An Optimized Protocol for the Generation of Alveolospheres from Wild-Type Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice and Tamoxifen Administration
2.2. Lung Dissociation and Fluorescence-Activated Cell Sorting
2.3. Alveolosphere Generation and Analysis
2.4. Induction of Cre Activity in Ex Vivo Alveolosphere Cultures
2.5. Whole-Mount Immunofluorescence
2.6. Image Analysis
2.7. RNA Extraction and Gene Expression Analysis
2.8. Statistical Analysis
3. Results
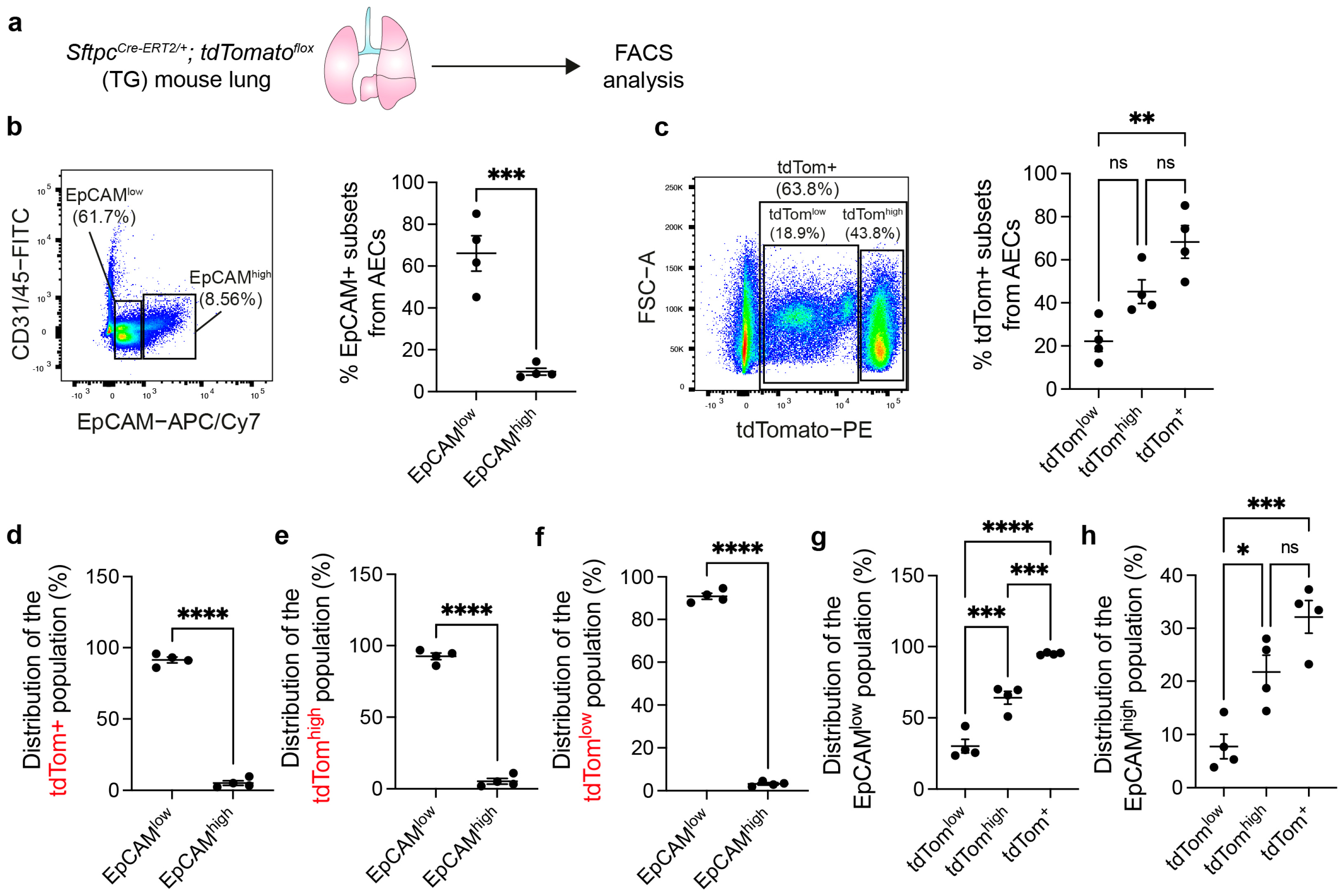
3.1. AEC2s Can Be Isolated from WT Mouse Lungs by Sorting CD45− CD31− EpCAMlow Cells
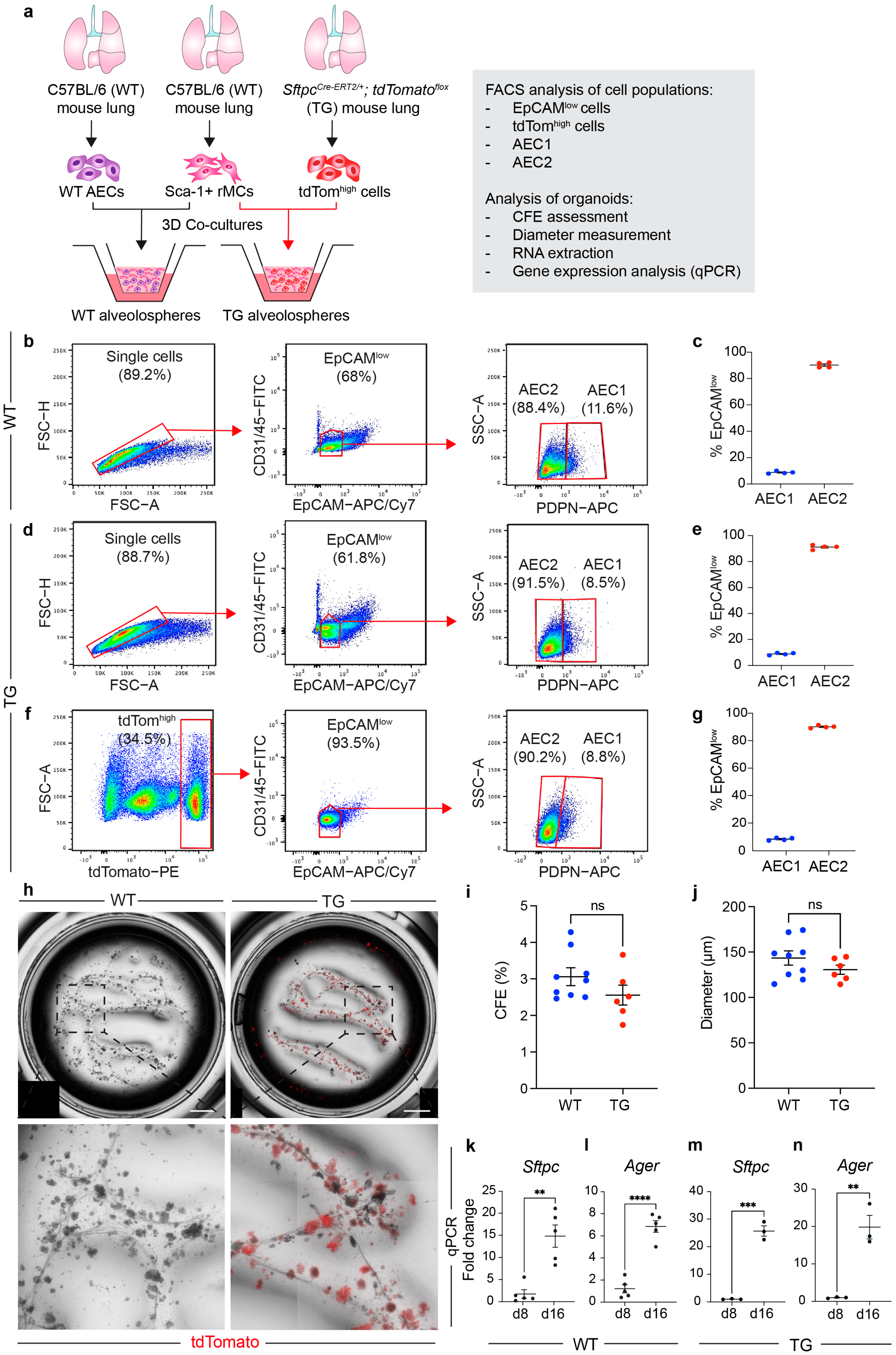
3.2. The EpCAMlow Population Robustly Generates Alveolospheres and It Is Comparable with Sorted Lineage-Traced AEC2s
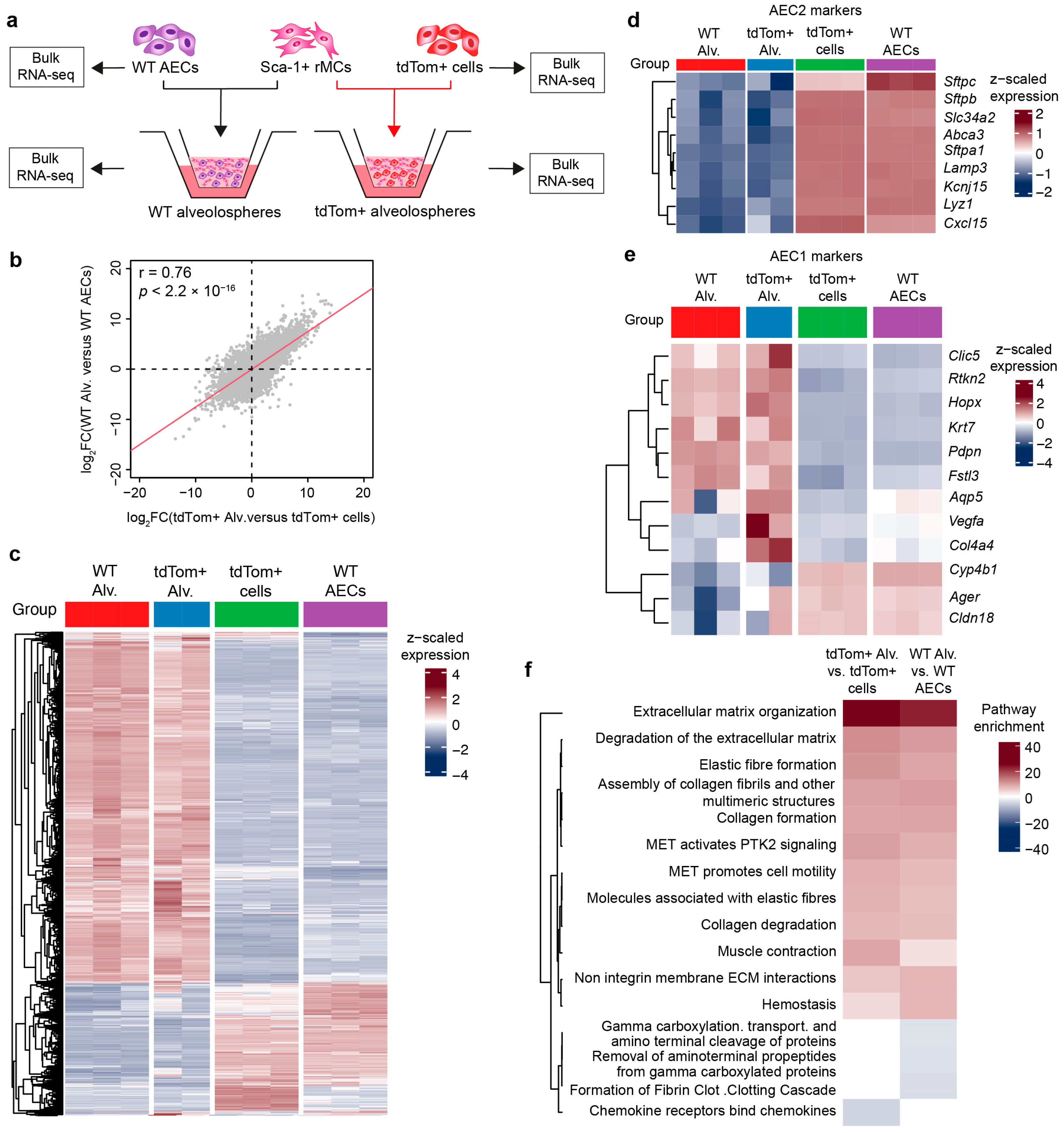
3.3. WT Alveolospheres Derive from Isolated AEC2s
3.4. Stimulation with Recombinant Murine IL-1β Alters Alveolosphere Formation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a Dish: Modeling Development and Disease Using Organoid Technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Chen, X.; Dowbaj, A.M.; Sljukic, A.; Bratlie, K.; Lin, L.; Fong, E.L.S.; Balachander, G.M.; Chen, Z.; Soragni, A. Organoids. Nat. Rev. Methods Primer 2022, 2, 94. [Google Scholar] [CrossRef]
- Vazquez-Armendariz, A.I.; Herold, S. From Clones to Buds and Branches: The Use of Lung Organoids to Model Branching Morphogenesis Ex Vivo. Front. Cell Dev. Biol. 2021, 9, 631579. [Google Scholar] [CrossRef] [PubMed]
- Rock, J.R.; Onaitis, M.W.; Rawlins, E.L.; Lu, Y.; Clark, C.P.; Xue, Y.; Randell, S.H.; Hogan, B.L. Basal Cells as Stem Cells of the Mouse Trachea and Human Airway Epithelium. Proc. Natl. Acad. Sci. USA 2009, 106, 12771–12775. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Armendariz, A.I.; Seeger, W.; Herold, S.; El Agha, E. Protocol for the Generation of Murine Bronchiolospheres. STAR Protoc. 2021, 2, 100594. [Google Scholar] [CrossRef] [PubMed]
- Moiseenko, A.; Vazquez-Armendariz, A.I.; Kheirollahi, V.; Chu, X.; Tata, A.; Rivetti, S.; Günther, S.; Lebrigand, K.; Herold, S.; Braun, T.; et al. Identification of a Repair-Supportive Mesenchymal Cell Population during Airway Epithelial Regeneration. Cell Rep. 2020, 33, 108549. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Tammela, T.; Hofree, M.; Choi, J.; Marjanovic, N.D.; Han, S.; Canner, D.; Wu, K.; Paschini, M.; Bhang, D.H.; et al. Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6. Cell 2017, 170, 1149–1163.e12. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Lingampally, A.; Moiseenko, A.; Kheirollahi, V.; Vazquez-Armendariz, A.I.; Koepke, J.; Khadim, A.; Kiliaris, G.; Shahriari Felordi, M.; Zabihi, M.; et al. GLI1+ Cells Are a Source of Repair-Supportive Mesenchymal Cells (RSMCs) during Airway Epithelial Regeneration. Cell. Mol. Life Sci. 2022, 79, 581. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Armendariz, A.I.; Heiner, M.; El Agha, E.; Salwig, I.; Hoek, A.; Hessler, M.C.; Shalashova, I.; Shrestha, A.; Carraro, G.; Mengel, J.P.; et al. Multilineage Murine Stem Cells Generate Complex Organoids to Model Distal Lung Development and Disease. EMBO J. 2020, 39, e103476. [Google Scholar] [CrossRef]
- Barkauskas, C.E.; Cronce, M.J.; Rackley, C.R.; Bowie, E.J.; Keene, D.R.; Stripp, B.R.; Randell, S.H.; Noble, P.W.; Hogan, B.L.M. Type 2 Alveolar Cells Are Stem Cells in Adult Lung. J. Clin. Investig. 2013, 123, 3025–3036. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Tata, A.; Konkimalla, A.; Katsura, H.; Lee, R.F.; Ou, J.; Banovich, N.E.; Kropski, J.A.; Tata, P.R. Persistence of a Regeneration-Associated, Transitional Alveolar Epithelial Cell State in Pulmonary Fibrosis. Nat. Cell Biol. 2020, 22, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Park, J.-E.; Tsagkogeorga, G.; Yanagita, M.; Koo, B.-K.; Han, N.; Lee, J.-H. Inflammatory Signals Induce AT2 Cell-Derived Damage-Associated Transient Progenitors That Mediate Alveolar Regeneration. Cell Stem Cell 2020, 27, 366–382.e7. [Google Scholar] [CrossRef] [PubMed]
- Strunz, M.; Simon, L.M.; Ansari, M.; Kathiriya, J.J.; Angelidis, I.; Mayr, C.H.; Tsidiridis, G.; Lange, M.; Mattner, L.F.; Yee, M.; et al. Alveolar Regeneration through a Krt8+ Transitional Stem Cell State That Persists in Human Lung Fibrosis. Nat. Commun. 2020, 11, 3559. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, W.J.; Frank, D.B.; Zepp, J.A.; Morley, M.P.; Alkhaleel, F.A.; Kong, J.; Zhou, S.; Cantu, E.; Morrisey, E.E. Regeneration of the Lung Alveolus by an Evolutionarily Conserved Epithelial Progenitor. Nature 2018, 555, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.-Q.; Cai, G.-F.; Zeng, P.-P.; Dhlamini, Q.; Chen, L.-F.; Chen, J.-J.; Lyu, H.-D.; Mossahebi-Mohammadi, M.; Ahmadvand, N.; Bellusci, S.; et al. FGF10 Therapeutic Administration Promotes Mobilization of Injury-Activated Alveolar Progenitors in a Mouse Fibrosis Model. Cells 2022, 11, 2396. [Google Scholar] [CrossRef]
- Ahmadvand, N.; Khosravi, F.; Lingampally, A.; Wasnick, R.; Vazquez-Armendariz, A.I.; Carraro, G.; Heiner, M.; Rivetti, S.; Lv, Y.; Wilhelm, J.; et al. Identification of a Novel Subset of Alveolar Type 2 Cells Enriched in PD-L1 and Expanded Following Pneumonectomy. Eur. Respir. J. 2021, 58, 2004168. [Google Scholar] [CrossRef]
- Van der Velden, J.L.; Bertoncello, I.; McQualter, J.L. LysoTracker Is a Marker of Differentiated Alveolar Type II Cells. Respir. Res. 2013, 14, 1–7. [Google Scholar] [CrossRef]
- Taghizadeh, S.; Heiner, M.; Vazquez-Armendariz, A.I.; Wilhelm, J.; Herold, S.; Chen, C.; Zhang, J.S.; Bellusci, S. Characterization in Mice of the Resident Mesenchymal Niche Maintaining AT2 Stem Cell Proliferation in Homeostasis and Disease. Stem Cells Dayt. Ohio 2021, 39, 1382–1394. [Google Scholar] [CrossRef] [PubMed]
- Quantius, J.; Schmoldt, C.; Vazquez-Armendariz, A.I.; Becker, C.; El Agha, E.; Wilhelm, J.; Morty, R.E.; Vadász, I.; Mayer, K.; Gattenloehner, S.; et al. Influenza Virus Infects Epithelial Stem/Progenitor Cells of the Distal Lung: Impact on Fgfr2b-Driven Epithelial Repair. PLoS Pathog. 2016, 12, e1005544. [Google Scholar] [CrossRef]
- Chung, M.-I.; Bujnis, M.; Barkauskas, C.E.; Kobayashi, Y.; Hogan, B.L.M. Niche-Mediated BMP/SMAD Signaling Regulates Lung Alveolar Stem Cell Proliferation and Differentiation. Development 2018, 145, dev163014. [Google Scholar] [CrossRef]
- McQualter, J.L.; Yuen, K.; Williams, B.; Bertoncello, I. Evidence of an Epithelial Stem/Progenitor Cell Hierarchy in the Adult Mouse Lung. Proc. Natl. Acad. Sci. USA 2010, 107, 1414–1419. [Google Scholar] [CrossRef] [PubMed]
- Chapman, H.A.; Li, X.; Alexander, J.P.; Brumwell, A.; Lorizio, W.; Tan, K.; Sonnenberg, A.; Wei, Y.; Vu, T.H. Integrin Alpha6beta4 Identifies an Adult Distal Lung Epithelial Population with Regenerative Potential in Mice. J. Clin. Investig. 2011, 121, 2855–2862. [Google Scholar] [CrossRef]
- Dekkers, J.F.; Alieva, M.; Wellens, L.M.; Ariese, H.C.R.; Jamieson, P.R.; Vonk, A.M.; Amatngalim, G.D.; Hu, H.; Oost, K.C.; Snippert, H.J.G.; et al. High-Resolution 3D Imaging of Fixed and Cleared Organoids. Nat. Protoc. 2019, 14, 1756–1771. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Armendariz, A.I.; Barroso, M.M.; El Agha, E.; Herold, S. 3D In Vitro Models: Novel Insights into Idiopathic Pulmonary Fibrosis Pathophysiology and Drug Screening. Cells 2022, 11, 1526. [Google Scholar] [CrossRef] [PubMed]
- Liberti, D.C.; Morrisey, E.E. Organoid Models: Assessing Lung Cell Fate Decisions and Disease Responses. Trends Mol. Med. 2021, 27, 1159–1174. [Google Scholar] [CrossRef] [PubMed]
- Evans, K.V.; Lee, J.-H. Alveolar Wars: The Rise of in Vitro Models to Understand Human Lung Alveolar Maintenance, Regeneration, and Disease. Stem Cells Transl. Med. 2020, 9, 867–881. [Google Scholar] [CrossRef] [PubMed]
- van der Vaart, J.; Clevers, H. Airway Organoids as Models of Human Disease. J. Intern. Med. 2021, 289, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Nabhan, A.N.; Brownfield, D.G.; Harbury, P.B.; Krasnow, M.A.; Desai, T.J. Single-Cell Wnt Signaling Niches Maintain Stemness of Alveolar Type 2 Cells. Science 2018, 359, 1118–1123. [Google Scholar] [CrossRef]
- Frank, D.B.; Peng, T.; Zepp, J.A.; Snitow, M.; Vincent, T.L.; Penkala, I.J.; Cui, Z.; Herriges, M.J.; Morley, M.P.; Zhou, S.; et al. Emergence of a Wave of Wnt Signaling That Regulates Lung Alveologenesis by Controlling Epithelial Self-Renewal and Differentiation. Cell Rep. 2016, 17, 2312–2325. [Google Scholar] [CrossRef]
- Ahmadvand, N.; Lingampally, A.; Khosravi, F.; Vazquez-Armendariz, A.I.; Rivetti, S.; Jones, M.R.; Wilhelm, J.; Herold, S.; Barreto, G.; Koepke, J.; et al. Fgfr2b Signaling Is Essential for the Maintenance of the Alveolar Epithelial Type 2 Lineage during Lung Homeostasis in Mice. Cell. Mol. Life Sci. 2022, 79, 302. [Google Scholar] [CrossRef] [PubMed]
- Chong, L.; Ahmadvand, N.; Noori, A.; Lv, Y.; Chen, C.; Bellusci, S.; Zhang, J.-S. Injury Activated Alveolar Progenitors (IAAPs): The Underdog of Lung Repair. Cell. Mol. Life Sci. 2023, 80, 145. [Google Scholar] [CrossRef]
- Liberti, D.C.; Kremp, M.M.; Liberti, W.A.; Penkala, I.J.; Li, S.; Zhou, S.; Morrisey, E.E. Alveolar Epithelial Cell Fate Is Maintained in a Spatially Restricted Manner to Promote Lung Regeneration after Acute Injury. Cell Rep. 2021, 35, 109092. [Google Scholar] [CrossRef] [PubMed]
- Brownfield, D.G.; de Arce, A.D.; Ghelfi, E.; Gillich, A.; Desai, T.J.; Krasnow, M.A. Alveolar Cell Fate Selection and Lifelong Maintenance of AT2 Cells by FGF Signaling. Nat. Commun. 2022, 13, 7137. [Google Scholar] [CrossRef]
- Yuan, T.; Volckaert, T.; Redente, E.F.; Hopkins, S.; Klinkhammer, K.; Wasnick, R.; Chao, C.-M.; Yuan, J.; Zhang, J.-S.; Yao, C.; et al. FGF10-FGFR2B Signaling Generates Basal Cells and Drives Alveolar Epithelial Regeneration by Bronchial Epithelial Stem Cells after Lung Injury. Stem Cell Rep. 2019, 12, 1041–1055. [Google Scholar] [CrossRef]
- Hein, R.F.C.; Wu, J.H.; Holloway, E.M.; Frum, T.; Conchola, A.S.; Tsai, Y.-H.; Wu, A.; Fine, A.S.; Miller, A.J.; Szenker-Ravi, E.; et al. R-SPONDIN2+ Mesenchymal Cells Form the Bud Tip Progenitor Niche during Human Lung Development. Dev. Cell 2022, 57, 1598–1614.e8. [Google Scholar] [CrossRef] [PubMed]
- Zepp, J.A.; Zacharias, W.J.; Frank, D.B.; Cavanaugh, C.A.; Zhou, S.; Morley, M.P.; Morrisey, E.E. Distinct Mesenchymal Lineages and Niches Promote Epithelial Self-Renewal and Myofibrogenesis in the Lung. Cell 2017, 170, 1134–1148.e10. [Google Scholar] [CrossRef]
- Chanda, D.; Rehan, M.; Smith, S.R.; Dsouza, K.G.; Wang, Y.; Bernard, K.; Kurundkar, D.; Memula, V.; Kojima, K.; Mobley, J.A.; et al. Mesenchymal Stromal Cell Aging Impairs the Self-Organizing Capacity of Lung Alveolar Epithelial Stem Cells. eLife 2021, 10, e68049. [Google Scholar] [CrossRef]
- Katsura, H.; Kobayashi, Y.; Tata, P.R.; Hogan, B.L.M. IL-1 and TNFα Contribute to the Inflammatory Niche to Enhance Alveolar Regeneration. Stem Cell Rep. 2019, 12, 657–666. [Google Scholar] [CrossRef]
- Konishi, S.; Tata, A.; Tata, P.R. Defined Conditions for Long-Term Expansion of Murine and Human Alveolar Epithelial Stem Cells in Three-Dimensional Cultures. STAR Protoc. 2022, 3, 101447. [Google Scholar] [CrossRef]


| Primary Antibody | Dilution | Secondary Antibody | Dilution |
|---|---|---|---|
| Rabbit α-Pro-SFTPC (Seven Hills Bioreagents, WRAB-9337, Cincinnati, OH, USA) | 1:800 | Donkey α-rabbit AF488 (Thermo Fisher Scientific, A-32790, Waltham, MA, USA) | 1:1000 |
| Mouse α-HOP (Santa Cruz, sc-398703, Santa Cruz, CA, USA) | 1:150 | Goat α-mouse AF647 (Abcam, ab150115, Cambridge, UK) | 1:500 |
| Rat α-KRT8 (Developmental Studies Hybridoma Bank (DSHB), TROMA-I-c, Iowa City, IA, USA) | 1:200 | Goat α-rat AF555 (Thermo Fisher Scientific, A-48263, Waltham, MA, USA) | 1:1000 |
| Gene Name | Forward Primer | Reverse Primer |
|---|---|---|
| Gapdh | 5′-CAT CAC TGC CAC CCA GAA GAC TG-3′ | 5′-ATG CCA GTG AGC TTC CCG TTC AG-3′ |
| Sftpc | 5′-GGT CCT GAT GGA GAG TCC AC-3′ | 5′-GAT GAG AAG GCG TTT GAG GT-3′ |
| Ager | 5′-GCC ACT GGA ATT GTC GAT GAG G-3′ | 5′-GCT GTG AGT TCA GAG GCA GGA T-3′ |
| Study | Mouse Strain | Epithelial Cells | Mesenchymal Cells | Epi:Mes Ratio | Set Up | CFE | Research Question |
|---|---|---|---|---|---|---|---|
| [10] | SftpcCre-ERT2; tdTomatoflox and PdgfraH2B-GFP | 5 × 103 SFTPC+ cells | 1 × 105 PDGFRαhigh cells (also shown to be LipidTOX+) | 1:20 | ALI in Matrigel (mixed 1:1) for 16–17 days | 2.3% ± 0.3% | Self-renewal and differentiation of AEC2s |
| [30] | SftpcCre-ERT2; EYFPflox and Axin2Cre-ERT2; tdTomatoflox | 5 × 103 EYFP+ or tdTom+ cells | 5 × 104 primary adult lung fibroblasts | 1:10 | ALI in Matrigel (mixed 1:1 with MTEC-SAGM) for 14 days | ~1.7% with total AEC2s versus ~2.6% with AXIN2+ AEC2s before passaging | Comparison between bulk and WNT-responsive AEC2s |
| [7] | SftpcCre-ERT2; tdTomatoflox, Lgr5Cre-ERT2; tdTomatoflox and Lgr6EGFP-Cre-ERT2; tdTomatoflox | 0.5–1 × 104 SFTPC+ cells | 0.5–1 × 105 LGR5+ or LGR6+ cells | 1:10 | ALI in Matrigel (mixed 1:1) for 14 days | ~4.8% with LGR5+ cells versus ~2.5% with LGR6+ cells before passaging | Comparison between LGR5+ and LGR6+ cells in terms of supporting bronchial, alveolar, and bronchioalveolar organoids |
| [7] | Scgb1a1Cre-ERT; YFPflox and Lgr5Cre-ERT2; EYFPflox * | 0.5–1 × 104 SCGB1A1+ cells | 0.5–1 × 105 LGR5+ cells | 1:10 | ALI in Matrigel (mixed 1:1) for 14 days | ~2.6% with most organoids being alveolospheres | Comparison between LGR5+ and LGR6+ cells in terms of supporting bronchial, alveolar, and bronchioalveolar organoids |
| [37] | SftpcCre-ERT2; EYFPflox, Axin2Cre-ERT2; EYFPflox, Wnt2Cre-ERT2; EYFPflox and PdgfraH2B-GFP | 5 × 103 SFTPC+ cells | 5 × 104 cells | 1:10 | ALI in Matrigel (mixed 1:1) for 21 days | Between ~1.2% and ~6.2% | Comparison between the alveolosphere-supportive ability of various mesenchymal subsets |
| [20] | SftpcCre-ERT2; tdTomatoflox and PdgfraH2B-GFP | 5 × 103 SFTPC+ cells | 5 × 104 cells | 1:10 | ALI in Matrigel (mixed 1:1) for 14 days | Up to ~9% | Investigating BMP/SMAD signaling in AEC2 renewal and differentiation |
| [12] | SftpcCre-ERT2; tdTomatoflox | 5 × 103 SFTPC+ cells | Cultured lung stromal cells | 1:5 | ALI in Matrigel (mixed 1:1) for 14 days | Between ~1.6% (without IL-1β) and ~3% (with IL-1β) | Investigate the effect of IL-1β stimulation on AEC2 self-renewal, transitional state, and differentiation |
| [16] | SftpcCre-ERT2; tdTomatoflox | 1 × 103 SFTPC+ cells | 2 × 104 rMCs (EpCAM− CD45− CD31− Sca-1+) from C57BL/6 lungs | 1:20 | ALI in Matrigel (mixed 1:1) for 14 days | ~1.2% | Assess the ability of AEC2 subsets to form alveolospheres |
| [38] | C57BL/6 | 5 × 103 AEC2 isolated via enzymatic digestion (Dispase II), cell-specific antibody labeling (biotinylated Ter-119, CD104, CD16/32, CD45, CD31), and magnetic separation (Anti-Biotin MicroBeads) | 1 × 105 lung mesenchymal stromal cells (L-MSCs) isolated by differential adhesion | 1:20 | ALI in Matrigel (mixed 1:1) for 9–12 days | Not directly stated by can be estimated to be ~0.6% | Assess the effect of aging on alveolosphere formation |
| [33] | SftpcCre-ERT2; tdTomatoflox and SftpcCre-ERT2; Fgfr2bflox/flox; tdTomatoflox | 5 × 103 SFTPC+ cells | 5 × 104 primary adult lung fibroblasts | 1:10 | ALI in 1:1 mixture of MTEC-SAGM:Matrigel (mixed 1:1) for 14 days | Between ~9% and ~11% | Assessing the role of FGFR2b signaling on alveolosphere formation |
| [18] | SftpcCre-ERT2; tdTomatoflox, Fgf10-lacZ, ob/ob and C57BL/6 | 5 × 103 EpCAM+ Lysotracker+ tdTom+ | 5 × 104 rMCs (EpCAM− CD45− CD31− Sca-1+) or EpCAM− CD45− CD31− Sca-1− cells; LipidTOX+ rMCs or LipidTOX− rMCs; FGF10+ rMCs/FGF10− rMCs; ob/ob rMCs from C57BL/6 lungs | 1:10 | ALI in Matrigel (mixed 1:1) for 14 days | Between ~2.2% and ~6.2% | Comparison of niche activity between various mesenchymal subsets |
| [39] | SftpcCre-ERT2; tdTomatoflox and C57BL/6 | 2–5 × 103 tdTom+ cells or Lysotracker+ EpCAM+ cells | 5 × 104 PDGFRα+ fibroblasts | 1:10 to 1:25 | Matrigel droplets (mixed 1:1) for 10–15 days | Between ~8% and 9% with tdTom+ cells | Assessing the impact of inflammation on alveolar regeneration |
| [40] | C57BL/6 | 2–3 × 103 CD45− CD31− EpCAM+ Lysotacker+ cells | None | N/A | Matrigel droplets (mixed 1:1) for 10–12 days | Information not available | Protocol to generate feeder-free alveolospheres ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zabihi, M.; Khadim, A.; Schäfer, T.M.; Alexopoulos, I.; Bartkuhn, M.; El Agha, E.; Vazquez-Armendariz, A.I.; Herold, S. An Optimized Protocol for the Generation of Alveolospheres from Wild-Type Mice. Cells 2024, 13, 922. https://doi.org/10.3390/cells13110922
Zabihi M, Khadim A, Schäfer TM, Alexopoulos I, Bartkuhn M, El Agha E, Vazquez-Armendariz AI, Herold S. An Optimized Protocol for the Generation of Alveolospheres from Wild-Type Mice. Cells. 2024; 13(11):922. https://doi.org/10.3390/cells13110922
Chicago/Turabian StyleZabihi, Mahsa, Ali Khadim, Theresa M. Schäfer, Ioannis Alexopoulos, Marek Bartkuhn, Elie El Agha, Ana I. Vazquez-Armendariz, and Susanne Herold. 2024. "An Optimized Protocol for the Generation of Alveolospheres from Wild-Type Mice" Cells 13, no. 11: 922. https://doi.org/10.3390/cells13110922
APA StyleZabihi, M., Khadim, A., Schäfer, T. M., Alexopoulos, I., Bartkuhn, M., El Agha, E., Vazquez-Armendariz, A. I., & Herold, S. (2024). An Optimized Protocol for the Generation of Alveolospheres from Wild-Type Mice. Cells, 13(11), 922. https://doi.org/10.3390/cells13110922







