Phosphoproteomics Reveals Selective Regulation of Signaling Pathways by Lysophosphatidic Acid Species in Macrophages
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Culture of Monocyte-Derived Macrophages (MDMs) from Healthy Donors
2.2. LPA and Recombinant Proteins
2.3. Measurement of Intracellular cAMP
2.4. RNA Sequencing
2.5. Sample Preparation for Mass Spectrometry (MS)
2.6. Mass Spectrometry (MS)
2.7. Immunoblotting and Quantification
2.8. Staining of Actin Filaments
2.9. Transwell Migration Assay
2.10. Macropinocytosis Assay
2.11. Functional Annotations
2.12. Other Statistical Analyses
3. Results
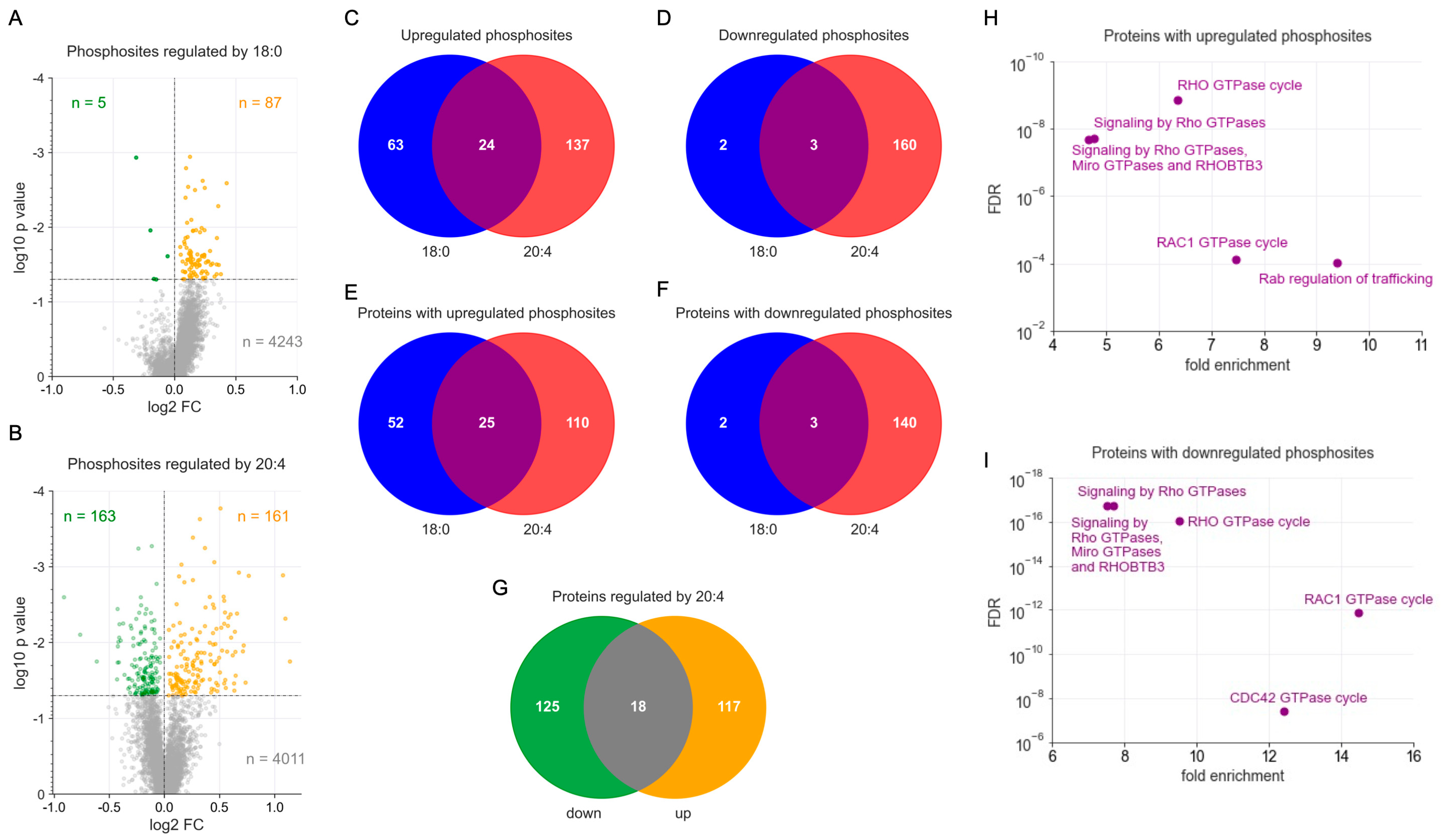
3.1. Phosphoproteomics Identifies LPA-Species-Selective Target Proteins in MDMs
3.2. Functional Annotation of 18:0 and 20:4 LPAs Target Proteins
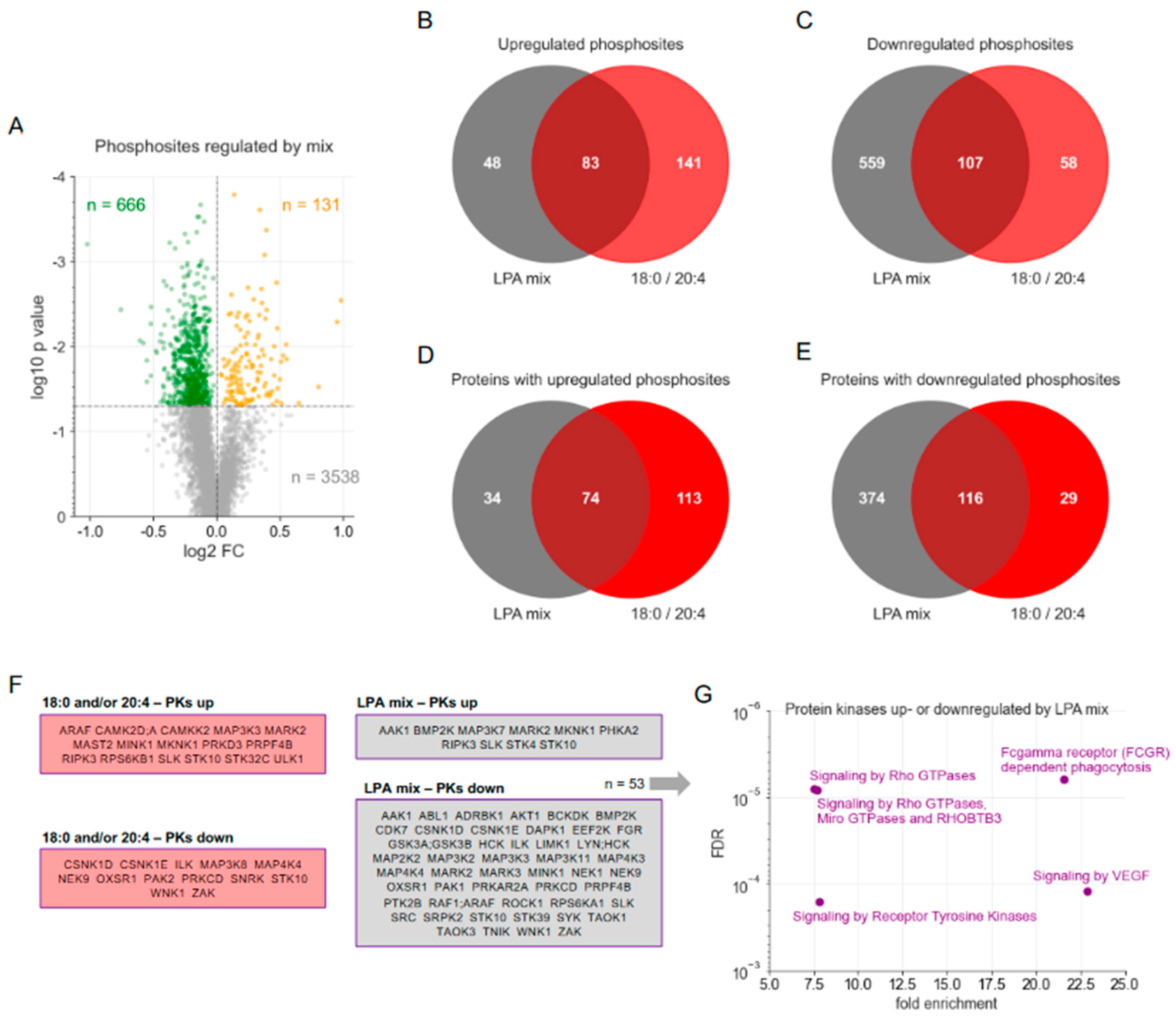
3.3. Phosphoproteomic Analysis of an Ascites-Emulating Mixture of LPA Species
3.4. LPA-Species-Selective Phosphorylation of MLC2, AKT, and p38 in MDMs
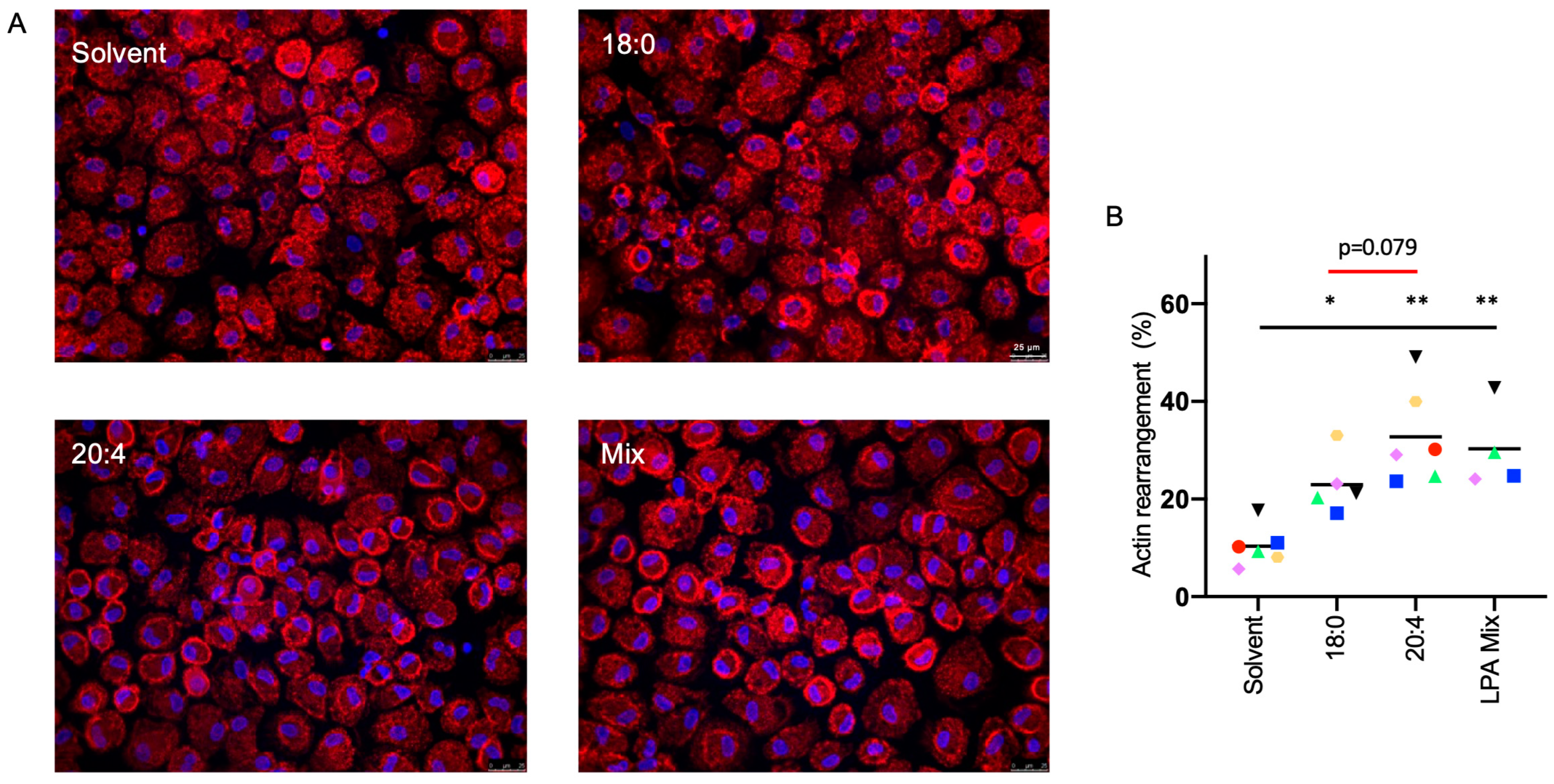
3.5. Effect of LPA Species on the Actin Filament Network and Migration of MDMs
3.6. Transcriptional Response of MDMs to 18:0 of 20:4 LPA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Xu, Y.; Gaudette, D.C.; Boynton, J.D.; Frankel, A.; Fang, X.J.; Sharma, A.; Hurteau, J.; Casey, G.; Goodbody, A.; Mellors, A.; et al. Characterization of an ovarian cancer activating factor in ascites from ovarian cancer patients. Clin. Cancer Res. 1995, 1, 1223–1232. [Google Scholar] [PubMed]
- Worzfeld, T.; Pogge von Strandmann, E.; Huber, M.; Adhikary, T.; Wagner, U.; Reinartz, S.; Müller, R. The Unique Molecular and Cellular Microenvironment of Ovarian Cancer. Front. Oncol. 2017, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Ford, C.E.; Werner, B.; Hacker, N.F.; Warton, K. The untapped potential of ascites in ovarian cancer research and treatment. Br. J. Cancer 2020, 123, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Balijepalli, P.; Sitton, C.C.; Meier, K.E. Lysophosphatidic Acid Signaling in Cancer Cells: What Makes LPA So Special? Cells 2021, 10, 2059. [Google Scholar] [CrossRef]
- Almeida-Nunes, D.L.; Mendes-Frias, A.; Silvestre, R.; Dinis-Oliveira, R.J.; Ricardo, S. Immune Tumor Microenvironment in Ovarian Cancer Ascites. Int. J. Mol. Sci. 2022, 23, 10692. [Google Scholar] [CrossRef] [PubMed]
- Mei, S.; Chen, X.; Wang, K.; Chen, Y. Tumor microenvironment in ovarian cancer peritoneal metastasis. Cancer Cell Int. 2023, 23, 11. [Google Scholar] [CrossRef]
- Cao, L.; Zhang, Y.; Fu, Z.; Dong, L.; Yang, S.; Meng, W.; Li, Y.; Zhang, W.; Zhang, J.; Zheng, C.; et al. Diagnostic value of plasma lysophosphatidic acid levels in ovarian cancer patients: A case-control study and updated meta-analysis. J. Obstet. Gynaecol. Res. 2015, 41, 1951–1958. [Google Scholar] [CrossRef] [PubMed]
- Jesionowska, A.; Cecerska-Heryc, E.; Matoszka, N.; Dolegowska, B. Lysophosphatidic acid signaling in ovarian cancer. J. Recept. Signal Transduct. Res. 2015, 35, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Reinartz, S.; Lieber, S.; Pesek, J.; Brandt, D.T.; Asafova, A.; Finkernagel, F.; Watzer, B.; Nockher, W.A.; Nist, A.; Stiewe, T.; et al. Cell-type-selective pathways and clinical associations of lysophosphatidic acid biosynthesis and signaling in the ovarian cancer microenvironment. Mol. Oncol. 2019, 23, 185–201. [Google Scholar] [CrossRef]
- Mills, G.B.; Moolenaar, W.H. The emerging role of lysophosphatidic acid in cancer. Nat. Rev. Cancer 2003, 3, 582–591. [Google Scholar] [CrossRef]
- Wallner, S.; Schmitz, G. Plasmalogens the neglected regulatory and scavenging lipid species. Chem. Phys. Lipids 2011, 164, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Houben, A.J.; Moolenaar, W.H. Autotaxin and LPA receptor signaling in cancer. Cancer Metastasis Rev. 2011, 30, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Hla, T.; Spiegel, S.; Moolenaar, W. (Eds.) Lysophospholipid Receptors: Signaling and Biochemistry; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013. [Google Scholar]
- Yung, Y.C.; Stoddard, N.C.; Chun, J. LPA receptor signaling: Pharmacology, physiology, and pathophysiology. J. Lipid Res. 2014, 55, 1192–1214. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Lin, Y.C.; Chen, C.C. Lysophosphatidic Acid Receptor Antagonists and Cancer: The Current Trends, Clinical Implications, and Trials. Cells 2021, 10, 1629. [Google Scholar] [CrossRef]
- Cui, R.; Bai, H.; Cao, G.; Zhang, Z. The Role of Lysophosphatidic Acid Receptors in Ovarian Cancer: A Minireview. Crit. Rev. Eukaryot. Gene Expr. 2020, 30, 265–272. [Google Scholar] [CrossRef]
- Aiello, S.; Casiraghi, F. Lysophosphatidic Acid: Promoter of Cancer Progression and of Tumor Microenvironment Development. A Promising Target for Anticancer Therapies? Cells 2021, 10, 1390. [Google Scholar] [CrossRef]
- Ojasalu, K.; Lieber, S.; Sokol, A.M.; Nist, A.; Stiewe, T.; Bullwinkel, I.; Finkernagel, F.; Reinartz, S.; Müller-Brüsselbach, S.; Grosse, R.; et al. The lysophosphatidic acid-regulated signal transduction network in ovarian cancer cells and its role in actomyosin dynamics, cell migration and entosis. Theranostics 2023, 13, 1921–1948. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Yang, H.; Li, M. Emerging Roles of Lysophosphatidic Acid in Macrophages and Inflammatory Diseases. Int. J. Mol. Sci. 2023, 24, 12524. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.S.; Lieberthal, W.; Heydrick, S.; Levine, J.S. Lysophosphatidic acid is a major serum noncytokine survival factor for murine macrophages which acts via the phosphatidylinositol 3-kinase signaling pathway. J. Clin. Investig. 1998, 102, 716–727. [Google Scholar] [CrossRef]
- Feng, Y.; Xiao, M.; Zhang, Z.; Cui, R.; Jiang, X.; Wang, S.; Bai, H.; Liu, C.; Zhang, Z. Potential interaction between lysophosphatidic acid and tumor-associated macrophages in ovarian carcinoma. J. Inflamm. 2020, 17, 23. [Google Scholar] [CrossRef]
- Ray, R.; Rai, V. Lysophosphatidic acid converts monocytes into macrophages in both mice and humans. Blood 2017, 129, 1177–1183. [Google Scholar] [CrossRef]
- Fransson, J.; Gomez-Conde, A.I.; Romero-Imbroda, J.; Fernandez, O.; Leyva, L.; de Fonseca, F.R.; Chun, J.; Louapre, C.; Van-Evercooren, A.B.; Zujovic, V.; et al. Activation of Macrophages by Lysophosphatidic Acid through the Lysophosphatidic Acid Receptor 1 as a Novel Mechanism in Multiple Sclerosis Pathogenesis. Mol. Neurobiol. 2021, 58, 470–482. [Google Scholar] [CrossRef]
- Zhang, D.; Shi, R.; Xiang, W.; Kang, X.; Tang, B.; Li, C.; Gao, L.; Zhang, X.; Zhang, L.; Dai, R.; et al. The Agpat4/LPA axis in colorectal cancer cells regulates antitumor responses via p38/p65 signaling in macrophages. Signal Transduct. Target. Ther. 2020, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.K.; Yun, S.J.; Do, K.H.; Kim, M.S.; Cho, M.; Suh, D.S.; Kim, C.D.; Kim, J.H.; Birnbaum, M.J.; Bae, S.S. Lysophosphatidic acid induces cell migration through the selective activation of Akt1. Exp. Mol. Med. 2008, 40, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Gustin, C.; Van Steenbrugge, M.; Raes, M. LPA modulates monocyte migration directly and via LPA-stimulated endothelial cells. Am. J. Physiol. Cell Physiol. 2008, 295, C905–C914. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, J.; Yang, X.; Liu, Y.; Deng, X.; Yu, C. Lysophosphatidic acid decreased macrophage foam cell migration correlated with downregulation of fucosyltransferase 8 via HNF1alpha. Atherosclerosis 2019, 290, 19–30. [Google Scholar] [CrossRef]
- Reinartz, S.; Finkernagel, F.; Adhikary, T.; Rohnalter, V.; Schumann, T.; Schober, Y.; Nockher, W.A.; Nist, A.; Stiewe, T.; Jansen, J.M.; et al. A transcriptome-based global map of signaling pathways in the ovarian cancer microenvironment associated with clinical outcome. Genome Biol. 2016, 17, 108. [Google Scholar] [CrossRef]
- Worzfeld, T.; Finkernagel, F.; Reinartz, S.; Konzer, A.; Adhikary, T.; Nist, A.; Stiewe, T.; Wagner, U.; Looso, M.; Graumann, J.; et al. Proteotranscriptomics Reveal Signaling Networks in the Ovarian Cancer Microenvironment. Mol. Cell Proteom. 2018, 17, 270–289. [Google Scholar] [CrossRef]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Dietze, R.; Hammoud, M.K.; Gomez-Serrano, M.; Unger, A.; Bieringer, T.; Finkernagel, F.; Sokol, A.M.; Nist, A.; Stiewe, T.; Reinartz, S.; et al. Phosphoproteomics identify arachidonic-acid-regulated signal transduction pathways modulating macrophage functions with implications for ovarian cancer. Theranostics 2021, 11, 1377–1395. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium, G. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium, G. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef] [PubMed]
- Kamburov, A.; Wierling, C.; Lehrach, H.; Herwig, R. ConsensusPathDB—A database for integrating human functional interaction networks. Nucleic Acids Res. 2009, 37, D623–D628. [Google Scholar] [CrossRef] [PubMed]
- Herwig, R.; Hardt, C.; Lienhard, M.; Kamburov, A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc. 2016, 11, 1889–1907. [Google Scholar] [CrossRef] [PubMed]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- Mosaddeghzadeh, N.; Ahmadian, M.R. The RHO Family GTPases: Mechanisms of Regulation and Signaling. Cells 2021, 10, 1831. [Google Scholar] [CrossRef]
- Geraldo, L.H.M.; Spohr, T.; Amaral, R.F.D.; Fonseca, A.; Garcia, C.; Mendes, F.A.; Freitas, C.; dosSantos, M.F.; Lima, F.R.S. Role of lysophosphatidic acid and its receptors in health and disease: Novel therapeutic strategies. Signal Transduct. Target. Ther. 2021, 6, 45. [Google Scholar] [CrossRef]
- Vicente-Manzanares, M.; Ma, X.; Adelstein, R.S.; Horwitz, A.R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009, 10, 778–790. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Canovas, B.; Nebreda, A.R. Diversity and versatility of p38 kinase signalling in health and disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 346–366. [Google Scholar] [CrossRef]
- Sit, S.T.; Manser, E. Rho GTPases and their role in organizing the actin cytoskeleton. J. Cell Sci. 2011, 124, 679–683. [Google Scholar] [CrossRef]
- Porter, F.D.; Herman, G.E. Malformation syndromes caused by disorders of cholesterol synthesis. J. Lipid Res. 2011, 52, 6–34. [Google Scholar] [CrossRef]
- Sharpe, L.J.; Brown, A.J. Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). J. Biol. Chem. 2013, 288, 18707–18715. [Google Scholar] [CrossRef]
- Willier, S.; Butt, E.; Grunewald, T.G. Lysophosphatidic acid (LPA) signalling in cell migration and cancer invasion: A focussed review and analysis of LPA receptor gene expression on the basis of more than 1700 cancer microarrays. Biol. Cell 2013, 105, 317–333. [Google Scholar] [CrossRef]
- Aikawa, S.; Hashimoto, T.; Kano, K.; Aoki, J. Lysophosphatidic acid as a lipid mediator with multiple biological actions. J. Biochem. 2015, 157, 81–89. [Google Scholar] [CrossRef]
- Canton, J. Macropinocytosis: New Insights into Its Underappreciated Role in Innate Immune Cell Surveillance. Front. Immunol. 2018, 9, 2286. [Google Scholar] [CrossRef]
- Aderem, A.; Underhill, D.M. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 1999, 17, 593–623. [Google Scholar] [CrossRef] [PubMed]
- Uribe-Querol, E.; Rosales, C. Phagocytosis: Our Current Understanding of a Universal Biological Process. Front. Immunol. 2020, 11, 1066. [Google Scholar] [CrossRef]
- Shao, N.; Qiu, H.; Liu, J.; Xiao, D.; Zhao, J.; Chen, C.; Wan, J.; Guo, M.; Liang, G.; Zhao, X.; et al. Targeting lipid metabolism of macrophages: A new strategy for tumor therapy. J. Adv. Res. 2024. [Google Scholar] [CrossRef] [PubMed]
- Goossens, P.; Rodriguez-Vita, J.; Etzerodt, A.; Masse, M.; Rastoin, O.; Gouirand, V.; Ulas, T.; Papantonopoulou, O.; Van Eck, M.; Auphan-Anezin, N.; et al. Membrane Cholesterol Efflux Drives Tumor-Associated Macrophage Reprogramming and Tumor Progression. Cell Metab. 2019, 29, 1376–1389. e4. [Google Scholar] [CrossRef]
- Hoppstadter, J.; Dembek, A.; Horing, M.; Schymik, H.S.; Dahlem, C.; Sultan, A.; Wirth, N.; Al-Fityan, S.; Diesel, B.; Gasparoni, G.; et al. Dysregulation of cholesterol homeostasis in human lung cancer tissue and tumour-associated macrophages. EBioMedicine 2021, 72, 103578. [Google Scholar] [CrossRef]
- Gruenbacher, G.; Thurnher, M. Mevalonate metabolism governs cancer immune surveillance. Oncoimmunology 2017, 6, e1342917. [Google Scholar] [CrossRef] [PubMed]
- Vassiliou, E.; Farias-Pereira, R. Impact of Lipid Metabolism on Macrophage Polarization: Implications for Inflammation and Tumor Immunity. Int. J. Mol. Sci. 2023, 24, 12032. [Google Scholar] [CrossRef]
- Akula, M.K.; Shi, M.; Jiang, Z.; Foster, C.E.; Miao, D.; Li, A.S.; Zhang, X.; Gavin, R.M.; Forde, S.D.; Germain, G.; et al. Control of the innate immune response by the mevalonate pathway. Nat. Immunol. 2016, 17, 922–929. [Google Scholar] [CrossRef]
- Reinartz, S.; Schumann, T.; Finkernagel, F.; Wortmann, A.; Jansen, J.M.; Meissner, W.; Krause, M.; Schwörer, A.M.; Wagner, U.; Müller-Brüsselbach, S.; et al. Mixed-polarization phenotype of ascites-associated macrophages in human ovarian carcinoma: Correlation of CD163 expression, cytokine levels and early relapse. Int. J. Cancer 2014, 134, 32–42. [Google Scholar] [CrossRef]
- Pustilnik, T.B.; Estrella, V.; Wiener, J.R.; Mao, M.; Eder, A.; Watt, M.A.; Bast, R.C., Jr.; Mills, G.B. Lysophosphatidic acid induces urokinase secretion by ovarian cancer cells. Clin. Cancer Res. 1999, 5, 3704–3710. [Google Scholar]
- Tigyi, G. Aiming drug discovery at lysophosphatidic acid targets. Br. J. Pharmacol. 2010, 161, 241–270. [Google Scholar] [CrossRef]
- Bandoh, K.; Aoki, J.; Taira, A.; Tsujimoto, M.; Arai, H.; Inoue, K. Lysophosphatidic acid (LPA) receptors of the EDG family are differentially activated by LPA species. Structure-activity relationship of cloned LPA receptors. FEBS Lett. 2000, 478, 159–165. [Google Scholar] [CrossRef]
- Noguchi, K.; Ishii, S.; Shimizu, T. Identification of p2y9/GPR23 as a novel G protein-coupled receptor for lysophosphatidic acid, structurally distant from the Edg family. J. Biol. Chem. 2003, 278, 25600–25606. [Google Scholar] [CrossRef]
- Williams, J.R.; Khandoga, A.L.; Goyal, P.; Fells, J.I.; Perygin, D.H.; Siess, W.; Parrill, A.L.; Tigyi, G.; Fujiwara, Y. Unique ligand selectivity of the GPR92/LPA5 lysophosphatidate receptor indicates role in human platelet activation. J. Biol. Chem. 2009, 284, 17304–17319. [Google Scholar] [CrossRef]
- Yanagida, K.; Masago, K.; Nakanishi, H.; Kihara, Y.; Hamano, F.; Tajima, Y.; Taguchi, R.; Shimizu, T.; Ishii, S. Identification and characterization of a novel lysophosphatidic acid receptor, p2y5/LPA6. J. Biol. Chem. 2009, 284, 17731–17741. [Google Scholar] [CrossRef]
- McIntyre, T.M.; Pontsler, A.V.; Silva, A.R.; St Hilaire, A.; Xu, Y.; Hinshaw, J.C.; Zimmerman, G.A.; Hama, K.; Aoki, J.; Arai, H.; et al. Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPA is a transcellular PPARgamma agonist. Proc. Natl. Acad. Sci. USA 2003, 100, 131–136. [Google Scholar] [CrossRef]
- Meizoso-Huesca, A.; Villegas-Comonfort, S.; Romero-Avila, M.T.; Garcia-Sainz, J.A. Free fatty acid receptor 4 agonists induce lysophosphatidic acid receptor 1 (LPA(1)) desensitization independent of LPA(1) internalization and heterodimerization. FEBS Lett. 2018, 592, 2612–2623. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dietze, R.; Szymanski, W.; Ojasalu, K.; Finkernagel, F.; Nist, A.; Stiewe, T.; Graumann, J.; Müller, R. Phosphoproteomics Reveals Selective Regulation of Signaling Pathways by Lysophosphatidic Acid Species in Macrophages. Cells 2024, 13, 810. https://doi.org/10.3390/cells13100810
Dietze R, Szymanski W, Ojasalu K, Finkernagel F, Nist A, Stiewe T, Graumann J, Müller R. Phosphoproteomics Reveals Selective Regulation of Signaling Pathways by Lysophosphatidic Acid Species in Macrophages. Cells. 2024; 13(10):810. https://doi.org/10.3390/cells13100810
Chicago/Turabian StyleDietze, Raimund, Witold Szymanski, Kaire Ojasalu, Florian Finkernagel, Andrea Nist, Thorsten Stiewe, Johannes Graumann, and Rolf Müller. 2024. "Phosphoproteomics Reveals Selective Regulation of Signaling Pathways by Lysophosphatidic Acid Species in Macrophages" Cells 13, no. 10: 810. https://doi.org/10.3390/cells13100810
APA StyleDietze, R., Szymanski, W., Ojasalu, K., Finkernagel, F., Nist, A., Stiewe, T., Graumann, J., & Müller, R. (2024). Phosphoproteomics Reveals Selective Regulation of Signaling Pathways by Lysophosphatidic Acid Species in Macrophages. Cells, 13(10), 810. https://doi.org/10.3390/cells13100810





