Subcellular Feature-Based Classification of α and β Cells Using Soft X-ray Tomography
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Model
2.2. Islet Isolation and Dissociation
2.3. Specimen Cryopreservation
2.4. Fluorescence Microscopy
2.5. Cryogenic Confocal Fluorescence Microscopy
2.6. Transmission Electron Microscopy
2.7. Soft X-ray Tomography Data Collection and Reconstruction
2.8. SXT Data Segmentation LAC Quantification
2.9. Quantification of Cellular and Subcellular Features and Statistical Analysis
2.9.1. Cellular and Organelle Volume Analysis
2.9.2. LAC Value Comparison
2.10. Statistical Analysis
2.11. UMAP Projection of Multidimensional Structural Data
2.12. Machine Learning Modeling and Validation Strategy
2.13. Interpretation of Feature Importances from Machine Learning
3. Results
3.1. α and β Cell Morphology Visualized by SXT
3.2. Analysis of Vesicle Properties in α and β Cells
3.3. Differentiating α and β Cells Using Machine Learning Models Based on Extracted Vesicle Characteristics
3.4. Displaying Distinguishing Features of Insulin and Glucagon Vesicles Using Structure-Based UMAP Visualizations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cabrera, O.; Berman, D.M.; Kenyon, N.S.; Ricordi, C.; Berggren, P.-O.; Caicedo, A. The Unique Cytoarchitecture of Human Pancreatic Islets Has Implications for Islet Cell Function. Proc. Natl. Acad. Sci. USA 2006, 103, 2334–2339. [Google Scholar] [CrossRef] [PubMed]
- Steiner, D.J.; Kim, A.; Miller, K.; Hara, M. Pancreatic Islet Plasticity: Interspecies Comparison of Islet Architecture and Composition. Islets 2010, 2, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Shuai, H.; Xu, Y.; Yu, Q.; Gylfe, E.; Tengholm, A. Fluorescent Protein Vectors for Pancreatic Islet Cell Identification in Live-Cell Imaging. Pflug. Arch. 2016, 468, 1765–1777. [Google Scholar] [CrossRef] [PubMed]
- Montet, X.; Lamprianou, S.; Vinet, L.; Meda, P.; Fort, A. Approaches for Imaging Pancreatic Islets: Recent Advances and Future Prospects. In Islets of Langerhans, 2nd ed.; Springer Netherlands: Dordrecht, The Netherlands, 2014; pp. 1–21. [Google Scholar]
- Wang, Z.; Gurlo, T.; Matveyenko, A.V.; Elashoff, D.; Wang, P.; Rosenberger, M.; Junge, J.A.; Stevens, R.C.; White, K.L.; Fraser, S.E.; et al. Live-Cell Imaging of Glucose-Induced Metabolic Coupling of β and α Cell Metabolism in Health and Type 2 Diabetes. Commun. Biol. 2021, 4, 594. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, S.; Menge, B.A.; Grüber, L.; Deacon, C.F.; Schmidt, W.E.; Veldhuis, J.D.; Holst, J.J.; Meier, J.J. Impaired Crosstalk between Pulsatile Insulin and Glucagon Secretion in Prediabetic Individuals. J. Clin. Endocrinol. Metab. 2012, 97, E791–E795. [Google Scholar] [CrossRef] [PubMed]
- Lang, D.A.; Matthews, D.R.; Burnett, M.; Turner, R.C. Brief, Irregular Oscillations of Basal Plasma Insulin and Glucose Concentrations in Diabetic Man. Diabetes 1981, 30, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, C.R.; Shomorony, A.; Aronova, M.A.; Zhang, G.; Cai, T.; Xu, H.; Notkins, A.L.; Leapman, R.D. Quantitative Analysis of Mouse Pancreatic Islet Architecture by Serial Block-Face SEM. J. Struct. Biol. 2015, 189, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Shomorony, A.; Pfeifer, C.R.; Aronova, M.A.; Zhang, G.; Cai, T.; Xu, H.; Notkins, A.L.; Leapman, R.D. Combining Quantitative 2D and 3D Image Analysis in the Serial Block Face SEM: Application to Secretory Organelles of Pancreatic Islet Cells. J. Microsc. 2015, 259, 155–164. [Google Scholar] [CrossRef]
- Müller, A.; Schmidt, D.; Xu, C.S.; Pang, S.; D’Costa, J.V.; Kretschmar, S.; Münster, C.; Kurth, T.; Jug, F.; Weigert, M.; et al. 3D FIB-SEM Reconstruction of Microtubule–Organelle Interaction in Whole Primary Mouse β Cells. J. Cell Biol. 2021, 220, e202010039. [Google Scholar] [CrossRef]
- de Boer, P.; Pirozzi, N.M.; Wolters, A.H.G.; Kuipers, J.; Kusmartseva, I.; Atkinson, M.A.; Campbell-Thompson, M.; Giepmans, B.N.G. Large-Scale Electron Microscopy Database for Human Type 1 Diabetes. Nat. Commun. 2020, 11, 2475. [Google Scholar] [CrossRef]
- Brereton, M.F.; Vergari, E.; Zhang, Q.; Clark, A. Alpha-, Delta- and PP-Cells. J. Histochem. Cytochem. 2015, 63, 575–591. [Google Scholar] [CrossRef] [PubMed]
- Pettway, Y.D.; Saunders, D.C.; Brissova, M. The Human α Cell in Health and Disease. J. Endocrinol. 2023, 258, e220298. [Google Scholar] [CrossRef] [PubMed]
- Ekman, A.; Chen, J.-H.; Vanslembrouck, B.; Loconte, V.; Larabell, C.A.; Le Gros, M.A.; Weinhardt, V. Extending Imaging Volume in Soft X-Ray Tomography. Adv. Photonics. Res. 2023, 4, 2200142. [Google Scholar] [CrossRef]
- Schneider, G.; Guttmann, P.; Heim, S.; Rehbein, S.; Mueller, F.; Nagashima, K.; Heymann, J.B.; Müller, W.G.; McNally, J.G. Three-Dimensional Cellular Ultrastructure Resolved by X-Ray Microscopy. Nat. Methods 2010, 7, 985–987. [Google Scholar] [CrossRef] [PubMed]
- Ekman, A.A.; Chen, J.; Guo, J.; McDermott, G.; Le Gros, M.A.; Larabell, C.A. Mesoscale Imaging with Cryo-light and X-rays: Larger than Molecular Machines, Smaller than a Cell. Biol. Cell 2017, 109, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Kirz, J.; Jacobsen, C.; Howells, M. Soft X-Ray Microscopes and Their Biological Applications. Q. Rev. Biophys. 1995, 28, 33–130. [Google Scholar] [CrossRef]
- Le Gros, M.A.; McDermott, G.; Larabell, C.A. X-Ray Tomography of Whole Cells. Curr. Opin. Struct. Biol. 2005, 15, 593–600. [Google Scholar] [CrossRef]
- McDermott, G.; Fox, D.M.; Epperly, L.; Wetzler, M.; Barron, A.E.; Le Gros, M.A.; Larabell, C.A. Visualizing and Quantifying Cell Phenotype Using Soft X-Ray Tomography. BioEssays 2012, 34, 320–327. [Google Scholar] [CrossRef]
- Larabell, C.A.; Nugent, K.A. Imaging Cellular Architecture with X-Rays. Curr. Opin. Struct. Biol. 2010, 20, 623–631. [Google Scholar] [CrossRef]
- Loconte, V.; White, K.L. The Use of Soft X-Ray Tomography to Explore Mitochondrial Structure and Function. Mol. Metab. 2022, 57, 101421. [Google Scholar] [CrossRef]
- Le Gros, M.A.; Clowney, E.J.; Magklara, A.; Yen, A.; Markenscoff-Papadimitriou, E.; Colquitt, B.; Myllys, M.; Kellis, M.; Lomvardas, S.; Larabell, C.A. Soft X-Ray Tomography Reveals Gradual Chromatin Compaction and Reorganization during Neurogenesis In Vivo. Cell Rep. 2016, 17, 2125–2136. [Google Scholar] [CrossRef] [PubMed]
- Azzarello, F.; Carli, F.; De Lorenzi, V.; Tesi, M.; Marchetti, P.; Beltram, F.; Raimondi, F.; Cardarelli, F. Machine-Learning-Guided Recognition of α and β Cells from Label-Free Infrared Micrographs of Living Human Islets of Langerhans. Preprints, 2024. [Google Scholar]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Zhu, S.; Larkin, D.; Lu, S.; Inouye, C.; Haataja, L.; Anjum, A.; Kennedy, R.; Castle, D.; Arvan, P. Monitoring C-Peptide Storage and Secretion in Islet β-Cells In Vitro and In Vivo. Diabetes 2016, 65, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Georgia, S.; Tschen, S.; Nakayama, K.; Nakayama, K.; Bhushan, A. Essential Role of Skp2-Mediated P27 Degradation in Growth and Adaptive Expansion of Pancreatic β Cells. J. Clin. Investig. 2007, 117, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-H.; Vanslembrouck, B.; Loconte, V.; Ekman, A.; Cortese, M.; Bartenschlager, R.; McDermott, G.; Larabell, C.A.; Le Gros, M.A.; Weinhardt, V. A Protocol for Full-Rotation Soft X-Ray Tomography of Single Cells. STAR Protoc. 2022, 3, 101176. [Google Scholar] [CrossRef] [PubMed]
- Le Gros, M.A.; McDermott, G.; Uchida, M.; Knoechel, C.G.; Larabell, C.A. High-Aperture Cryogenic Light Microscopy. J. Microsc. 2009, 235, 1–8. [Google Scholar] [CrossRef] [PubMed]
- McDermott, G.; Le Gros, M.A.; Knoechel, C.G.; Uchida, M.; Larabell, C.A. Soft X-Ray Tomography and Cryogenic Light Microscopy: The Cool Combination in Cellular Imaging. Trends Cell Biol. 2009, 19, 587–595. [Google Scholar] [CrossRef]
- Smith, E.A.; Cinquin, B.P.; Do, M.; McDermott, G.; Le Gros, M.A.; Larabell, C.A. Correlative Cryogenic Tomography of Cells Using Light and Soft X-Rays. Ultramicroscopy 2014, 143, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.A.; McDermott, G.; Do, M.; Leung, K.; Panning, B.; Le Gros, M.A.; Larabell, C.A. Quantitatively Imaging Chromosomes by Correlated Cryo-Fluorescence and Soft X-Ray Tomographies. Biophys. J. 2014, 107, 1988–1996. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Le Gros, M.A.; McDermott, G.; Cinquin, B.P.; Smith, E.A.; Do, M.; Chao, W.L.; Naulleau, P.P.; Larabell, C.A. Biological Soft X-Ray Tomography on Beamline 2.1 at the Advanced Light Source. J. Synchrotron Radiat. 2014, 21, 1370–1377. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, D.Y.; Epperly, L.R.; McDermott, G.; Le Gros, M.A.; Boudreau, R.M.; Larabell, C.A. Nanoimaging Cells Using Soft X-Ray Tomography. In Nanoimaging. Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; pp. 457–481. [Google Scholar]
- Parkinson, D.Y.; Knoechel, C.; Yang, C.; Larabell, C.A.; Le Gros, M.A. Automatic Alignment and Reconstruction of Images for Soft X-Ray Tomography. J. Struct. Biol. 2012, 177, 259–266. [Google Scholar] [CrossRef]
- White, K.L.; Singla, J.; Loconte, V.; Chen, J.-H.; Ekman, A.; Sun, L.; Zhang, X.; Francis, J.P.; Li, A.; Lin, W.; et al. Visualizing Subcellular Rearrangements in Intact β Cells Using Soft X-Ray Tomography. Sci. Adv. 2020, 6, eabc8262. [Google Scholar] [CrossRef]
- Lösel, P.D.; van de Kamp, T.; Jayme, A.; Ershov, A.; Faragó, T.; Pichler, O.; Tan Jerome, N.; Aadepu, N.; Bremer, S.; Chilingaryan, S.A.; et al. Introducing Biomedisa as an Open-Source Online Platform for Biomedical Image Segmentation. Nat. Commun. 2020, 11, 5577. [Google Scholar] [CrossRef]
- Erozan, A.A.; Lösel, P.D.; Heuveline, V.; Weinhardt, V. Automated 3D Cytoplasm Segmentation in Soft X-Ray Tomography. iScience, 2024; 27, 109856. [Google Scholar] [CrossRef]
- Loconte, V.; Singla, J.; Li, A.; Chen, J.-H.; Ekman, A.; McDermott, G.; Sali, A.; Le Gros, M.; White, K.L.; Larabell, C.A. Soft X-Ray Tomography to Map and Quantify Organelle Interactions at the Mesoscale. Structure 2022, 30, 510–521. [Google Scholar] [CrossRef] [PubMed]
- In’t Veld, P.; Marichal, M. Microscopic Anatomy of the Human Islet of Langerhans. In Advances in Experimental Medicine and Biology; Springer: Dordrecht, The Netherlands, 2010; pp. 1–19. [Google Scholar]
- Greider, M.H.; Howell, S.L.; Lacy, P.E. Isolation and properties of secretory granules from rat islets of langerhans. J. Cell Biol. 1969, 41, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Reetz, A.; Solimena, M.; Matteoli, M.; Folli, F.; Takei, K.; De Camilli, P. GABA and Pancreatic Beta-Cells: Colocalization of Glutamic Acid Decarboxylase (GAD) and GABA with Synaptic-like Microvesicles Suggests Their Role in GABA Storage and Secretion. EMBO J. 1991, 10, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Faisal, M.; Zamzami, E.M. Sutarman Comparative Analysis of Inter-Centroid K-Means Performance Using Euclidean Distance, Canberra Distance and Manhattan Distance. J. Phys. Conf. Ser. 2020, 1566, 012112. [Google Scholar] [CrossRef]
- ’Jurman, G.; ’Riccadonna, S.; ’Visintainer, R.; ’Furlanello, C. Canberra Distance on Ranked Lists. In Proceedings of the Advances in Ranking NIPS 09 Workshop, Vancouver, BC, Canada, 7–10 December 2009; pp. 22–27. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Müller, A.; Nothman, J.; Louppe, G.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2012, 12, 2825–2830. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 13 August 2016; ACM: New York, NY, USA; pp. 785–794. [Google Scholar]
- Greener, J.G.; Kandathil, S.M.; Moffat, L.; Jones, D.T. A Guide to Machine Learning for Biologists. Nat. Rev. Mol. Cell. Biol. 2022, 23, 40–55. [Google Scholar] [CrossRef]
- Snoek, J.; Larochelle, H.; Adams, R.P. Practical Bayesian Optimization of Machine Learning Algorithms. arXiv 2012, arXiv:arXiv:1206.2944. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Cho, N.H.; Cheveralls, K.C.; Brunner, A.-D.; Kim, K.; Michaelis, A.C.; Raghavan, P.; Kobayashi, H.; Savy, L.; Li, J.Y.; Canaj, H.; et al. OpenCell: Endogenous Tagging for the Cartography of Human Cellular Organization. Science 2022, 375, eabi6983. [Google Scholar] [CrossRef] [PubMed]
- Androvic, P.; Schifferer, M.; Perez Anderson, K.; Cantuti-Castelvetri, L.; Jiang, H.; Ji, H.; Liu, L.; Gouna, G.; Berghoff, S.A.; Besson-Girard, S.; et al. Spatial Transcriptomics-Correlated Electron Microscopy Maps Transcriptional and Ultrastructural Responses to Brain Injury. Nat. Commun. 2023, 14, 4115. [Google Scholar] [CrossRef] [PubMed]
- Dominguez Gutierrez, G.; Xin, Y.; Okamoto, H.; Kim, J.; Lee, A.-H.; Ni, M.; Adler, C.; Yancopoulos, G.D.; Murphy, A.J.; Gromada, J. Gene Signature of Proliferating Human Pancreatic α Cells. Endocrinology 2018, 159, 3177–3186. [Google Scholar] [CrossRef] [PubMed]
- Autin, L.; Barbaro, B.; Jewett, A.I.; Ekman, A.; Verma, S.; Olson, A.J.; Goodsell, D.S. Integrative Structural Modeling and Visualization of a Cellular Organelle. QRB Discov. 2022, 3, e11. [Google Scholar] [CrossRef] [PubMed]
- Leiter, E.H.; Gapp, D.A.; Eppig, J.J.; Coleman, D.L. Utrastructural and Morphometric Studies of Delta Cells in Pancreatic Islets from C57BL/Ks Diabetes Mice. Diabetologia 1979, 17, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.-M.; Furnes, M.W.; Stenström, B.; Kulseng, B.; Chen, D. Characterization of Obestatin- and Ghrelin-Producing Cells in the Gastrointestinal Tract and Pancreas of Rats: An Immunohistochemical and Electron-Microscopic Study. Cell Tissue Res. 2008, 331, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Castorina, S.; Barresi, V.; Luca, T.; Privitera, G.; De Geronimo, V.; Lezoche, G.; Cosentini, I.; Di Vincenzo, A.; Barbatelli, G.; Giordano, A.; et al. Gastric Ghrelin Cells in Obese Patients Are Hyperactive. Int. J. Obes. 2021, 45, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Wierup, N.; Sundler, F. Ultrastructure of Islet Ghrelin Cells in the Human Fetus. Cell Tissue Res. 2005, 319, 423–428. [Google Scholar] [CrossRef]
- Arrojo e Drigo, R.; Jacob, S.; García-Prieto, C.F.; Zheng, X.; Fukuda, M.; Nhu, H.T.T.; Stelmashenko, O.; Peçanha, F.L.M.; Rodriguez-Diaz, R.; Bushong, E.; et al. Structural Basis for Delta Cell Paracrine Regulation in Pancreatic Islets. Nat. Commun. 2019, 10, 3700. [Google Scholar] [CrossRef]
- Rorsman, P.; Huising, M.O. The Somatostatin-Secreting Pancreatic δ-Cell in Health and Disease. Nat. Rev. Endocrinol. 2018, 14, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Babajide Mustapha, I.; Saeed, F. Bioactive Molecule Prediction Using Extreme Gradient Boosting. Molecules 2016, 21, 983. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.J.; Taylor, S.; Contrepois, K.; Kim, M.; Bravo, J.I.; Ellenberger, M.; Sampathkumar, N.K.; Benayoun, B.A. Multi-Omic Profiling of Primary Mouse Neutrophils Predicts a Pattern of Sex- and Age-Related Functional Regulation. Nat. Aging 2021, 1, 715–733. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Chiou, J.; Zeng, C.; Miller, M.; Matta, I.; Han, J.Y.; Kadakia, N.; Okino, M.-L.; Beebe, E.; Mallick, M.; et al. Inte-grating Genetics with Single-Cell Multiomic Measurements across Disease States Identifies Mechanisms of Beta Cell Dysfunction in Type 2 Diabetes. Nat. Genet 2023, 55, 984–994. [Google Scholar] [CrossRef] [PubMed]
- Miranda, M.A.; Macias-Velasco, J.F.; Lawson, H.A. Pancreatic β-Cell Heterogeneity in Health and Diabetes: Classes, Sources, and Subtypes. Am. J. Physiol.-Endocrinol. Metab. 2021, 320, E716–E731. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, G.D.; Gromada, J.; Sussel, L. Heterogeneity of the Pancreatic Beta Cell. Front. Genet. 2017, 8, 248456. [Google Scholar] [CrossRef]
- Benninger, R.K.P.; Kravets, V. The Physiological Role of β-Cell Heterogeneity in Pancreatic Islet Function. Nat. Rev. Endocrinol. 2022, 18, 9–22. [Google Scholar] [CrossRef]
- Singla, J.; White, K.L. A Community Approach to Whole-Cell Modeling. Curr. Opin. Syst. Biol. 2021, 26, 33–38. [Google Scholar] [CrossRef]
- Singla, J.; McClary, K.M.; White, K.L.; Alber, F.; Sali, A.; Stevens, R.C. Opportunities and Challenges in Building a Spatiotemporal Multi-Scale Model of the Human Pancreatic β Cell. Cell 2018, 173, 11–19. [Google Scholar] [CrossRef]
- Loconte, V.; Chen, J.; Vanslembrouck, B.; Ekman, A.A.; McDermott, G.; Le Gros, M.A.; Larabell, C.A. Soft X-ray Tomograms Provide a Structural Basis for Whole-cell Modeling. FASEB J. 2023, 37, e22681. [Google Scholar] [CrossRef]
- Raveh, B.; Sun, L.; White, K.L.; Sanyal, T.; Tempkin, J.; Zheng, D.; Bharath, K.; Singla, J.; Wang, C.; Zhao, J.; et al. Bayesian Metamodeling of Complex Biological Systems across Varying Representations. Proc. Natl. Acad. Sci. USA 2021, 118, e2104559118. [Google Scholar] [CrossRef] [PubMed]

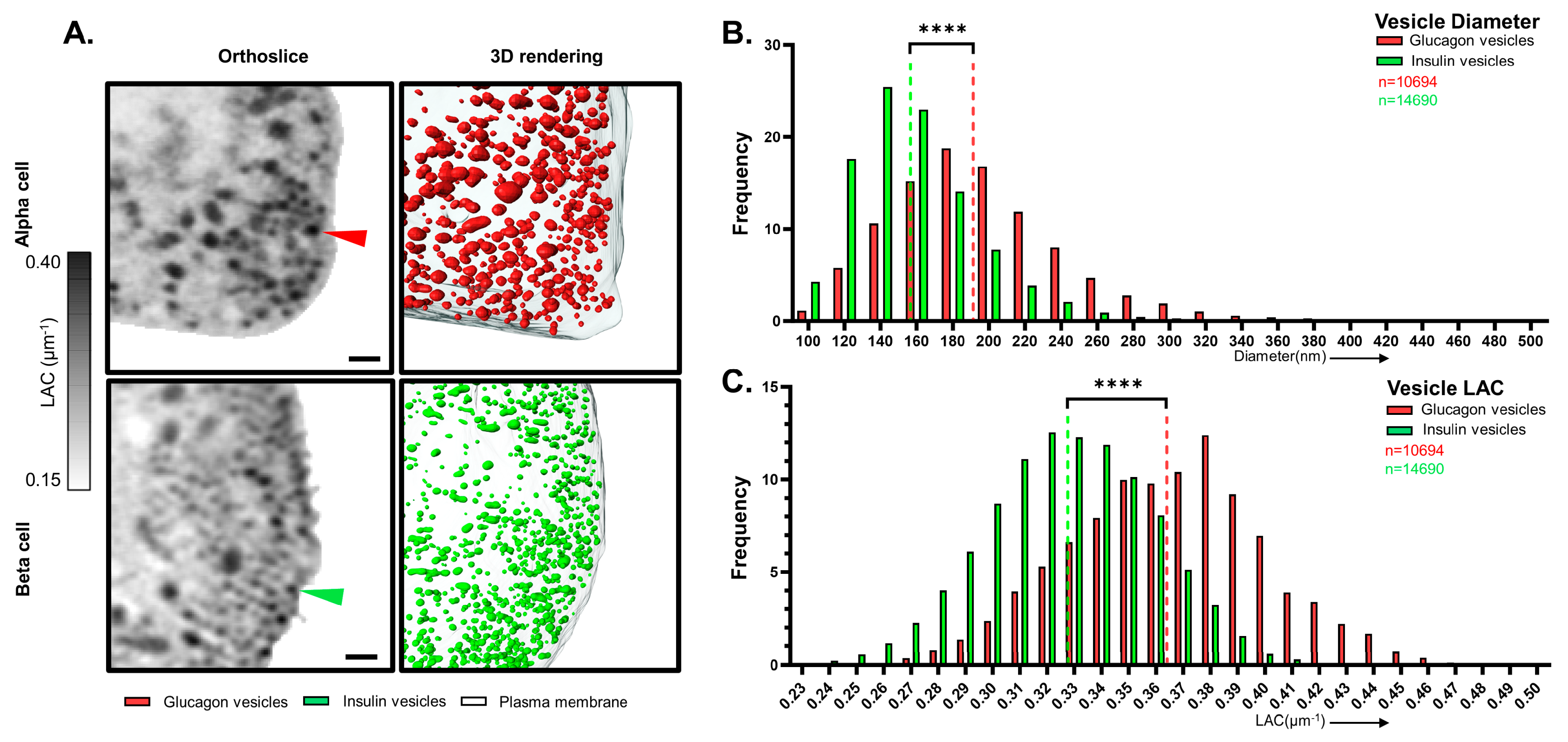
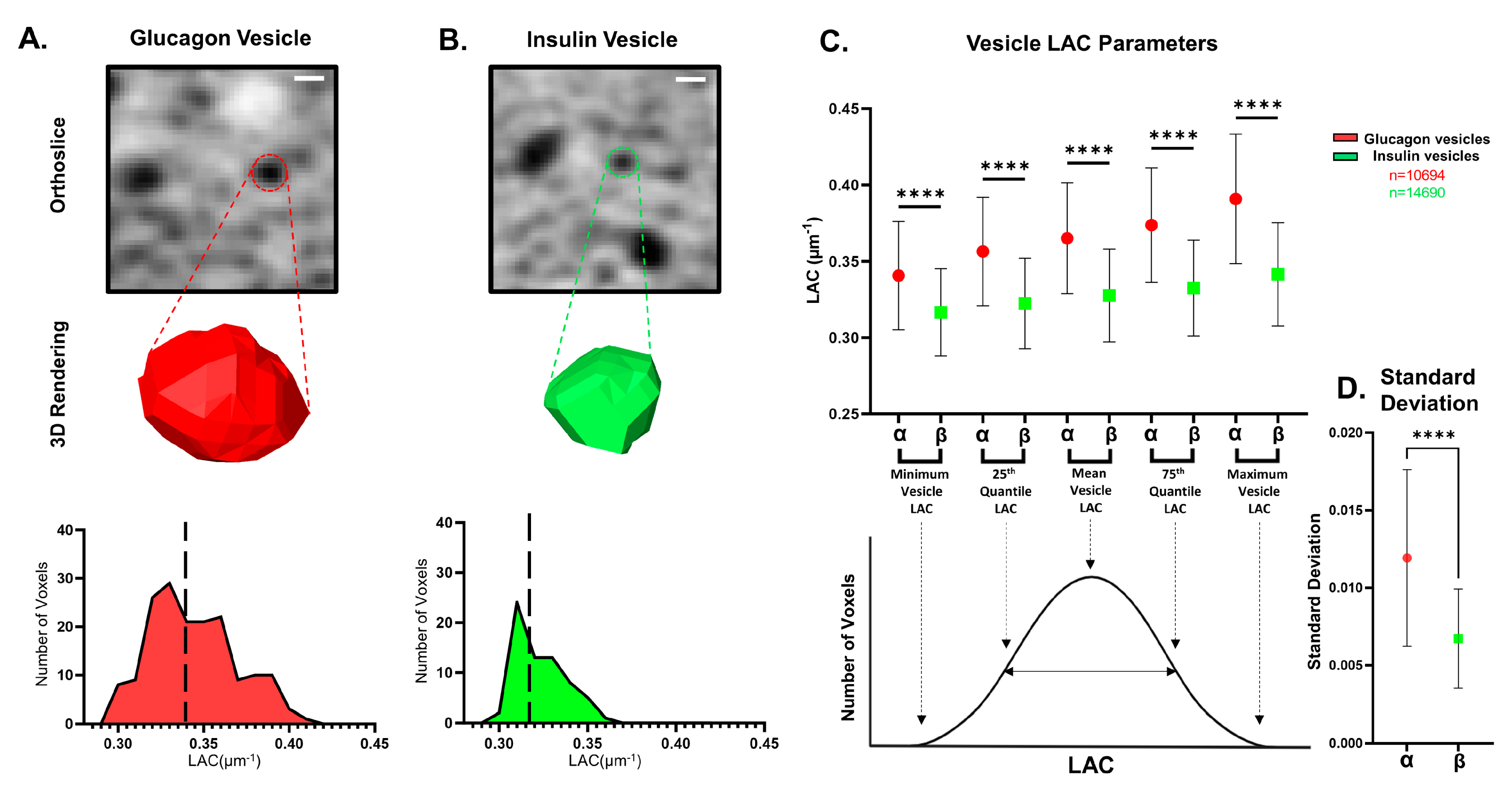
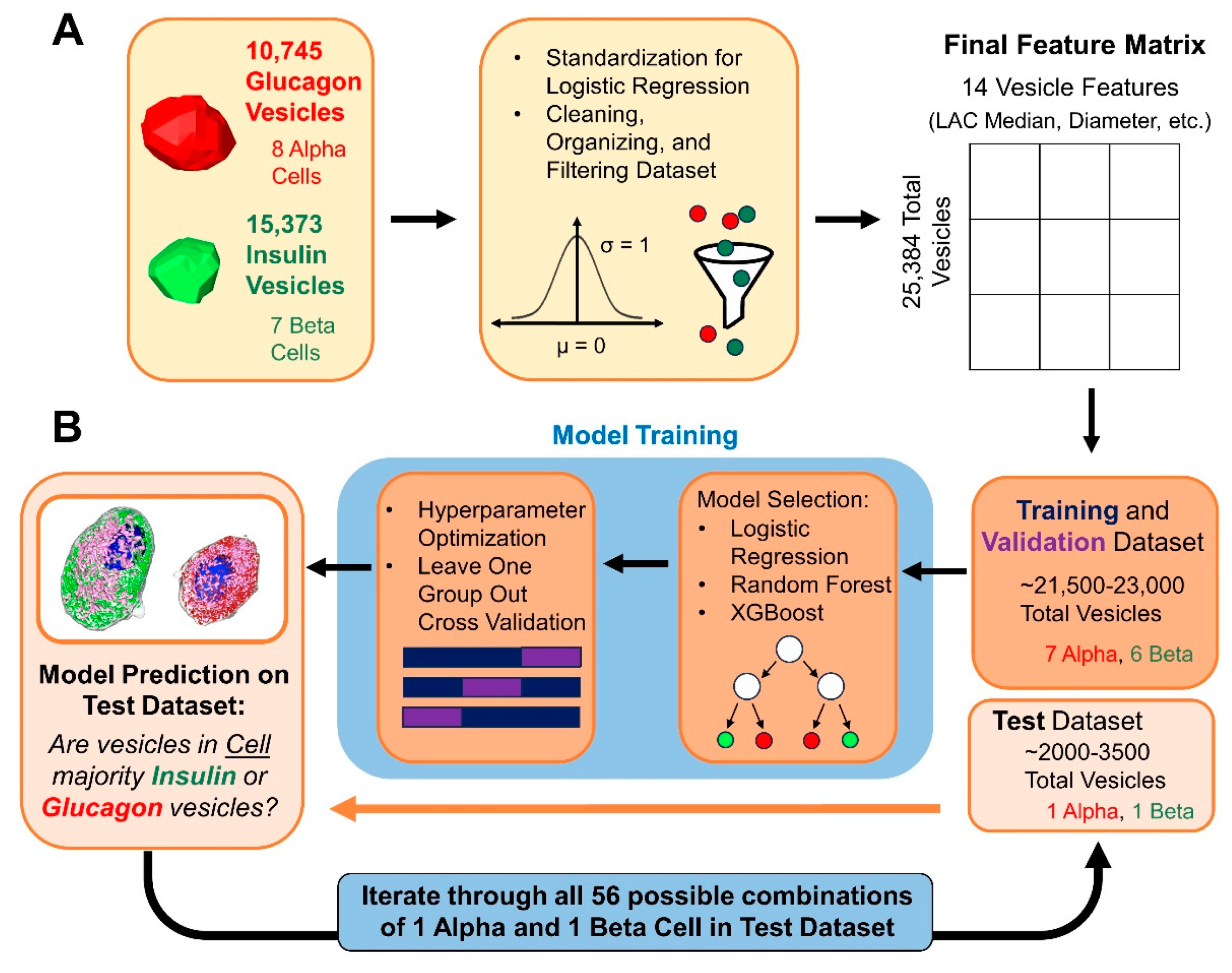
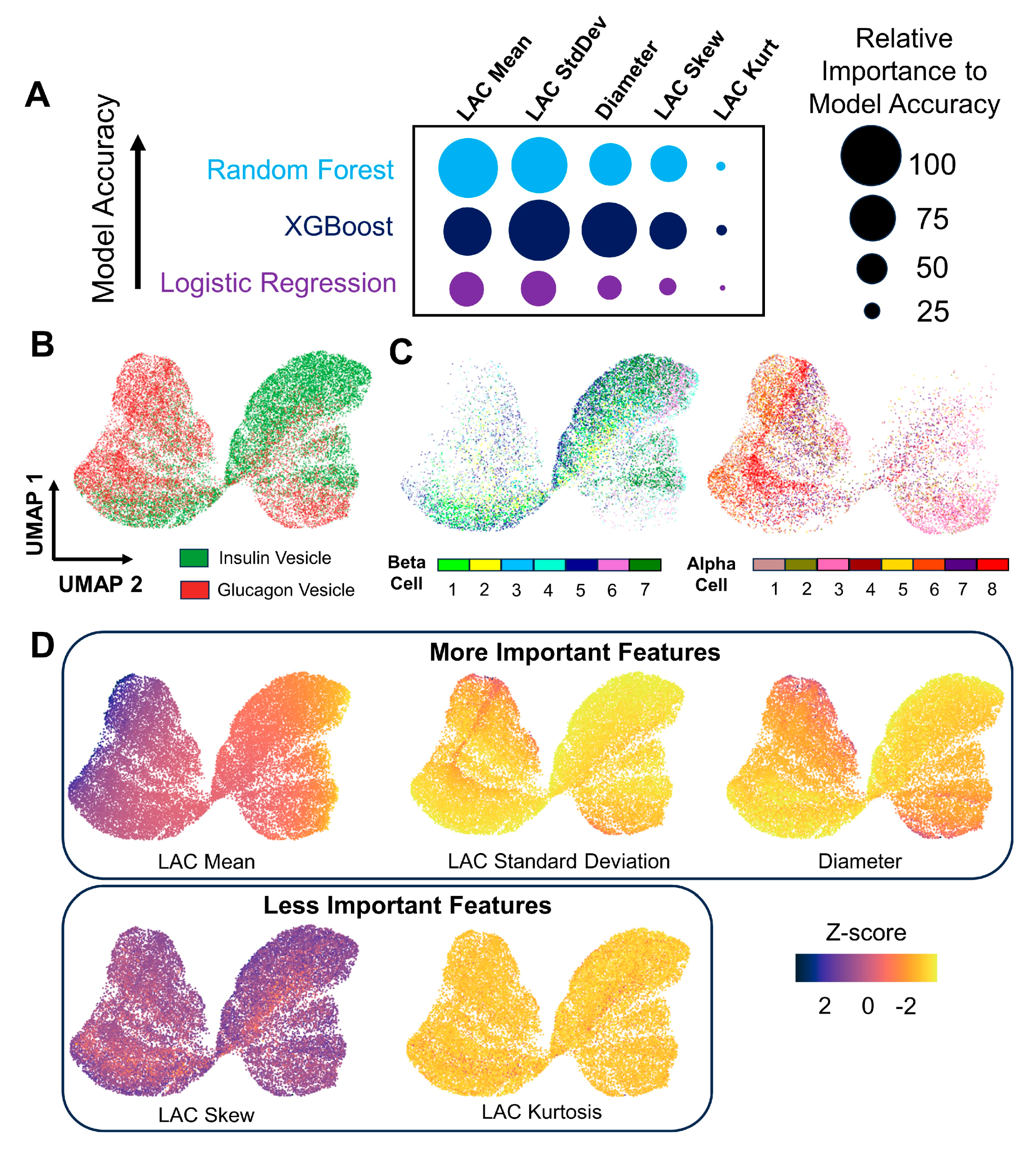
| Cell Type | α-Cell | β-Cell |
|---|---|---|
| Number of Cells | 8 | 7 |
| Cell Volume (µm3) | 579 ± 247 | 1191 ± 277 *** |
| Nucleus Volume (µm3) | 112 ± 32 | 118 ± 42 |
| Nucleus Volume (%) | 21 ± 5 | 10 ± 3 *** |
| Vesicle Volume (µm3) | 6 ± 2 | 5 ± 2 |
| Vesicle Volume (%) | 1.1 ± 0.4 | 0.4 ± 0.1 |
| Vesicle Number | 1337 ± 480 | 2099 ± 710 * |
| Vesicle Diameter (nm) | 213 ± 21 | 163 ± 13 *** |
| Vesicle LAC (µm−1) | 0.375 ± 0.03 | 0.334 ± 0.02 ** |
| Nucleus LAC (µm−1) | 0.24 ± 0.02 | 0.21 ± 0.02 |
| Mitochondria LAC (µm−1) | 0.357 ± 0.03 | 0.335 ± 0.03 |
| Cytosol LAC (µm−1) | 0.263 ± 0.02 | 0.237 ± 0.02 * |
| Vesicle Type | Glucagon Vesicle | Insulin Vesicle |
|---|---|---|
| Number of Vesicles | 10,964 | 14,960 |
| Mean LAC (µm−1) | 0.365 ± 0.04 | 0.328 ± 0.03 † |
| Median LAC (µm−1) | 0.364 ± 0.04 | 0.327 ± 0.03 † |
| Mode LAC (µm−1) | 0.365 ± 0.04 | 0.328 ± 0.03 † |
| Maximum LAC (µm−1) | 0.391 ± 0.04 | 0.342 ± 0.03 † |
| Minimum LAC (µm−1) | 0.341 ± 0.04 | 0.317 ± 0.03 † |
| Standard Deviation (µm−1) | 0.012 ± 0.005 | 0.007 ± 0.003 † |
| 25th Quantile LAC (µm−1) | 0.356 ± 0.04 | 0.322 ± 0.03 † |
| 75th Quantile LAC (µm−1) | 0.374 ± 0.04 | 0.332 ± 0.03 † |
| Skewness | 0.085 ± 0.37 | 0.288 ± 0.44 † |
| Kurtosis | −0.506 ± 0.49 | −0.617 ± 0.55 † |
| Diameter (nm) | 194 ± 49 | 157 ± 35 † |
| Model | Accuracy | F1 Score | ROC AUC |
|---|---|---|---|
| Logistic Regression | 0.75 ± 0.11 | 0.68 ± 0.12 | 0.82 ± 0.13 |
| Random Forest | 0.77 ± 0.11 | 0.71 ± 0.09 | 0.85 ± 0.10 |
| XGBoost | 0.75 ± 0.12 | 0.70 ± 0.11 | 0.83 ± 0.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deshmukh, A.; Chang, K.; Cuala, J.; Vanslembrouck, B.; Georgia, S.; Loconte, V.; White, K.L. Subcellular Feature-Based Classification of α and β Cells Using Soft X-ray Tomography. Cells 2024, 13, 869. https://doi.org/10.3390/cells13100869
Deshmukh A, Chang K, Cuala J, Vanslembrouck B, Georgia S, Loconte V, White KL. Subcellular Feature-Based Classification of α and β Cells Using Soft X-ray Tomography. Cells. 2024; 13(10):869. https://doi.org/10.3390/cells13100869
Chicago/Turabian StyleDeshmukh, Aneesh, Kevin Chang, Janielle Cuala, Bieke Vanslembrouck, Senta Georgia, Valentina Loconte, and Kate L. White. 2024. "Subcellular Feature-Based Classification of α and β Cells Using Soft X-ray Tomography" Cells 13, no. 10: 869. https://doi.org/10.3390/cells13100869
APA StyleDeshmukh, A., Chang, K., Cuala, J., Vanslembrouck, B., Georgia, S., Loconte, V., & White, K. L. (2024). Subcellular Feature-Based Classification of α and β Cells Using Soft X-ray Tomography. Cells, 13(10), 869. https://doi.org/10.3390/cells13100869








