Senotherapeutic Peptide 14 Suppresses Th1 and M1 Human T Cell and Monocyte Subsets In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptide Synthesis
2.2. Isolation and Culture of Peripheral Blood Mononuclear Cells (PBMCs)
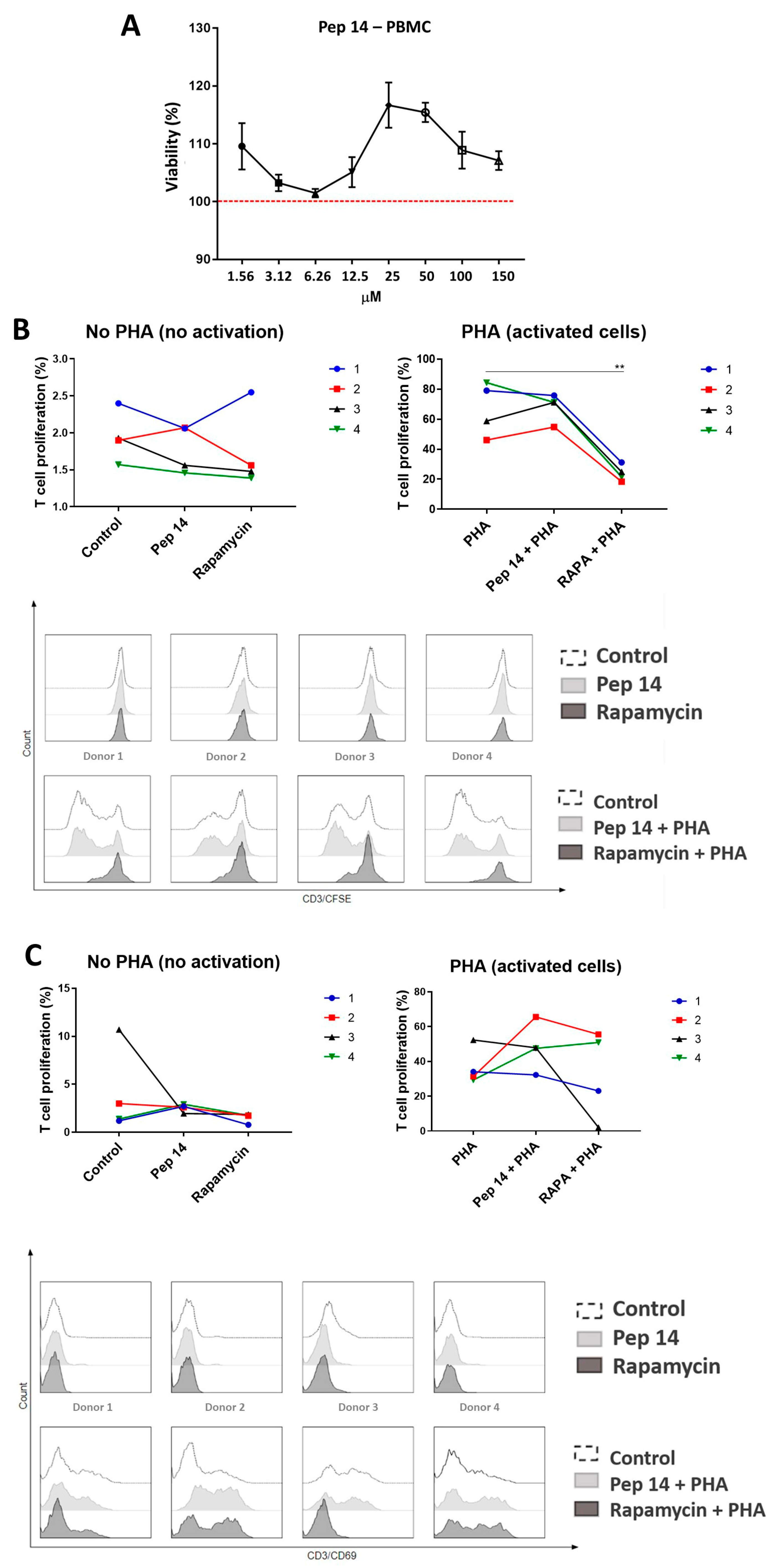
2.3. PBMC Viability Assay
2.4. PBMC Proliferation Assay
2.5. T Cell Activation Assay
2.6. Human Monocyte Cell Cultures
2.7. U-937 Cell Viability Assay
2.8. U-937 Macrophage Differentiation
2.9. Nitric Oxide Release
2.10. Enzyme-Linked Immunosorbent Assay (ELISA)
2.11. Real-Time PCR
2.12. Cell Migration Assay
2.13. Statistical Analysis
3. Results
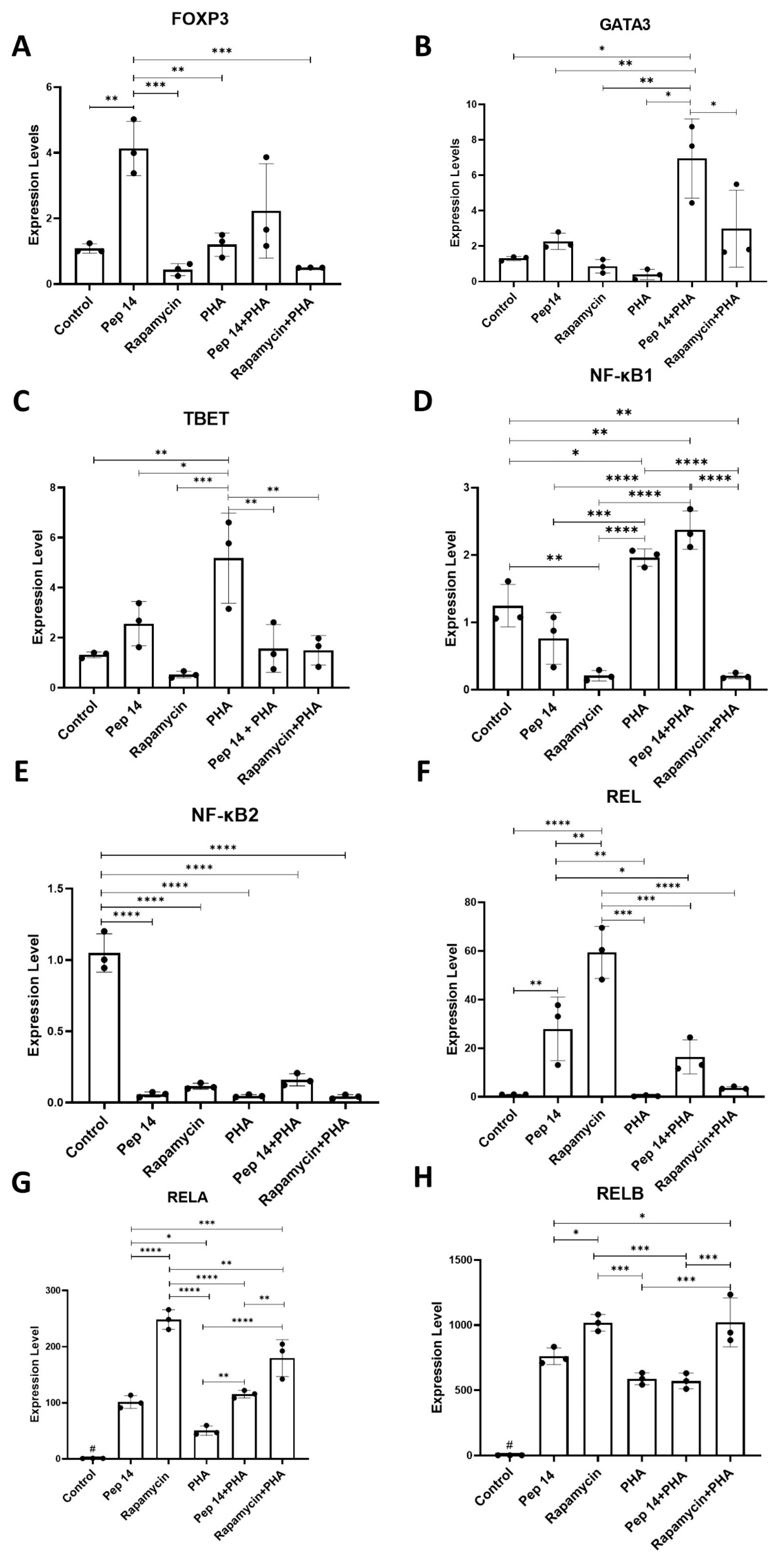
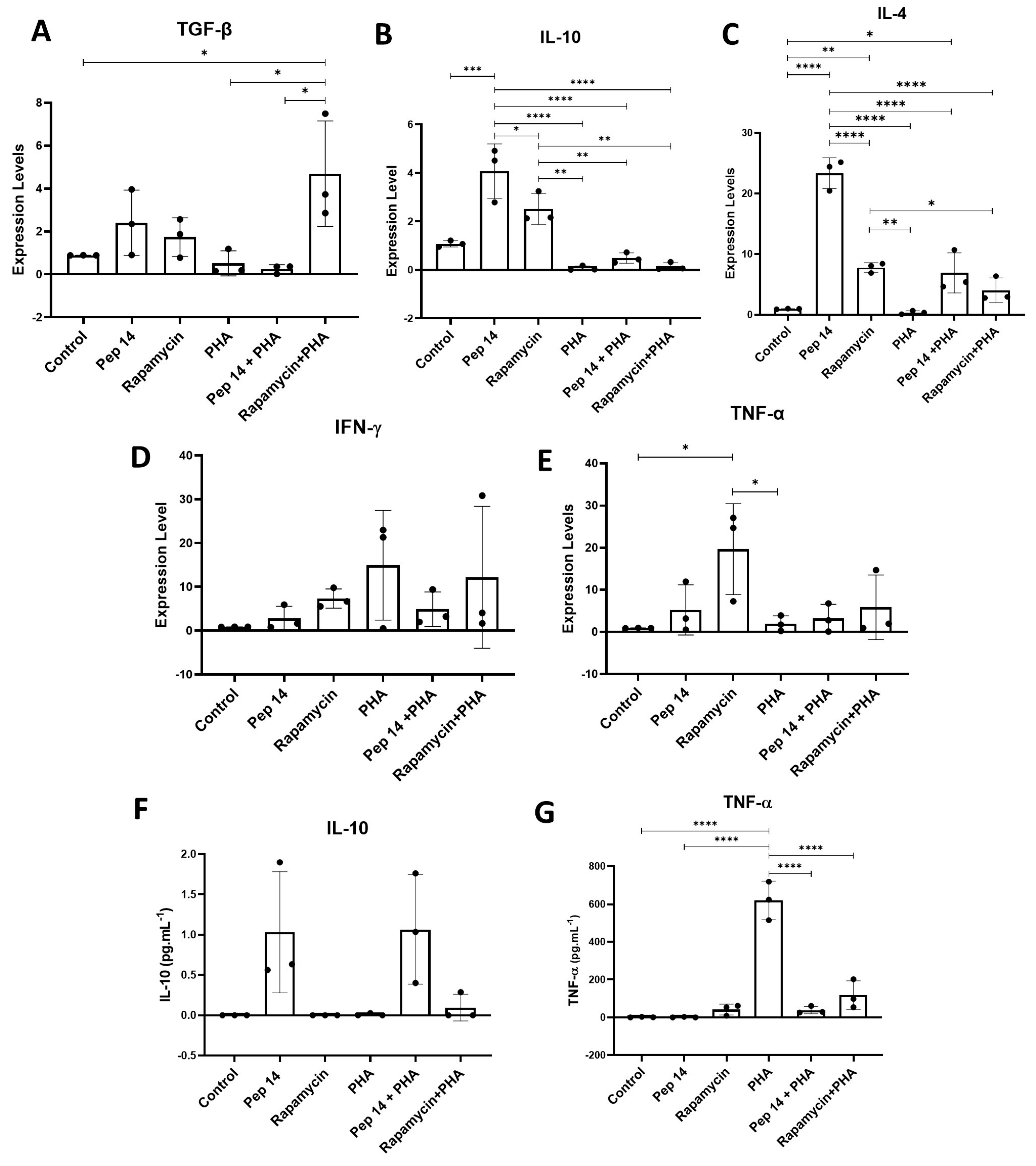
3.1. Pep 14 Promotes Th2 and Treg mRNA and Cytokine Signature in Phytohemagglutinin-Treated PBMCs
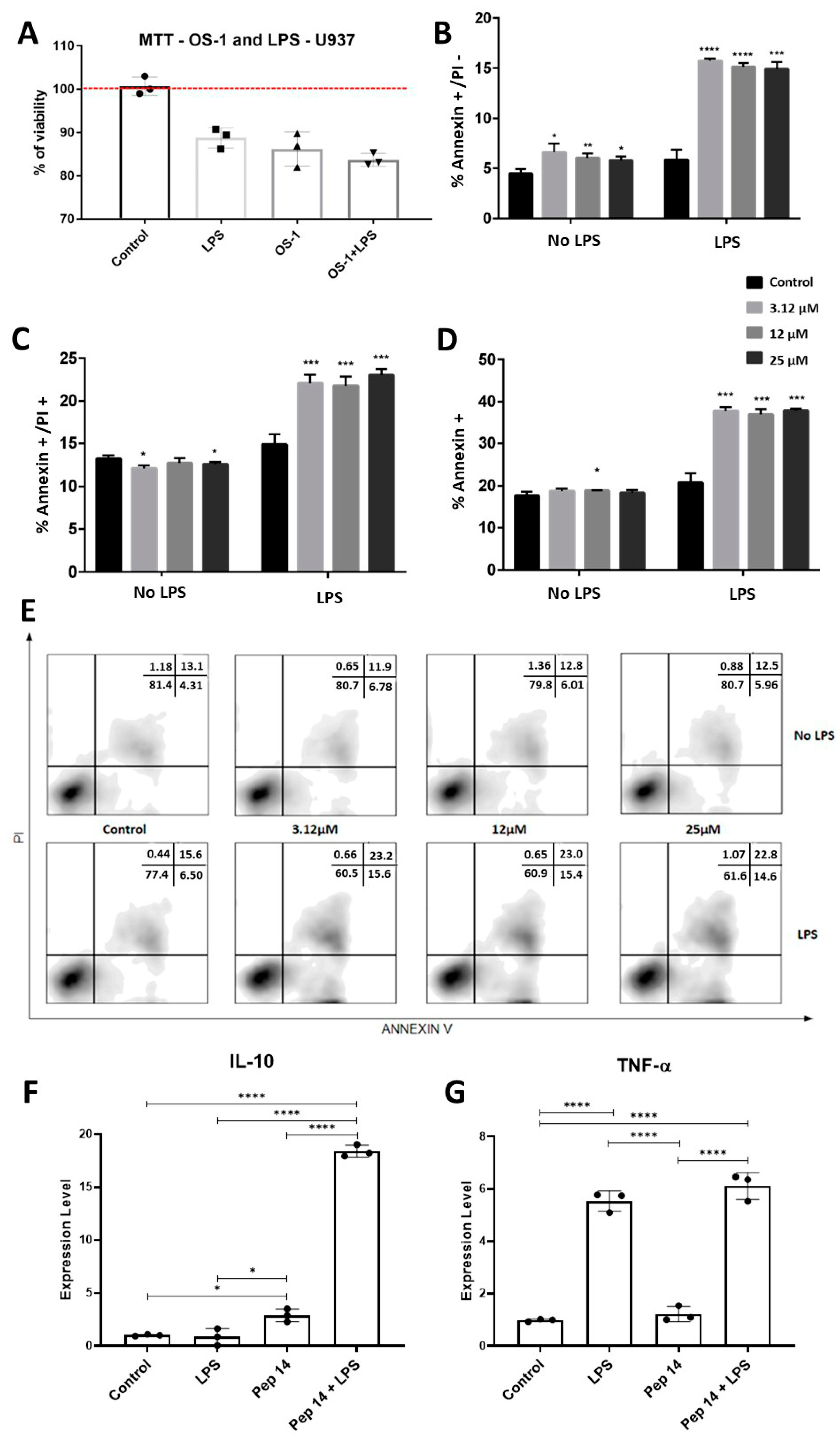
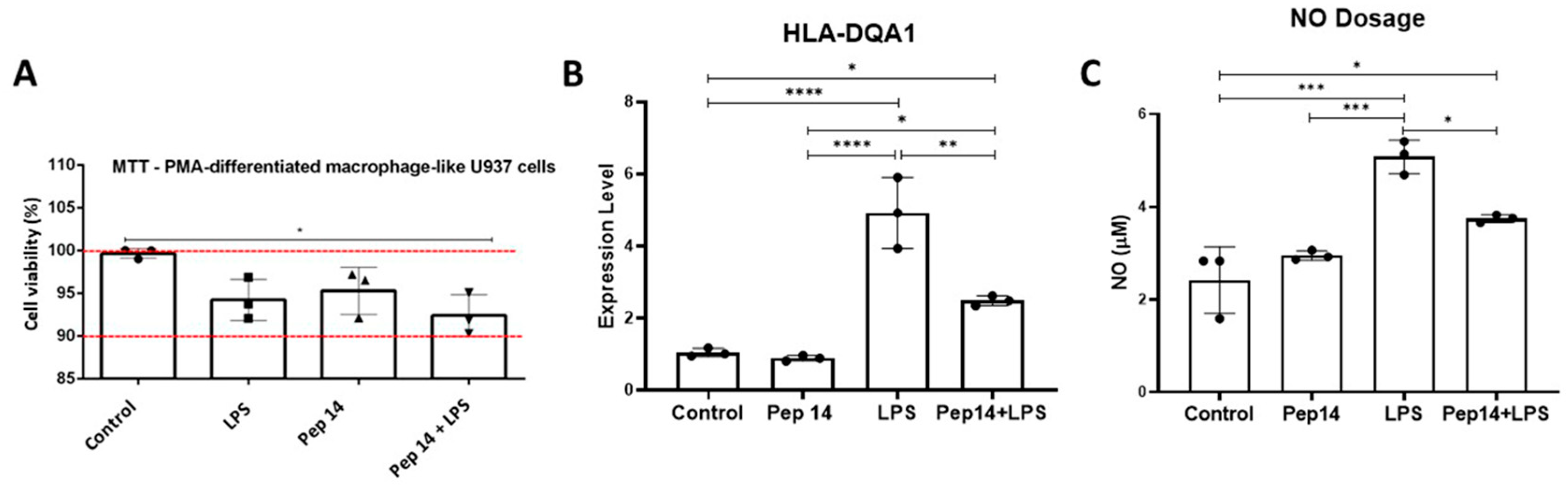
3.2. Pep 14 Influences LPS-Stimulated U-937 Monocytes by Inducing Apoptosis of Activated Cells and IL-10 Expression
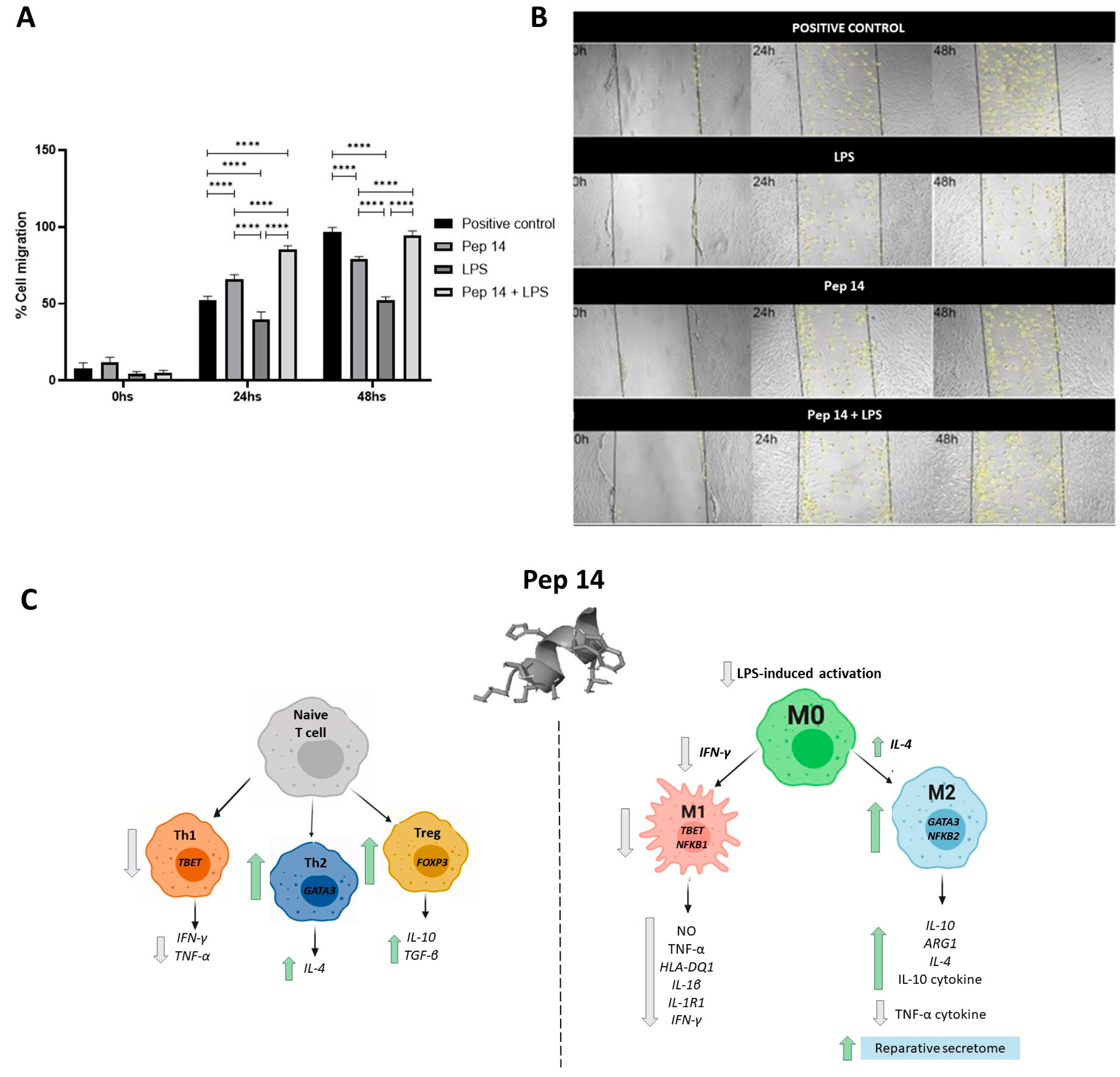
3.3. Pep14 Prevents LPS-Induced Activation and Promotes Regenerative M2 Polarization in U-937-Derived Macrophages
3.4. The Secretome of LPS- and Pep 14-Treated Macrophages Promotes Fibroblast Migration and Is Compatible with Functional M2 Macrophage Differentiation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rodrigues, L.P.; Teixeira, V.R.; Alencar-Silva, T.; Simonassi-Paiva, B.; Pereira, R.W.; Pogue, R.; Carvalho, J.L. Hallmarks of aging and immunosenescence: Connecting the dots. Cytokine Growth Factor Rev. 2021, 59, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Pawelec, G. Hallmarks of human “immunosenescence”: Adaptation or dysregulation? Immun. Ageing 2012, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, G.R.; Almeida, P.P.; de Oliveira Santos, L.; Rodrigues, L.P.; de Carvalho, J.L.; Boroni, M. Hallmarks of Aging in Macrophages: Consequences to Skin Inflammaging. Cells 2021, 10, 1323. [Google Scholar] [CrossRef] [PubMed]
- Wikby, A.; Nilsson, B.-O.; Forsey, R.; Thompson, J.; Strindhall, J.; Löfgren, S.; Ernerudh, J.; Pawelec, G.; Ferguson, F.; Johansson, B. The immune risk phenotype is associated with IL-6 in the terminal decline stage: Findings from the Swedish NONA immune longitudinal study of very late life functioning. Mech. Ageing Dev. 2006, 127, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, V.; Rink, L.; Uciechowski, P. The Th17/Treg balance is disturbed during aging. Exp. Gerontol. 2013, 48, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Bruunsgaard, H.; Skinhøj, P.; Pedersen, A.N.; Schroll, M.; Pedersen, B.K. Ageing, tumour necrosis factor-alpha (TNF-α) and atherosclerosis. Clin. Exp. Immunol. 2008, 121, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Zonari, A.; Brace, L.E.; Alencar-Silva, T.; Porto, W.F.; Foyt, D.; Guiang, M.; Cruz, E.A.O.; Franco, O.L.; Oliveira, C.R.; Boroni, M.; et al. In vitro and in vivo toxicity assessment of the senotherapeutic Peptide 14. Toxicol. Rep. 2022, 9, 1632–1638. [Google Scholar] [CrossRef] [PubMed]
- Zonari, A.; Brace, L.E.; Al-Katib, K.; Porto, W.F.; Foyt, D.; Guiang, M.; Cruz, E.A.O.; Marshall, B.; Gentz, M.; Guimarães, G.R.; et al. Senotherapeutic peptide treatment reduces biological age and senescence burden in human skin models. NPJ Aging 2023, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.M.; Kalim, U.U.; Khan, M.H.; Lahesmaa, R. PP2A and Its Inhibitors in Helper T-Cell Differentiation and Autoimmunity. Front. Immunol. 2022, 12, 786857. [Google Scholar] [CrossRef] [PubMed]
- Serejo, T.R.T.; Silva-Carvalho, A.É.; Braga, L.D.D.C.F.; Neves, F.D.A.R.; Pereira, R.W.; Carvalho, J.L.D.; Saldanha-Araujo, F. Assessment of the Immunosuppressive Potential of INF-γ Licensed Adipose Mesenchymal Stem Cells, Their Secretome and Extracellular Vesicles. Cells 2019, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Pinto, S.M.; Kim, H.; Subbannayya, Y.; Giambelluca, M.S.; Bösl, K.; Ryan, L.; Sharma, A.; Kandasamy, R.K. Comparative Proteomic Analysis Reveals Varying Impact on Immune Responses in Phorbol 12-Myristate-13-Acetate-Mediated THP-1 Monocyte-to-Macrophage Differentiation. Front. Immunol. 2021, 12, 679458. [Google Scholar] [CrossRef] [PubMed]
- Green, L.C.; Wagner, D.A.; Glogowski, J.; Skipper, P.L.; Wishnok, J.S.; Tannenbaum, S.R. Analysis of nitrate, nitrite, and [15N] nitrate in biological fluids. Anal. Biochem. 1982, 126, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.-C.; Park, A.Y.; Guan, J.-L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Terada, N.; Lucas, J.J.; Szepesi, A.; Franklin, R.A.; Domenico, J.; Gelfand, E.W. Rapamycin blocks cell cycle progression of activated T cells prior to events characteristic of the middle to late G1 phase of the cycle. J. Cell Physiol. 1993, 154, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.; Ghosh, S. NF-κB: Roles and regulation in different CD4(+) T-cell subsets. Immunol. Rev. 2013, 252, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Hövelmeyer, N.; Schmidt-Supprian, M.; Ohnmacht, C. NF-κB in control of regulatory T cell development, identity, and function. J. Mol. Med. 2022, 100, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lozano-Ruiz, B.; Yang, F.M.; Fan, D.D.; Shen, L.; González-Navajas, J.M. The Multifaceted Role of Th1, Th9, and Th17 Cells in Immune Checkpoint Inhibition Therapy. Front. Immunol. 2021, 12, 625667. [Google Scholar] [CrossRef] [PubMed]
- Franceschelli, S.; Pesce, M.; Ferrone, A.; De Lutiis, M.A.; Patruno, A.; Grilli, A.; Felaco, M.; Speranza, L. Astaxanth in treatment confers protection against oxidative stress in U937 cells stimulated with lipopolysaccharide reducing O2− production. PLoS ONE 2014, 9, e88359. [Google Scholar] [CrossRef] [PubMed]
- Viola, A.; Munari, F.; Sánchez-Rodríguez, R.; Scolaro, T.; Castegna, A. The Metabolic Signature of Macrophage Responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chen, Y.; Zhou, L.; Ren, J.; Tian, M.; Yang, Q.; Wang, L.; Wu, Y.; Wen, J.; Yang, Q. M2a macrophages regulate fibrosis and affect the outcome after stroke via PU.1/mTOR pathway in fibroblasts. Neurochem. Int. 2024, 173, 105674. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Bratlie, K.M. Fibroblasts treated with macrophage conditioned medium results in phenotypic shifts and changes in collagen organization. Mater. Sci. Eng. C 2021, 122, 111915. [Google Scholar] [CrossRef] [PubMed]
- Salimi, S.; Shardell, M.D.; Seliger, S.L.; Bandinelli, S.; Guralnik, J.M.; Ferrucci, L. Inflammation and Trajectory of Renal Function in Community-Dwelling Older Adults. J. Am. Geriatr. Soc. 2018, 66, 804–811. [Google Scholar] [CrossRef]
- Ferrucci, L.; Fabbri, E. Inflammageing: Chronic inflammation in ageing, cardiovascular disease, and frailty. Nat. Rev. Cardiol. 2018, 15, 505–522. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Van Hoecke, L.; Vandenbroucke, R.E. The Impact of Systemic Inflammation on Alzheimer’s Disease Pathology. Front. Immunol. 2022, 12, 796867. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.S.; Wang, H.; Maggio, D.; Kovach, J.S.; Zhang, Q.; Song, Q.; Marincola, F.M.; Heiss, J.D.; Gilbert, M.R.; Lu, R.; et al. Pharmacologic inhibition of protein phosphatase-2A achieves durable immune-mediated antitumor activity when combined with PD-1 blockade. Nat. Commun. 2018, 9, 2126. [Google Scholar] [CrossRef] [PubMed]
- Mangan, D.F.; Mergenhagen, S.E.; Wahl, S.M. Apoptosis in human monocytes: Possible role in chronic inflammatory diseases. J. Periodontol. 1993, 64 (Suppl. S5), 461–466. [Google Scholar] [PubMed]
- Boirivant, M.; Marini, M.; Di Felice, G.; Pronio, A.M.; Montesani, C.; Tersigni, R.; Strober, W. Lamina propria T cells in Crohn’s disease and other gastrointestinal inflammation show defective CD2 pathway-induced apoptosis. Gastroenterology 1999, 116, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Ina, K.; Itoh, J.; Fukushima, K.; Kusugami, K.; Yamaguchi, T.; Kyokane, K.; Imada, A.; Binion, D.G.; Musso, A.; West, G.A.; et al. Resistance of Crohn’s disease T cells to multiple apoptotic signals is associated with a Bcl-2/Bax mucosal imbalance. J. Immunol. 1999, 163, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Boumaza, A.; Gay, L.; Mezouar, S.; Bestion, E.; Diallo, A.B.; Michel, M.; Desnues, B.; Raoult, D.; La Scola, B.; Halfon, P.; et al. Monocytes and Macrophages, Targets of Severe Acute Respiratory Syndrome Coronavirus 2: The Clue for Coronavirus Disease 2019 Immunoparalysis. J. Infect. Dis. 2021, 224, 395–406. [Google Scholar] [CrossRef]
- Zheng, W.; Song, H.; Luo, Z.; Wu, H.; Chen, L.; Wang, Y.; Cui, H.; Zhang, Y.; Wang, B.; Li, W.; et al. Acetylcholine ameliorates colitis by promoting IL-10 secretion of monocytic myeloid-derived suppressor cells through the nAChR/ERK pathway. Proc. Natl. Acad. Sci. USA 2021, 118, e2017762118. [Google Scholar] [CrossRef] [PubMed]
- Hortelano, S.; Zeini, M.; Castrillo, A.; Alvarez, A.M.; Boscá, L. Induction of apoptosis by nitric oxide in macrophages is independent of apoptotic volume decreas. Cell Death Differ. 2002, 9, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Hu, Y.; Li, Y.; Zhou, Q.; Lv, X. Involvement of JNK signaling in IL4-induced M2 macrophage polarization. Exp. Cell Res. 2017, 357, 155–162. [Google Scholar] [CrossRef]
- Nair, M.G.; Guild, K.J.; Artis, D. Novel Effector Molecules in Type 2 Inflammation: Lessons Drawn from Helminth Infection and Allergy. J. Immunol. 2006, 177, 1393–1399. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Shu, D.; Zheng, M.; Wang, J.; Luo, C.; Wang, Y.; Guo, F.; Zou, X.; Lv, X.; Li, Y.; et al. Microbial metabolite butyrate facilitates M2 macrophage polarization and function. Sci. Rep. 2016, 6, 24838. [Google Scholar] [CrossRef] [PubMed]
- Ploeger, D.T.; Hosper, N.A.; Schipper, M.; Koerts, J.A.; de Rond, S.; Bank, R.A. Cell plasticity in wound healing: Paracrine factors of M1/M2 polarized macrophages influence the phenotypical state of dermal fibroblasts. Cell Commun. Signal. 2013, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Weichhart, T. mTOR as Regulator of Lifespan, Aging, and Cellular Senescence: A Mini-Review. Gerontology 2018, 64, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Yalcin, G.; Kim, J.; Seo, D.; Lee, C.K. FPR1 is essential for rapamycin-induced lifespan extension in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2023, 653, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.; Salmon, A.; Strong, R.; Fernandez, E.; Javors, M.; Richardson, A.; Tardif, S. Metabolic consequences of long-term rapamycin exposure on common marmoset monkeys (Callithrix jacchus). Aging 2015, 7, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Phillips, E.J.; Simons, M.J.P. Rapamycin not dietary restriction improves resilience against pathogens: A meta-analysis. GeroScience 2023, 45, 1263–1270. [Google Scholar] [CrossRef] [PubMed]
- Mannick, J.B.; Teo, G.; Bernardo, P.; Quinn, D.; Russell, K.; Klickstein, L.; Marshall, W.; Shergill, S. Targeting the biology of ageing with mTOR inhibitors to improve immune function in older adults: Phase 2b and phase 3 randomised trials. Lancet Health Longev. 2021, 2, e250–e262. [Google Scholar] [CrossRef] [PubMed]
- Taffs, R.E.; Redegeld, F.A.; Sitkovsky, M.V. Modulation of cytolytic T lymphocyte functions by an inhibitor of serine/threonine phosphatase, okadaic acid. Enhancement of cytolytic T lymphocyte-mediated cytotoxicity. J. Immunol. 1991, 147, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Eitelhuber, A.C.; Warth, S.; Schimmack, G.; Düwel, M.; Hadian, K.; Demski, K.; Beisker, W.; Shinohara, H.; Kurosaki, T.; Heissmeyer, V.; et al. Dephosphorylation of Carma1 by PP2A negatively regulates T-cell activation. EMBO J. 2011, 30, 594–605. [Google Scholar] [CrossRef]
- Peterson, R.T.; Desai, B.N.; Hardwick, J.S.; Schreiber, S.L. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12–rapamycin associated protein. Proc. Natl. Acad. Sci. USA 1999, 96, 4438–4442. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Brautigan, D.L. Protein phosphatase 2A suppresses MAP kinase signalling and ectopic protein expression. Cell Signal. 1999, 11, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Yu, A.; Tsokos, G.C.; Malek, T.R. CD25 and Protein Phosphatase 2A Cooperate to Enhance IL-2R Signaling in Human Regulatory T Cells. J. Immunol. 2019, 203, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Batra, L. Protein Phosphatase 2A: Role in T Cells and Diseases. J. Immunol. Res. 2023, 2023, 4522053. [Google Scholar] [CrossRef] [PubMed]
- Mondal, I.; Das, O.; Lu, R.; Ho, W. Protein Phosphatase 2A Inhibition in Macrophages Enhanced Type I Interferon Production and M1 Polarization. Neurosurgery 2020, 67 (Suppl. S1), 285. [Google Scholar] [CrossRef]
- Pournajaf, S.; Dargahi, L.; Javan, M.; Pourgholami, M.H. Molecular Pharmacology and Novel Potential Therapeutic Applications of Fingolimod. Front. Pharmacol. 2022, 13, 807639. [Google Scholar] [CrossRef] [PubMed]


| Transcript | Primer Forward (5′–3′) | Primer Reverse (5′–3′) | T (°C) |
|---|---|---|---|
| ARG1 | ATTGAGAAAGGCTGGTCTGC | CATTAGGGATGTCAGCAAAGG | 60 |
| FOXP3 | TCATCCGCTGGGCCATCCTG | GTGGAAACCTCACTTCTTGGTC | 58 |
| GAPDH | ACATCGCTCAGACACCATG | TGTAGTTGAGGTCAATGAAGGG | 60 |
| GATA3 | AGCCTGTCCTTTGGACCACA | CATCGAGCAGGGCTCTAACC | 60 |
| HLA-DQ1 | CTCTGTTCCGCAGATTTAGAAGA | GGAGACTTGGAAAACACTGTGACC | 60 |
| IFN-γ | ACTGTCGCCAGCAGCTAAAA | TATTGCAGGCAGGACAACCA | 60 |
| IL-10 | GGCACCCAGTCTGAGAACAG | ACTCTGCTGAAGGCATCTCG | 60 |
| IL-1R1 | CCTCCAGGATTCATCAACAC | AAAACTCCATATAAGGGCACAC | 60 |
| IL1β | ATGATGGCTTATTACAGTGGCAA | GTCGGAGATTCGTAGCTGGA | 60 |
| IL-4 | TTGTCCACGGACACAAGTGC | TGTTACGGTCAACTCGGTGC | 60 |
| NF-κB1 | GTGGTGCCTCACTGCTAACT | GGATGCACTTCAGCTTCTGT | 58 |
| NF-κB2 | TAGCCACAGAGATGGAGGAG | CCGAGTCGCTATCAGAGGT | 60 |
| REL | AAAGACTGCAGAGACGGCT | CTCACCACATTGAGGTCACA | 58 |
| RELA | GCACAGATACCACCAAGACC | TCAGCCTCATAGAAGCCATC | 58 |
| RELB | CATTGAGCGGAAGATTCAAC | GCAGCTCTGATGTGTTTGTG | 56 |
| TBET | CAACACGCATATCTTTACTTTCCA | AGCTGAGTAATCTCGGCATTCTG | 65 |
| TGF- β1 | GCTGTATTTAAGGACACCGTGC | TGACACAGAGATCCGCAGTC | 60 |
| TNF-α | CACAGTGAAGTGCTGGCAAC | GATCAAAGCTGTAGGCCCCA | 58 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alencar-Silva, T.; Barcelos, S.M.d.; Silva-Carvalho, A.; Sousa, M.G.d.C.; Rezende, T.M.B.; Pogue, R.; Saldanha-Araújo, F.; Franco, O.L.; Boroni, M.; Zonari, A.; et al. Senotherapeutic Peptide 14 Suppresses Th1 and M1 Human T Cell and Monocyte Subsets In Vitro. Cells 2024, 13, 813. https://doi.org/10.3390/cells13100813
Alencar-Silva T, Barcelos SMd, Silva-Carvalho A, Sousa MGdC, Rezende TMB, Pogue R, Saldanha-Araújo F, Franco OL, Boroni M, Zonari A, et al. Senotherapeutic Peptide 14 Suppresses Th1 and M1 Human T Cell and Monocyte Subsets In Vitro. Cells. 2024; 13(10):813. https://doi.org/10.3390/cells13100813
Chicago/Turabian StyleAlencar-Silva, Thuany, Stefhani Martins de Barcelos, Amandda Silva-Carvalho, Mauricio Gonçalves da Costa Sousa, Taia Maria Berto Rezende, Robert Pogue, Felipe Saldanha-Araújo, Octávio Luiz Franco, Mariana Boroni, Alessandra Zonari, and et al. 2024. "Senotherapeutic Peptide 14 Suppresses Th1 and M1 Human T Cell and Monocyte Subsets In Vitro" Cells 13, no. 10: 813. https://doi.org/10.3390/cells13100813
APA StyleAlencar-Silva, T., Barcelos, S. M. d., Silva-Carvalho, A., Sousa, M. G. d. C., Rezende, T. M. B., Pogue, R., Saldanha-Araújo, F., Franco, O. L., Boroni, M., Zonari, A., & Carvalho, J. L. (2024). Senotherapeutic Peptide 14 Suppresses Th1 and M1 Human T Cell and Monocyte Subsets In Vitro. Cells, 13(10), 813. https://doi.org/10.3390/cells13100813







