. Rep. 2023, 50, 815–828. [Google Scholar] [CrossRef]
- Popescu, A.; German, M. Vitamin K2 holds promise for Alzheimer’s prevention and treatment. Nutrients 2021, 13, 2206. [Google Scholar] [CrossRef]
- Shandilya, S.; Kesari, K.K.; Ruokolainen, J. Vitamin K2 Modulates Organelle Damage and Tauopathy Induced by Streptozotocin and Menadione in SH-SY5Y Cells. Antioxidants 2021, 10, 983. [Google Scholar] [CrossRef]
- Booth, S.L.; Shea, M.K.; Barger, K.; Leurgans, S.E.; James, B.D.; Holland, T.M.; Agarwal, P.; Fu, X.; Wang, J.; Matuszek, G.; et al. Association of vitamin K with cognitive decline and neuropathology in community-dwelling older persons. Alzheimers Dement. 2022, 8, e12255. [Google Scholar] [CrossRef]
- Chatterjee, K.; Mazumder, P.M.; Banerjee, S. Vitamin K2 protects against aluminium chloride-mediated neurodegeneration. Inflammopharmacology 2023, 31, 2675–2684. [Google Scholar] [CrossRef] [PubMed]
- Theuwissen, E.; Cranenburg, E.C.; Knapen, M.H.; Magdeleyns, E.J.; Teunissen, K.J.; Schurgers, L.J.; Smit, E.; Vermeer, C. Low-dose menaquinone-7 supplementation improved extra-hepatic vitamin K status, but had no effect on thrombin generation in healthy subjects. Br. J. Nutr. 2012, 108, 1652–1657. [Google Scholar] [CrossRef] [PubMed]
- Schulman, S.; Furie, B. How I treat poisoning with vitamin K antagonists. Blood 2015, 125, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Spahr, J.E.; Maul, J.S.; Rodgers, G.M. Superwarfarin poisoning: A Report of Two Cases and Review of the Literature. Am. J. Hematol. 2007, 82, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Suttie, J.W. The importance of menaquinones in human nutrition. Annu. Rev. Nutr. 1995, 15, 399–417. [Google Scholar] [CrossRef]
- Walther, B.; Karl, J.P.; Booth, S.L.; Boyaval, P. Menaquinones, bacteria, and the food supply: The relevance of dairy and fermented food products to vitamin K requirements. Adv. Nutr. 2013, 4, 463–473. [Google Scholar] [CrossRef]
- Collins, M.D.; Jones, D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol. Rev. 1981, 45, 316–354. [Google Scholar] [CrossRef]
- Thijssen, H.H.; Drittij-Reijnders, M.J. Vitamin K status in human tissues: Tissue-specific accumulation of phylloquinone and menaquinone-4. Br. J. Nutr. 1996, 75, 121–127. [Google Scholar] [CrossRef]
- Hirota, Y.; Tsugawa, N.; Nakagawa, K.; Suhara, Y.; Tanaka, K.; Uchino, Y.; Takeuchi, A.; Sawada, N.; Kamao, M.; Wada, A.; et al. Menadione (vitamin K3) is a catabolic product of oral phylloquinone (vitamin K1) in the intestine and a circulating precursor of tissue menaquinone-4 (vitamin K2) in rats. J. Biol. Chem. 2013, 288, 33071–33080. [Google Scholar] [CrossRef]
- Shearer, M.J.; Newman, P. Metabolism and cell biology of vitamin K. Thromb. Haemost. 2008, 100, 530–547. [Google Scholar]
- Shearer, M.J.; Newman, P. Recent trends in the metabolism and cell biology of vitamin K with special reference to vitamin K cycling and MK-4 biosynthesis. J. Lipid Res. 2014, 55, 345–362. [Google Scholar] [CrossRef] [PubMed]
- Conly, J.M.; Stein, K. The production of menaquinones (vitamin K2) by intestinal bacteria and their role in maintaining coagulation homeostasis. Prog. Food Nutr. Sci. 1992, 16, 307–343. [Google Scholar] [PubMed]
- Stafford, D.W. The vitamin K cycle. J. Thromb. Haemost. 2005, 3, 1873–1878. [Google Scholar] [CrossRef] [PubMed]
- Lurie, Y.; Loebstein, R.; Kurnik, D.; Almog, S.; Halkin, H. Warfarin and vitamin K intake in the era of pharmacogenetics. Br. J. Clin. Pharmacol. 2010, 70, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Schurgers, L.J.; Teunissen, K.J.; Hamulyák, K.; Knapen, M.H.; Vik, H.; Vermeer, C. Vitamin K-containing dietary supplements: Comparison of synthetic vitamin K1 and natto-derived menaquinone-7. Blood 2007, 109, 3279–3283. [Google Scholar] [CrossRef] [PubMed]
- Maresz, K. Growing evidence of a proven mechanism shows Vitamin K2 can impact health conditions beyond bone and cardiovascular. Integr. Med. 2021, 20, 34–38. [Google Scholar]
- Gorgels, T.G.; Waarsing, J.H.; Herfs, M.; Versteeg, D.; Schoensiegel, F.; Sato, T.; Schlingemann, R.O.; Ivandic, B.; Vermeer, C.; Schurgers, L.J.; et al. Vitamin K supplementation increases vitamin K tissue levels but fails to counteract ectopic calcification in a mouse model for pseudoxanthoma elasticum. J. Mol. Med. 2011, 89, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Iqbal, M.; Tariq, M.; Baig, S.M.; Abbas, W. Epigenetic regulation of HIV-1 latency: Focus on polycomb group (PcG) proteins. Clin. Epigenet. 2018, 10, 14. [Google Scholar] [CrossRef]
- Feinberg, A.P. Phenotypic plasticity and the epigenetics of human disease. Nature 2007, 447, 433–440. [Google Scholar] [CrossRef]
- Herceg, Z. Epigenetics and cancer: Towards an evaluation of the impact of environmental and dietary factors. Mutagenesis 2007, 22, 91–103. [Google Scholar] [CrossRef]
- Choi, S.W.; Friso, S. Epigenetics: A New Bridge between Nutrition and Health. Adv. Nutr. 2010, 1, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Carlos-Reyes, Á.; López-González, J.S.; Meneses-Flores, M.; Gallardo-Rincón, D.; Ruíz-García, E.; Marchat, L.A.; Astudillo-de la Vega, H.; Hernández de la Cruz, O.N.; López-Camarillo, C. Dietary compounds as epigenetic modulating agents in cancer. Front. Genet. 2019, 10, 79. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, M.; Srinivasan, A. Epigenetic gene regulation by dietary compounds in cancer prevention. Adv. Nutr. 2019, 10, 1012–1028. [Google Scholar] [CrossRef] [PubMed]
- Friso, S.; De Santis, D.; Pizzolo, F.; Udali, S. Chapter 30—Vitamins and epigenetics. In Molecular Nutrition; Patel, V.B., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 633–650. [Google Scholar] [CrossRef]
- Yu, Y.X.; Li, Y.P.; Gao, F.; Hu, Q.S.; Zhang, Y.; Chen, D.; Wang, G.H. Vitamin K2 suppresses rotenone-induced microglial activation in vitro. Acta Pharmacol. Sin. 2016, 37, 1178–2289. [Google Scholar] [CrossRef] [PubMed]
- Setoguchi, S.; Watase, D.; Matsunaga, K.; Matsubara, M.; Kubo, Y.; Kusuda, M.; Nagata-Akaho, N.; Enjoji, M.; Nakashima, M.; Takeshita, M.; et al. Enhanced antitumor effects of novel intracellular delivery of an active form of menaquinone-4, menahydroquinone-4, into hepatocellular carcinoma. Cancer Prev. Res. 2015, 8, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Ma, P.; Kong, L.; Wang, X.; Wang, Y.; Jiang, L. Vitamin K2 inhibits hepatocellular carcinoma cell proliferation by binding to 17β-Hydroxysteroid Dehydrogenase 4. Front. Oncol. 2021, 11, 757603. [Google Scholar] [CrossRef]
- Saputra, W.D.; Aoyama, N.; Komai, M.; Shirakawa, H. Menaquinone-4 suppresses lipopolysaccharide-induced inflammation in MG6 Mouse Microglia-Derived Cells by inhibiting the NF-Κb signaling pathway. Int. J. Mol. Sci. 2019, 20, 2317. [Google Scholar] [CrossRef]
- Huang, S.H.; Fang, S.T.; Chen, Y.C. Molecular mechanism of Vitamin K2 protection against amyloid-β-induced cytotoxicity. Biomolecules 2021, 11, 423. [Google Scholar] [CrossRef]
- Rishipal, S.; Alka, P.; Mojeer, H.; Bibhu Prasad, P. Development of a Rapid HPLC-UV Method for Analysis of Menaquinone-7 in Soy Nutraceutical. Pharm. Anal. Acta 2016, 7, 12. [Google Scholar] [CrossRef]
- Raia, T.; Armeli, F.; Cavallaro, R.A.; Ferraguti, G.; Businaro, R.; Lucarelli, M.; Fuso, A. Perinatal S-Adenosylmethionine supplementation represses PSEN1 expression by the cellular epigenetic memory of CpG and Non-CpG methylation in adult TgCRD8 Mice. Int. J. Mol. Sci. 2023, 24, 11675. [Google Scholar] [CrossRef]
- Fuso, A.; Ferraguti, G.; Scarpa, S.; Ferrer, I.; Lucarelli, M. Disclosing bias in bisulfite assay: MethPrimers underestimate high DNA methylation. PLoS ONE 2015, 10, e0118318. [Google Scholar] [CrossRef] [PubMed]
- Fuso, A.; Iyer, A.M.; van Scheppingen, J.; Maccarrone, M.; Scholl, T.; Hainfellner, J.A.; Feucht, M.; Jansen, F.E.; Spliet, W.G.; Krsek, P.; et al. Promoter-specific hypomethylation correlates with IL-1β overexpression in tuberous sclerosis complex (TSC). J. Mol. Neurosci. 2016, 59, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Dinicola, S.; Proietti, S.; Cucina, A.; Bizzarri, M.; Fuso, A. Alpha-Lipoic Acid downregulates IL-1β and IL-6 by DNA hypermethylation in SK-N-BE neuroblastoma cells. Antioxidants 2017, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Monti, N.; Cavallaro, R.A.; Stoccoro, A.; Nicolia, V.; Scarpa, S.; Kovacs, G.G.; Fiorenza, M.T.; Lucarelli, M.; Aronica, E.; Ferrer, I.; et al. CpG and non-CpG Presenilin1 methylation pattern in course of neurodevelopment and neurodegeneration is associated with gene expression in human and murine brain. Epigenetics 2020, 15, 781–799. [Google Scholar] [CrossRef]
- Sato, T.; Schurgers, L.J.; Uenishi, K. Comparison of Menaquinone-4 and Menaquinone-7 bioavailability in healthy women. Nutr. J. 2012, 11, 93. [Google Scholar] [CrossRef]
- Lin, X.; Wen, X.; Wei, Z.; Guo, K.; Shi, F.; Huang, T.; Wang, W.; Zheng, J. Vitamin K2 protects against Aβ42-induced neurotoxicity by activating autophagy and improving mitochondrial function in Drosophila. Neuroreport 2021, 32, 431–437. [Google Scholar] [CrossRef]
- Ramazani, E.; Fereidoni, M.; Tayarani-Najaran, Z. Protective effects of vitamin K2 on 6-OHDA-induced apoptosis in PC12 cells through modulation bax and caspase-3 activation. Mol. Biol. Rep. 2019, 46, 5777–5783. [Google Scholar] [CrossRef]
- Fuso, A.; Nicolia, V.; Cavallaro, R.A.; Ricceri, L.; D’Anselmi, F.; Coluccia, P.; Calamandrei, G.; Scarpa, S. B-vitamin deprivation induces hyperhomocysteinemia and brain S-adenosylhomocysteine, depletes brain S-adenosylmethionine, and enhances PS1 and BACE expression and amyloid-beta deposition in mice. Mol. Cell Neurosci. 2008, 37, 731–746. [Google Scholar] [CrossRef]
- Fuso, A.; Nicolia, V.; Ricceri, L.; Cavallaro, R.A.; Isopi, E.; Mangia, F.; Fiorenza, M.T.; Scarpa, S. S-adenosylmethionine reduces the progress of the Alzheimer-like features induced by B-vitamin deficiency in mice. Neurobiol. Aging 2012, 33, 1482.e1-16. [Google Scholar] [CrossRef]
- Hooper, N.M.; Turner, A.J. The search for alpha-secretase and its potential as a therapeutic approach to Alzheimer s disease. Curr. Med. Chem. 2002, 9, 1107–1119. [Google Scholar] [CrossRef]
- Qian, M.; Shen, X.; Wang, H. The distinct role of ADAM17 in APP proteolysis and microglial activation related to Alzheimer’s Disease. Cell Mol. Neurobiol. 2016, 36, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Glass, C.K.; Saijo, K.; Winner, B.; Marchetto, M.C.; Gage, F.H. Mechanisms underlying inflammation in neurodegeneration. Cell 2010, 140, 918–934. [Google Scholar] [CrossRef] [PubMed]
- Fuso, A.; Cavallaro, R.A.; Zampelli, A.; D’Anselmi, F.; Piscopo, P.; Confaloni, A.; Scarpa, S. Gamma-Secretase is differentially modulated by alterations of homocysteine cycle in neuroblastoma and glioblastoma cells. J. Alzheimers Dis. 2007, 11, 275–290. [Google Scholar] [CrossRef] [PubMed]
- Nicolia, V.; Cavallaro, R.A.; López-González, I.; Maccarrone, M.; Scarpa, S.; Ferrer, I.; Fuso, A. DNA methylation profiles of selected pro-inflammatory cytokines in Alzheimer Disease. J. Neuropathol. Exp. Neurol. 2017, 76, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Heaney, R.P. The nutrient problem. Nutr. Rev. 2012, 70, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Shao, A.; Drewnowski, A.; Willcox, D.C.; Krämer, L.; Lausted, C.; Eggersdorfer, M.; Mathers, J.; Bell, J.D.; Randolph, R.K.; Witkamp, R.; et al. Optimal nutrition and the ever- changing dietary landscape: A conference report. Eur. J. Nutr. 2017, 56, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Darnton-Hill, I. Public health aspects in the prevention and control of Vitamin deficiencies. Curr. Dev. Nutr. 2019, 3, nzz075. [Google Scholar] [CrossRef]
- Shenkin, A. Micronutrients in health and disease. Postgrad Med. J. 2006, 82, 559–567. [Google Scholar] [CrossRef]
- Woteki, C.E.; Thomas, P.R. Eat for Life: The Food and Nutrition Board’s Guide to Reducing Your Risk of Chronic Disease; Institute of Medicine (US) Committee on Diet and Health, Ed.; National Academies Press (US): Washington, DC, USA, 1992.
- Kurutas, E.B. The importance of antioxidants which play the role in cellular response against oxidative/nitrosative stress: Current state. Nutr. J. 2016, 15, 71. [Google Scholar] [CrossRef]
- Sies, H.; Stahl, W.; Sundquist, A.R. Antioxidant functions of vitamins. Vitamins E and C, beta-carotene, and other carotenoids. Ann. N. Y. Acad. Sci. 1992, 669, 7–20. [Google Scholar] [CrossRef]
- Rai, S.N.; Singh, P.; Steinbusch, H.W.M.; Vamanu, E.; Ashraf, G.; Singh, M.P. The role of Vitamins in neurodegenerative disease: An Update. Biomedicines 2021, 9, 1284. [Google Scholar] [CrossRef] [PubMed]
- Grimm, M.O.; Mett, J.; Hartmann, T. The Impact of Vitamin E and Other Fat-Soluble Vitamins on Alzheimer’s Disease. Int. J. Mol. Sci. 2016, 17, 1785. [Google Scholar] [CrossRef] [PubMed]
- Sandeep; Sahu, M.R.; Rani, L.; Kharat, A.S.; Mondal, A.C. Could Vitamins Have a Positive Impact on the Treatment of Parkinson’s Disease? Brain Sci. 2023, 13, 272. [Google Scholar] [CrossRef] [PubMed]
- Kola, A.; Nencioni, F.; Valensin, D. Bioinorganic Chemistry of Micronutrients Related to Alzheimer’s and Parkinson’s Diseases. Molecules 2023, 28, 5467. [Google Scholar] [CrossRef]



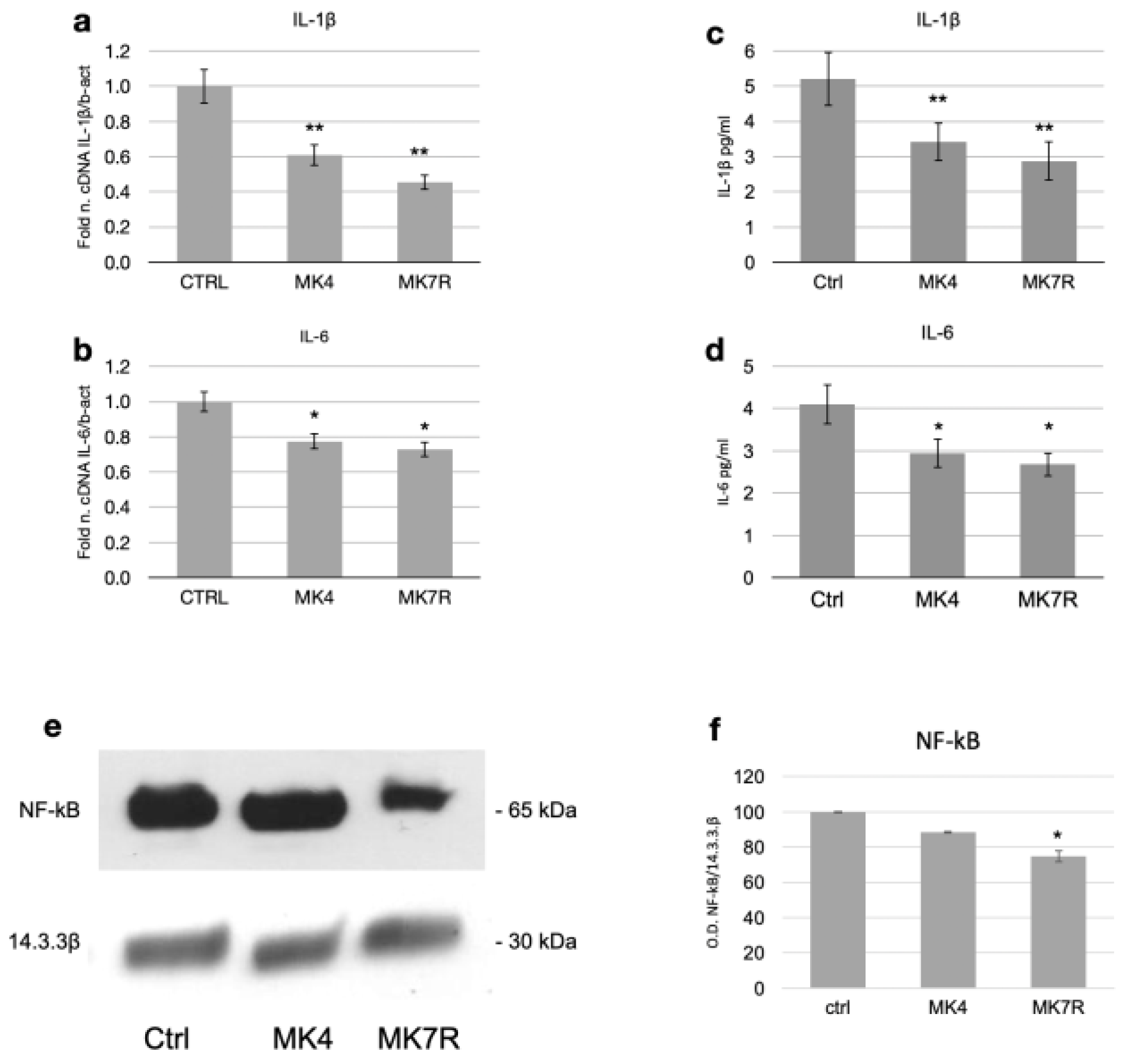


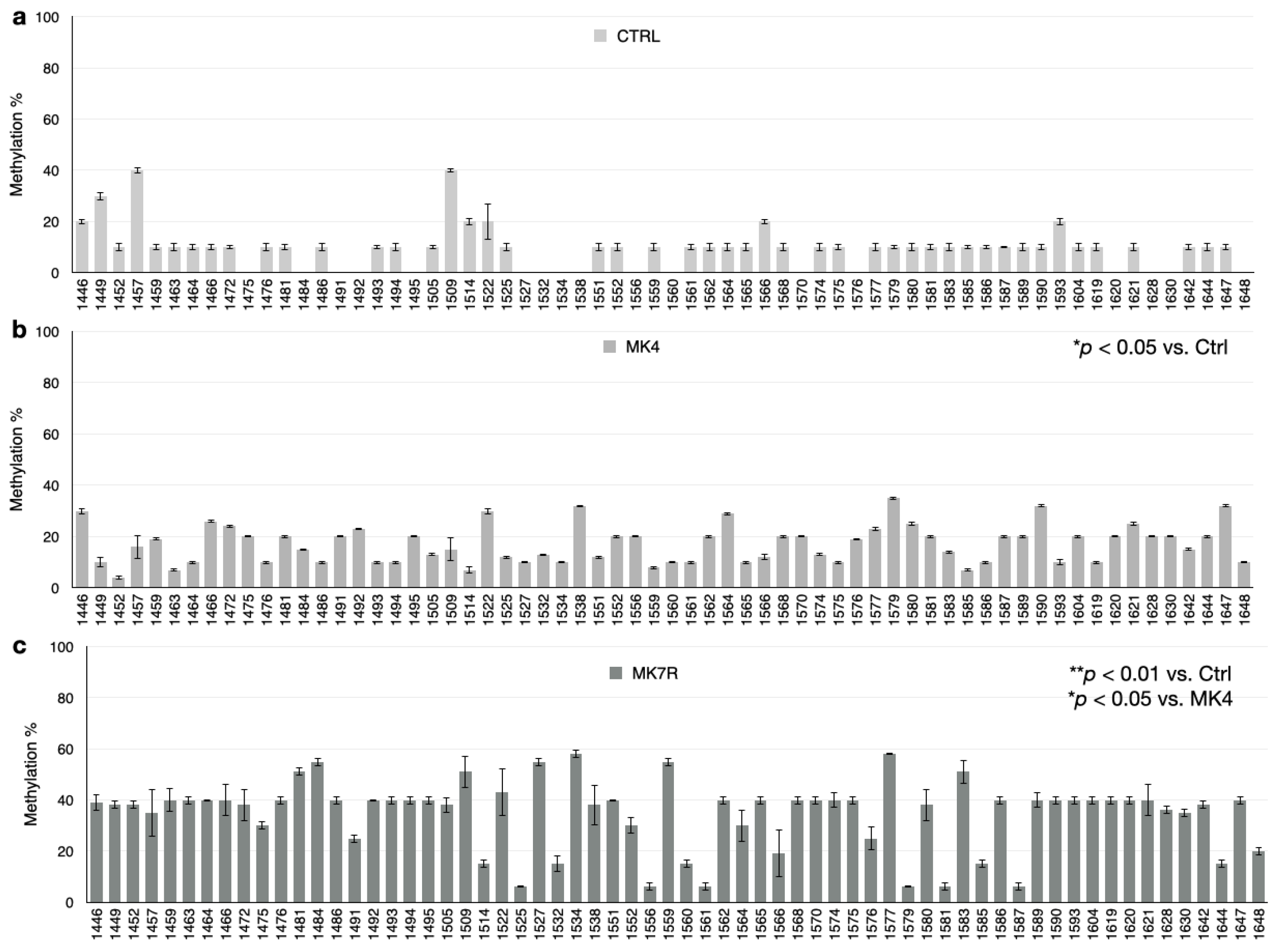

| Gene Name | Primer | Primer Sequence (5′-3′) | Primer Length (bp) | Amplicon Size (bp) |
|---|---|---|---|---|
| β-actin | Forward | CAACCGCGAGAAGATGACC | 19 | 94 |
| Reverse | AGAGGCGTACAGGGATAGCA | 20 | ||
| PSEN 1 | Forward | GGTCGTGGCTACCATTAAGTC | 21 | 94 |
| Reverse | GCCCACAGTCTCGGTATCTT | 20 | ||
| BACE 1 | Forward | AACGAATTGGCTTTGCTGTC | 20 | 102 |
| Reverse | AGCCACAGTCTTCCATGTCC | 20 | ||
| APP | Forward | GAACTACATCACCGCTCTGC | 20 | 77 |
| Reverse | CGCGGACATACTTCTTTAGC | 20 | ||
| ADAM10 | Forward | TGCCATGTATGCTGTATGAAGA | 22 | 109 |
| Reverse | ATCCAGGTTGCAGGGTGAT | 19 | ||
| ADAM17 | Forward | TCAAGAATGTTTCACGTTTGC | 21 | 81 |
| Reverse | ACCCTTTTGGGAGCAACTCT | 20 | ||
| IL-1β | Forward | TCCCCAGCCCTTTTGTTGA | 19 | 332 |
| Reverse | TTAGAACCAAATGTGGCCGTG | 21 | ||
| IL-6 | Forward | GGCACTGGCAGAAAACAACC | 20 | 257 |
| Reverse | GCAAGTCTCCTCATTGAATCC | 21 |
| Protein | Antibody | Manufacturer | Band Size | |
|---|---|---|---|---|
| APP | MAB348 | Monoclonal | Chemicon International, Temecula, CA, USA | 110 kDa |
| PSEN1 | MAB5232 | Monoclonal | Chemicon International, Temecula, CA, USA | 18–20 kDa |
| BACE 1 | sc-10055 | Polyclonal | Santa Cruz Byotechnology, Santa Cruz, CA, USA | 70 kDa |
| ADAM10 | AB19026 | Polyclonal | Chemicon International, Temecula, CA, USA | 60–85 kDa |
| ADAM17 | sc-6416 | Polyclonal | Santa Cruz Byotechnology, Santa Cruz, CA, USA | 85 kDa |
| sAPPα | 11088 | Monoclonal | IBL, Gunma, Japan | 80–100 kDa |
| NF-kB | D14E12 | Monoclonal | Cell Signaling Technology | 65 kDa |
| 14-3-3β | sc-629 | Polyclonal | Santa Cruz Byotechnology, Santa Cruz, CA, USA | 30 kDa |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orticello, M.; Cavallaro, R.A.; Antinori, D.; Raia, T.; Lucarelli, M.; Fuso, A. Amyloidogenic and Neuroinflammatory Molecular Pathways Are Contrasted Using Menaquinone 4 (MK4) and Reduced Menaquinone 7 (MK7R) in Association with Increased DNA Methylation in SK-N-BE Neuroblastoma Cell Line. Cells 2024, 13, 58. https://doi.org/10.3390/cells13010058
Orticello M, Cavallaro RA, Antinori D, Raia T, Lucarelli M, Fuso A. Amyloidogenic and Neuroinflammatory Molecular Pathways Are Contrasted Using Menaquinone 4 (MK4) and Reduced Menaquinone 7 (MK7R) in Association with Increased DNA Methylation in SK-N-BE Neuroblastoma Cell Line. Cells. 2024; 13(1):58. https://doi.org/10.3390/cells13010058
Chicago/Turabian StyleOrticello, Michela, Rosaria A. Cavallaro, Daniele Antinori, Tiziana Raia, Marco Lucarelli, and Andrea Fuso. 2024. "Amyloidogenic and Neuroinflammatory Molecular Pathways Are Contrasted Using Menaquinone 4 (MK4) and Reduced Menaquinone 7 (MK7R) in Association with Increased DNA Methylation in SK-N-BE Neuroblastoma Cell Line" Cells 13, no. 1: 58. https://doi.org/10.3390/cells13010058
APA StyleOrticello, M., Cavallaro, R. A., Antinori, D., Raia, T., Lucarelli, M., & Fuso, A. (2024). Amyloidogenic and Neuroinflammatory Molecular Pathways Are Contrasted Using Menaquinone 4 (MK4) and Reduced Menaquinone 7 (MK7R) in Association with Increased DNA Methylation in SK-N-BE Neuroblastoma Cell Line. Cells, 13(1), 58. https://doi.org/10.3390/cells13010058












