Altered Extracellular Vesicle-Derived Protein and microRNA Signatures in Bronchoalveolar Lavage Fluid from Patients with Chronic Obstructive Pulmonary Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Collection and Processing of Bronchoalveolar Lavage
2.3. Isolation of EVs from BAL Fluid
2.4. Cryo-TEM Imaging
2.5. Protein Concentration Measurement
2.6. Untargeted Proteomics Analysis
2.7. RNA Isolation
2.8. Small RNA Sequencing
2.9. Statistical Analysis
2.9.1. Proteomics Analysis
2.9.2. Ingenuity Pathway Analysis
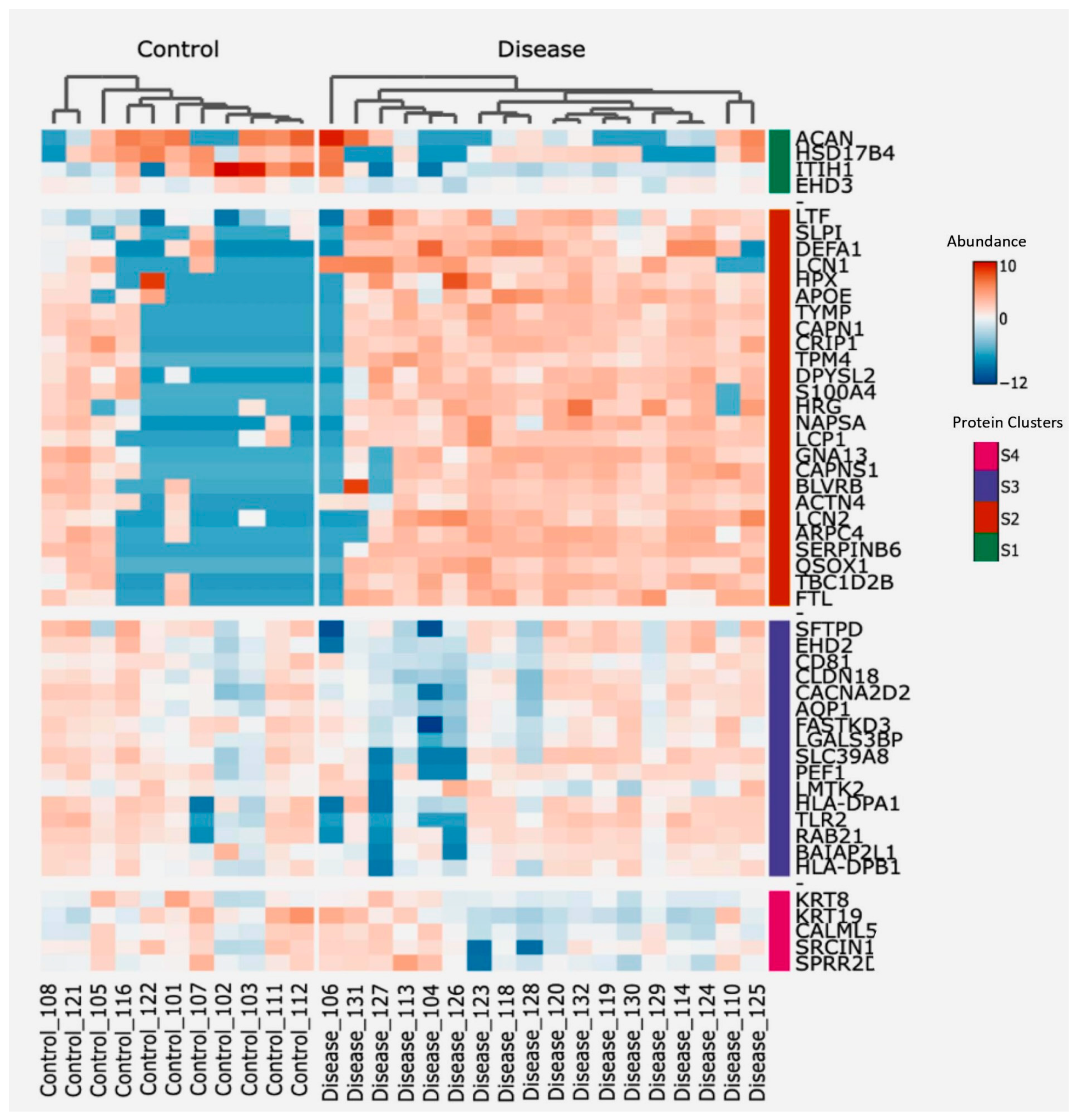
2.9.3. Protein Clustering and Functional Annotation
2.9.4. Correlations of Protein Abundance with Macrophage and Neutrophil Cell Number in BALF
2.9.5. microRNA Expression Profiling
3. Results
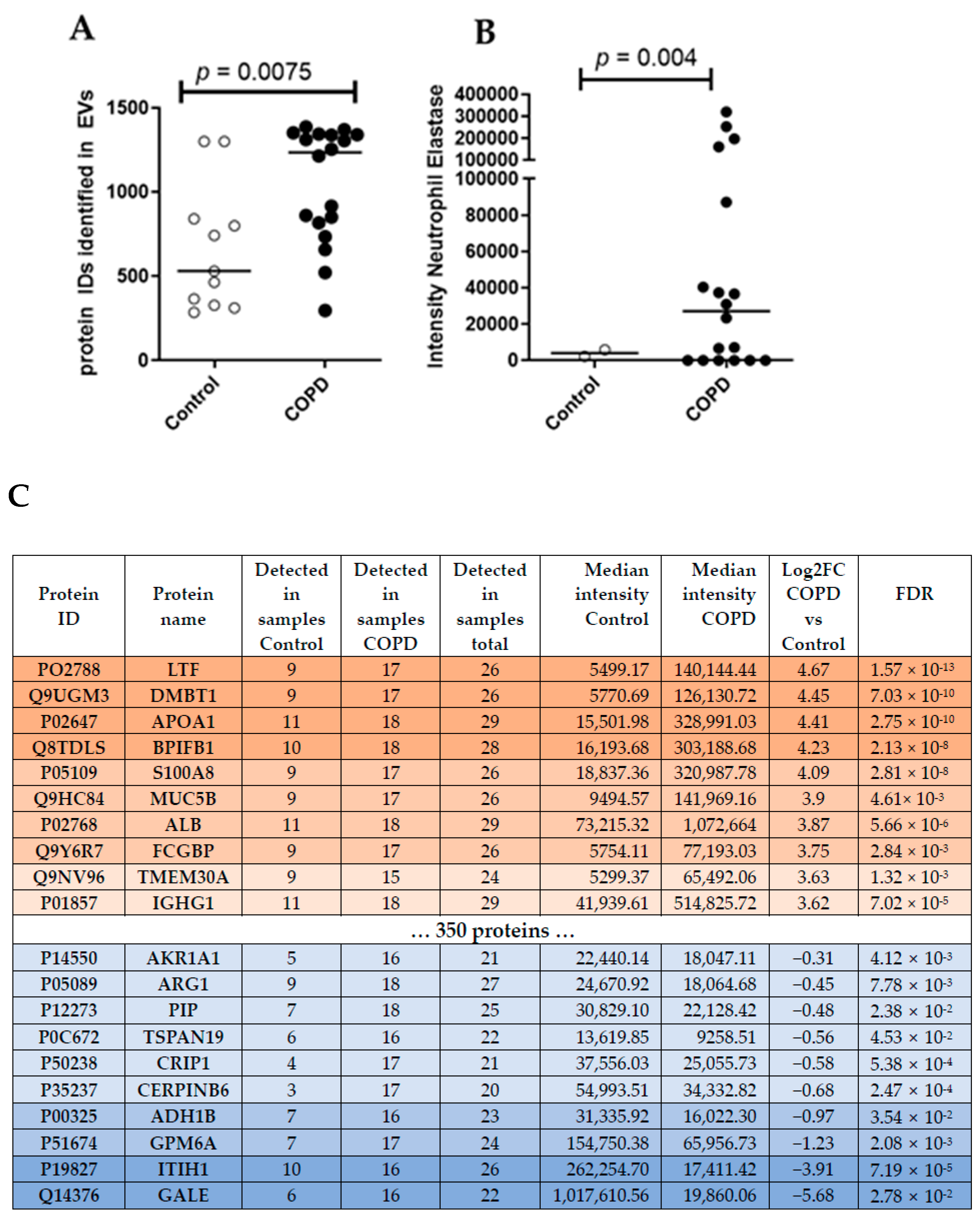
3.1. Small EVs Can Successfully Be Isolated from Frozen BALF Samples
3.2. Proteomics Analysis of BALF EVs
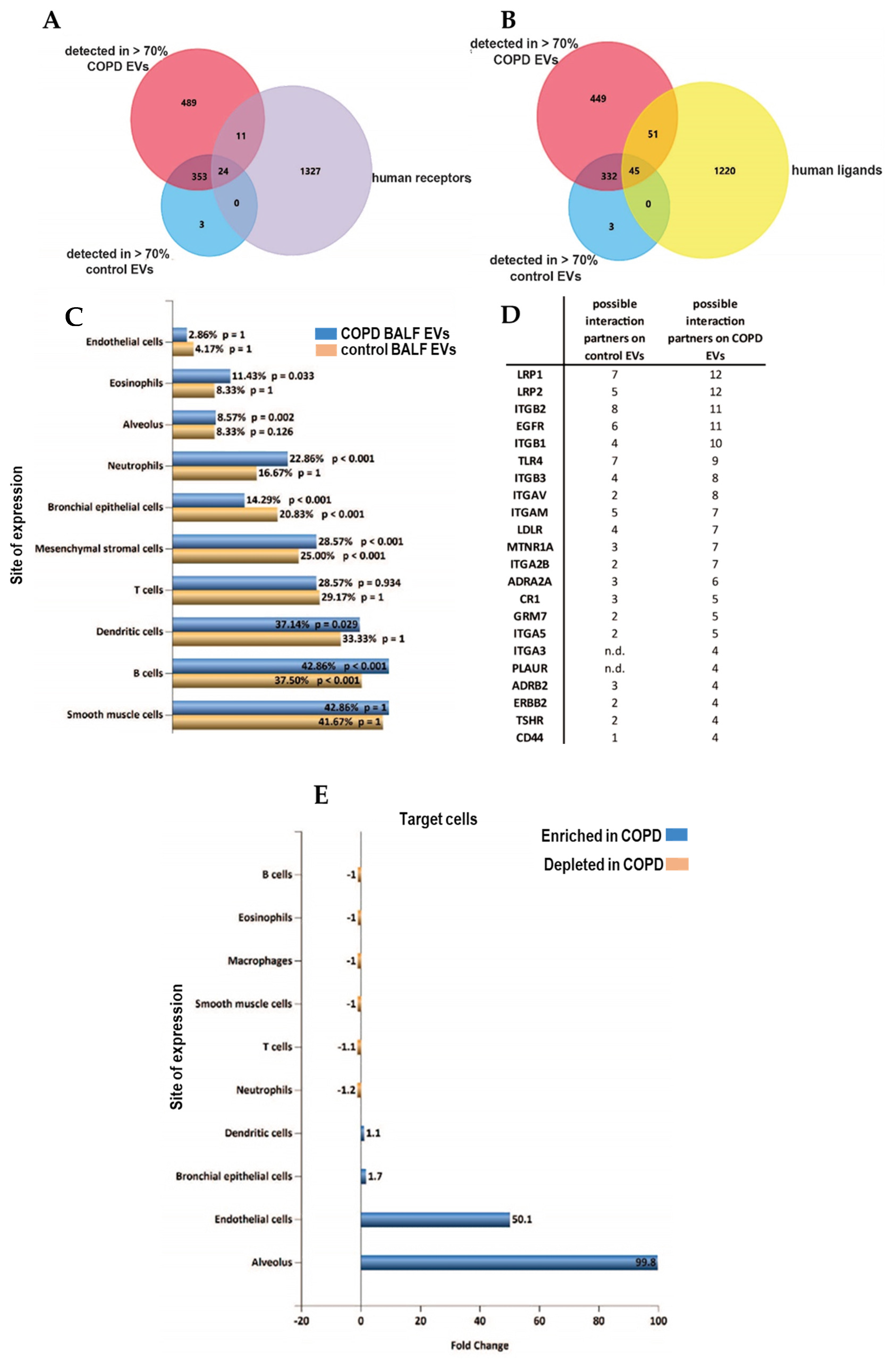
3.3. Altered Receptor/Ligand Pattern on BALF EVs in COPD
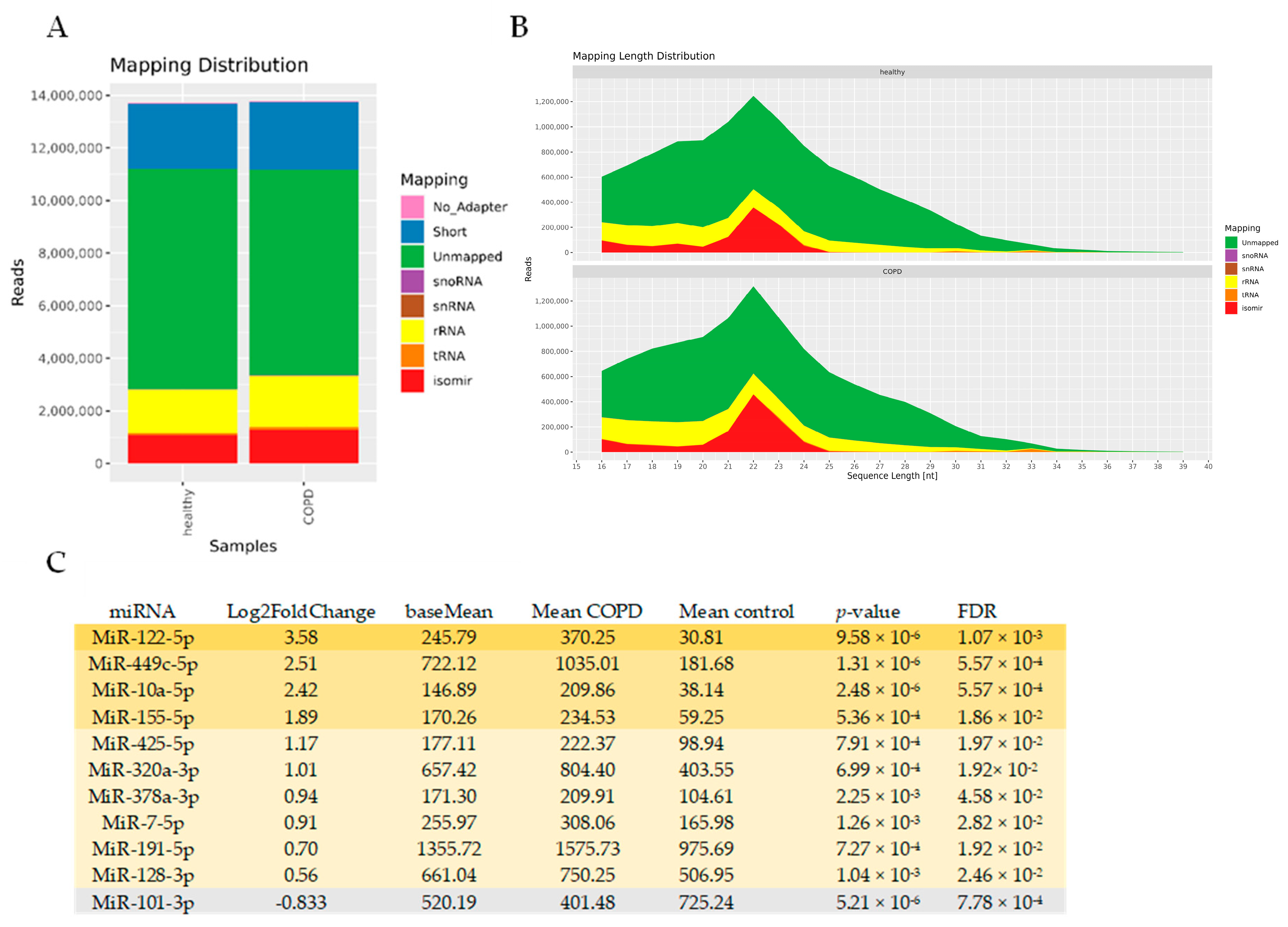
3.4. miRNAseq Analysis of BALF EVs
4. Discussion
5. Summary and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cosio, M.G.; Saetta, M.; Agusti, A. Immunologic Aspects of Chronic Obstructive Pulmonary Disease. N. Engl. J. Med. 2009, 360, 2445–2454. [Google Scholar] [CrossRef]
- Sundar, I.K.; Li, D.; Rahman, I. Small RNA-sequence analysis of plasma-derived extracellular vesicle miRNAs in smokers and patients with chronic obstructive pulmonary disease as circulating biomarkers. J. Extracell. Vesicles 2019, 8, 1684816. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Gutiérrez-Vázquez, C.; Villarroya-Beltri, C.; González, S.; Sánchez-Cabo, F.; González, M.Á.; Bernad, A.; Sánchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef]
- French, K.C.; Antonyak, M.A.; Cerione, R.A. Extracellular vesicle docking at the cellular port: Extracellular vesicle binding and uptake. Semin. Cell Dev. Biol. 2017, 67, 48–55. [Google Scholar] [CrossRef]
- Bartel, S.; Deshane, J.; Wilkinson, T.; Gabrielsson, S. Extracellular Vesicles as Mediators of Cellular Cross Talk in the Lung Microenvironment. Front. Med. 2020, 7, 326. [Google Scholar] [CrossRef]
- Genschmer, K.R.; Russell, D.W.; Lal, C.; Szul, T.; Bratcher, P.E.; Noerager, B.D.; Abdul Roda, M.; Xu, X.; Rezonzew, G.; Viera, L.; et al. Activated PMN Exosomes: Pathogenic Entities Causing Matrix Destruction and Disease in the Lung. Cell 2019, 176, 113–126. [Google Scholar] [CrossRef]
- Monsel, A.; Zhu, Y.G.; Gudapati, V.; Lim, H.; Lee, J.W. Mesenchymal stem cell derived secretome and extracellular vesicles for acute lung injury and other inflammatory lung diseases. Expert Opin. Biol. Ther. 2016, 16, 859–871. [Google Scholar] [CrossRef]
- Rutgers, S.R.; Timens, W.; Kaufmann, H.F.; van der Mark, T.W.; Koëter, G.H.; Postma, D.S. Comparison of induced sputum with bronchial wash, bronchoalveolar lavage and bronchial biopsies in COPD. Eur. Respir. J. 2000, 15, 109–115. [Google Scholar] [CrossRef]
- Wegrzyn, A.B.; Herzog, K.; Gerding, A.; Kwiatkowski, M.; Wolters, J.C.; Dolga, A.M.; van Lint, A.E.M.; Wanders, R.J.A.; Waterham, H.R.; Bakker, B.M. Fibroblast-specific genome-scale modelling predicts an imbalance in amino acid metabolism in Refsum disease. FEBS J. 2020, 287, 5096–5113. [Google Scholar] [CrossRef]
- Bahmer, T.; Krauss-Etschmann, S.; Buschmann, D.; Behrends, J.; Watz, H.; Kirsten, A.M.; Pedersen, F.; Waschki, B.; Fuchs, O.; Pfaffl, M.W.; et al. RNA-seq–based profiling of extracellular vesicles in plasma reveals a potential role of miR-122-5p in asthma. Allergy Eur. J. Allergy Clin. Immunol. 2021, 76, 366–371. [Google Scholar] [CrossRef]
- Akhmedov, M.; Martinelli, A.; Geiger, R.; Kwee, I. Omics Playground: A comprehensive self-service platform for visualization, analytics and exploration of Big Omics Data. NAR Genom. Bioinform. 2020, 2, lqz019. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y. Btrim: A fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 2011, 98, 152–153. [Google Scholar] [CrossRef] [PubMed]
- RNAcentral Consortium. RNAcentral 2021: Secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 2021, 8, D212–D220. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2011; Available online: http://www.R-project.org/ (accessed on 5 September 2021).
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Stam, J.; Bartel, S.; Bischoff, R.; Wolters, J.C. Isolation of extracellular vesicles with combined enrichment methods. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2021, 1169, 122604. [Google Scholar] [CrossRef]
- Shao, X.; Liao, J.; Li, C.; Lu, X.; Cheng, J.; Fan, X. CellTalkDB: A manually curated database of ligand–receptor interactions in humans and mice. Brief. Bioinform. 2021, 22, bbaa269. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Dubin, R.F.; Robinson, S.K.; Widdicombe, J.H. Secretion of lactoferrin and lysozyme by cultures of human airway epithelium. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2004, 286, L750–L755. [Google Scholar] [CrossRef]
- Thompson, A.B.; Bohling, T.; Payvandi, F.; Rennard, S.I. Lower respiratory tract lactoferrin and lysozyme arise primarily in the airways and are elevated in association with chronic bronchitis. J. Lab. Clin. Med. 1990, 115, 148–158. [Google Scholar]
- Pierce, L.M.; Kurata, W.E. Priming With Toll-Like Receptor 3 Agonist Poly(I:C) Enhances Content of Innate Immune Defense Proteins but Not MicroRNAs in Human Mesenchymal Stem Cell-Derived Extracellular Vesicles. Front. Cell Dev. Biol. 2021, 9, 676356. [Google Scholar] [CrossRef]
- Müller, H.; Schmiedl, A.; Weiss, C.; Ai, M.; Jung, S.; Renner, M. DMBT1 is upregulated in lung epithelial cells after hypoxia and changes surfactant ultrastructure. Pediatr. Pulmonol. 2020, 55, 2964–2969. [Google Scholar] [CrossRef]
- Müller, H.; Nagel, C.; Weiss, C.; Mollenhauer, J.; Poeschl, J. Deleted in malignant brain tumors 1 (DMBT1) elicits increased VEGF and decreased IL-6 production in type II lung epithelial cells. BMC Pulm. Med. 2015, 15, 32. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Rao, S.S.; Ren, L.; Hu, X.K.; Tan, Y.J.; Hu, Y.; Luo, J.; Liu, Y.W.; Yin, H.; Huang, J.; et al. Exosomal DMBT1 from human urine-derived stem cells facilitates diabetic wound repair by promoting angiogenesis. Theranostics 2018, 8, 1607–1623. [Google Scholar] [CrossRef] [PubMed]
- Tóth, E.Á.; Turiák, L.; Visnovitz, T.; Cserép, C.; Mázló, A.; Sódar, B.W.; Försönits, A.I.; Petővári, G.; Sebestyén, A.; Komlósi, Z.; et al. Formation of a protein corona on the surface of extracellular vesicles in blood plasma. J. Extracell. Vesicles 2021, 10, e12140. [Google Scholar] [CrossRef] [PubMed]
- Titz, B.; Sewer, A.; Schneider, T.; Elamin, A.; Martin, F.; Dijon, S.; Luettich, K.; Guedj, E.; Vuillaume, G.; Ivanov, N.V.; et al. Alterations in the sputum proteome and transcriptome in smokers and early-stage COPD subjects. J. Proteom. 2015, 128, 306–320. [Google Scholar] [CrossRef]
- Kim, C.; Lee, J.M.; Park, S.W.; Kim, K.S.; Lee, M.W.; Paik, S.; Jang, A.S.; Kim, D.J.; Uh, S.; Kim, Y.; et al. Attenuation of cigarette smoke-induced emphysema in mice by apolipoprotein A-1 overexpression. Am. J. Respir. Cell Mol. Biol. 2016, 54, 91–102. [Google Scholar] [CrossRef]
- Monteleone, M.C.; Billi, S.C.; Brocco, M.A.; Frasch, A.C. Neural glycoprotein M6a is released in extracellular vesicles and modulated by chronic stressors in blood. Sci. Rep. 2017, 7, 9788. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, S.; DeLeo, A.M.; Sethi, M.K.; Yukawa-Takamatsu, K.; Yang, Z.; Ko, J.; Hogan, J.D.; Ruan, Z.; You, Y.; Wang, Y.; et al. Proteomic and biological profiling of extracellular vesicles from Alzheimer’s disease human brain tissues. Alzheimer’s Dement. 2020, 16, 896–907. [Google Scholar] [CrossRef] [PubMed]
- The Human Protein Atlas [Internet]. Available online: https://www.proteinatlas.org/ (accessed on 4 November 2021).
- Kaur, G.; Maremanda, K.P.; Campos, M.; Chand, H.S.; Li, F.; Hirani, N.; Haseeb, M.A.; Li, D.; Rahman, I. Distinct exosomal mirna profiles from balf and lung tissue of copd and ipf patients. Int. J. Mol. Sci. 2021, 22, 11830. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Zhou, S.; Xu, A.; Sun, L.; Li, M.; Jiang, H.; Zhang, B.; Zeng, D.; Fei, G.; Wang, R. Microbiota Imbalance Contributes to COPD Deterioration by Enhancing IL-17a Production via miR-122 and miR-30a. Mol. Ther.-Nucleic Acids 2020, 22, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Cerón-Pisa, N.; Iglesias, A.; Shafiek, H.; Martín-Medina, A.; Esteva-Socias, M.; Muncunill, J.; Fleischer, A.; Verdú, J.; Cosío, B.G.; Sauleda, J. Hsa-Mir-320c, Hsa-Mir-200c-3p, and Hsa-Mir-449c-5p as Potential Specific miRNA Biomarkers of COPD: A Pilot Study. Pathophysiology 2022, 29, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.J.; Tsai, Y.C.; Chang, W.A.; Lin, Y.S.; Tsai, P.H.; Sheu, C.C.; Kuo, P.L.; Hsu, Y.L. Deducting microRNA-mediated changes common in bronchial epithelial cells of asthma and chronic obstructive pulmonary disease—A next-generation sequencing-guided bioinformatic approach. Int. J. Mol. Sci. 2019, 20, 553. [Google Scholar] [CrossRef] [PubMed]
- De Smet, E.G.; Van Eeckhoutte, H.P.; Avila Cobos, F.; Blomme, E.; Verhamme, F.M.; Provoost, S.; Verleden, S.E.; Venken, K.; Maes, T.; Joos, G.F.; et al. The role of miR-155 in cigarette smoke-induced pulmonary inflammation and COPD. Mucosal Immunol. 2020, 13, 423–436. [Google Scholar] [CrossRef]
- Xu, G.; Xia, Z.; Deng, F.; Liu, L.; Wang, Q.; Yu, Y.; Wang, F.; Zhu, C.; Liu, W.; Cheng, Z.; et al. Inducible LGALS3BP/90K activates antiviral innate immune responses by targeting TRAF6 and TRAF3 complex. PLoS Pathog. 2019, 15, e1008002. [Google Scholar] [CrossRef] [PubMed]
- Quan, Y.; Kan, B.; Yang, B. Aquaporins in Immune System. Adv. Exp. Med. Biol. 2023, 398, 195–202. [Google Scholar] [CrossRef]
- Tyteca, D.; Nishino, T.; Debaix, H.; Van Der Smissen, P.; N’Kuli, F.; Hoffmann, D.; Cnops, Y.; Rabolli, V.; van Loo, G.; Beyaert, R.; et al. Regulation of macrophage motility by the water channel aquaporin-1: Crucial role of M0/M2 phenotype switch. PLoS ONE 2015, 26, e0117398. [Google Scholar] [CrossRef]
- Chan, Y.R.; Liu, J.S.; Pociask, D.A.; Zheng, M.; Mietzner, T.A.; Berger, T.; Mak, T.W.; Clifton, M.C.; Strong, R.K.; Ray, P.; et al. Lipocalin 2 is required for pulmonary host defense against Klebsiella infection. J. Immunol. 2009, 182, 4947–4956. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Chen, S.; Hou, C.; Lin, J.; Xiong, W.; Shen, Y.; Zhou, T. Multi-omics analysis unravels dysregulated lysosomal function and lipid metabolism involved in sub-chronic particulate matter-induced pulmonary injury. Sci. Total Environ. 2022, 836, 155642. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Zieren, R.C.; Horie, K.; Kim, C.J.; Mallick, E.; Jing, Y.; Feng, M.; Kuczler, M.D.; Green, J.; Amend, S.R.; et al. Comprehensive evaluation of methods for small extracellular vesicles separation from human plasma, urine and cell culture medium. J. Extracell. Vesicles 2020, 10, e12044. [Google Scholar] [CrossRef] [PubMed]
- Mizenko, R.R.; Brostoff, T.; Rojalin, T.; Koster, H.J.; Swindell, H.S.; Leiserowitz, G.S.; Wang, A.; Carney, R.P. Tetraspanins are unevenly distributed across single extracellular vesicles and bias sensitivity to multiplexed cancer biomarkers. J. Nanobiotechnol. 2021, 19, 250. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; Hewapathirana, S.; García-Seisdedos, D.; Kamatchinathan, S.; Kundu, D.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A Hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]


| COPD (n = 18) | Controls (n-11) | |
|---|---|---|
| Age (years) | 62 ± 8 | 58 ± 8 |
| Female/Male | 4/14 | 3/8 |
| Smoking status–ex-smokers (%) | 84 | 100 |
| Pack years | 28 ± 22 | 25 ± 15 |
| FEV1 (% pred) * | 59 ± 13 | 104 ± 11 |
| FEV1/FVC * | 66 ± 11 | 97 ± 8 |
| Bronchoalveolar lavage wash | 18 | 11 |
| Concentration of cells in lavage fluid | ||
| Total cell count per mL | 0.061 (0.01–0.6) | 0.10 (0.03–0.2) |
| Macrophages (%) | 86 (13.3–94.2) | 92 (70.4–96.5) |
| Neutrophils (%) | 1.7 (0.3–85.5) | 1.1 (0.1–3.0) |
| Lymphocytes (%) | 6.9 (1.3–26.4) | 7.1 (2.5–26.5) |
| Eosinophils (%) | 0.4 (0–1.7) | 0.2 (0–0.45) |
| MISEV Category | Protein | Detected in Control Samples (Median Intensity) | Detected in COPD Samples (Median Intensity) |
|---|---|---|---|
| 1—transmembrane or GPI-anchored proteins | |||
| 1a—non-tissue specific | CD63 | 11/11 (15,509.93) | 18/18 (92,003.89) |
| CD81 | 11/11 (94,397.15) | 18/18 (126,942.64) | |
| CD82 | 10/11 (13,925.97) | 18/18 (45,828.48) | |
| CD47 | 8/11 (30,198.34) | 17/18 (128,907.36) | |
| GNA | 0/11 | 0/18 | |
| HLA-A | 9/11 (23,105.60) | 15/18 (66,913.24) | |
| H2-K/D/Q | 0/11 | 0/18 | |
| ITGA3 | 3/11 (16,334.62) | 15/18 (17,816.25) | |
| ITGB2 | 5/11 (28,874.69) | 16/18 (43,729.40) | |
| TFR2 | 0/11 | 0/18 | |
| LAMP1 | 3/11 (31,599.47) | 15/18 (28,388.38) | |
| SDC | 0/11 | 0/18 | |
| BSG | 0/11 | 0/18 | |
| ADAM10 | 4/11 (24,082.33) | 14/18 (47,027.64) | |
| CD73 | 0/11 | 0/18 | |
| CD55 | 11/11 (29,733.75) | 18/18 (162,760.10) | |
| CD59 | 11/11 (109,809.55) | 18/18 (363,903.25) | |
| 1b—tissue specific | CD9 | 11/11 (185,367.57) | 18/18 (612,304.21) |
| EPCAM | 5/11 (9394.25) | 11/18 (9267.61) | |
| TSPAN8 | 8/11 (13,506.14) | 17/18 (27,948.92) | |
| CD14 | 3/11 (18,100.96) | 14/18 (20,410.54) | |
| 2—cytosolic proteins recovered in EVs | |||
| 2a—lipid or membrane-binding ability | ESCRT-I/II/II | 0/11 | 0/18 |
| TSG101 | 11/11 (30,495.01) | 18/18 (68,322.18) | |
| ALIX (PDCD6IP) | 11/11 (26,746.72) | 18/18 (134,175.34) | |
| ARRDC1 | 8/11 (22,419.05) | 17/18 (114,076.60) | |
| FLOT1 | 7/11 (10,344.42) | 16/18 (35,016.16) | |
| CAV | 0/11 | 0/18 | |
| ANXA1 | 11/11 (49,761.63) | 18/18 (406,865.34) | |
| HSC70 | 0/11 | 0/18 | |
| HSP84 | 0/11 | 0/18 | |
| ARF6 | 2/11 (45,850.05) | 13/18 (38,333.01) | |
| SDCBP | 11/11 (49,047.33) | 18/18 (322,430.85) | |
| MAPT | 0/11 | 0/18 | |
| 2b—promiscuous incorporation into EVs | HSP70 | 0/11 | 0/18 |
| ACT | 0/11 | 0/18 | |
| TUB | 0/11 | 0/18 | |
| GAPDH | 11/11 (26,518.46) | 18/18 (87,466.69) | |
| 3—major components of non-EV co-isolated structures | |||
| 3a—lipoproteins | APOA1 | 11/11 (15,501.97) | 18/18 (328,991.03) |
| APOB | 8/11 (81,382.46) | 17/18 (19,999.87) | |
| APOB100 | 0/11 | 0/18 | |
| ALB | 11/11 (73,215.32) | 18/18 (1,072,664.00) | |
| 3b—protein–protein/nucleic acid aggregates | UMOD | 0/11 | 0/18 |
| 4—Transmembrane, lipid-bound soluble proteins associated with other intracellular compartments than PM/endosomes | |||
| 4a—nucleus | HIST1HC | 5/11 (16,701.19) | 13/18 (27,412.26) |
| LMNA | 4/11 (9030.91) | 14/18 (21,075.30) | |
| 4b—mitochondria | IMMT | 0/11 | 0/18 |
| CYC1 | 0/11 | 0/18 | |
| TOMM20 | 0/11 | 0/18 | |
| 4c—secretory pathway (endoplasmatic riticulum, Golgi apparatus) | CANX | 4/11 (1663.54) | 6/18 (16,269.11) |
| HSP90B1 | 3/11 (3891.60) | 8/18 (8363.01) | |
| BIP | 0/11 | 0/18 | |
| GM130 | 0/11 | 0/18 | |
| 4d—autophagosome, cytoskeleton | ATG9A | 0/11 | 0/18 |
| ACTN1 | 6/11 (14,611.46) | 17/18 (42,763.41) | |
| KRT18 | 0/11 | 0/18 | |
| 5—Secreted proteins recovered with EVs | |||
| 5a—cytokines and growth factors | TGFB1/2 | 0/11 | 0/18 |
| IFNG | 0/11 | 0/18 | |
| VEGFA | 0/11 | 0/18 | |
| FGF1/2 | 0/11 | 0/18 | |
| PDGF | 0/11 | 0/18 | |
| EGF | 0/11 | 0/18 | |
| IL | 0/11 | 0/18 | |
| 5b—adhesion and extracellular matrix proteins | FN1 | 4/11 (27,088.45) | 16/18 (19,794.65) |
| COL12A1 | 0/11 | 9/18 (12,138.44) | |
| MFGE8 | 0/11 | 0/18 | |
| LGAL3BP | 0/11 | 0/18 | |
| CD5L | 6/11 (10,115.46) | 16/18 (16,718.95) | |
| AHSG | 5/11 (17,509.83) | 16/18 (27,092.54) | |
| Annotations | S1 | S2 | S3 | S4 |
|---|---|---|---|---|
| GO_POSITIVE_REGULATION_OF_IMMUNE_SYSTEM_PROCESS | 0 | 0.44 | 0.36 | 0 |
| GO_REGULATION_OF_IMMUNE_RESPONSE | 0 | 0.41 | 0.37 | 0 |
| Innate Immune System_Homo sapiens_R-HSA-168249 | 0 | 0.34 | 0.17 | 0 |
| GO_INNATE_IMMUNE_RESPONSE | 0 | 0.53 | 0.31 | 0 |
| GO_POSITIVE_REGULATION_OF_IMMUNE_RESPONSE | 0 | 0.43 | 0.32 | 0 |
| GO_IMMUNE_EFFECTOR_PROCESS | 0 | 0.51 | 0 | 0 |
| neutrophil activation involved in immune response (GO_000228) | 0 | 0.53 | 0.62 | −0.27 |
| GO_ACTIVATION_OF_IMMUNE_RESPONSE | 0 | 0.23 | 0.36 | 0 |
| GO_REGULATION_OF_IMMUNE_EFFECTOR_PROCESS | 0 | 0.30 | 0 | 0 |
| GO_NEGATIVE_REGULATION_OF_IMMUNE_SYSTEM_PROCESS | 0 | 0.34 | 0.34 | 0 |
| GO_REGULATION_OF_INNATE_IMMUNE_RESPONSE | 0 | 0.48 | 0.24 | 0 |
| GO_ADAPTIVE_IMMUNE_RESPONSE | 0 | 0 | 0 | 0 |
| GO_POSITIVE_REGULATION_OF_INNATE_IMMUNE_RESPONSE | 0 | 0.43 | 0.29 | 0 |
| Hepatitis, Autoimmune C0241910 mouse GSE867 sample 230 (down) | 0 | 0.56 | 0 | 0 |
| GO_ACTIVATION_OF_INNATE_IMMUNE_RESPONSE | 0 | 0 | 0.35 | 0 |
| GO_HUMORAL_IMMUNE_RESPONSE | 0 | 0.67 | 0 | 0 |
| innate immune response activating cell surface receptor signaling pathway. | 0 | 0 | 0 | 0 |
| Parameter | Protein | Correlation | p-Value |
|---|---|---|---|
| Lung function | |||
| Controls | LCN2 | 0.60 | 0.03 |
| COPD | APOE | −0.45 | 0.05 |
| LTF | −0.50 | 0.04 | |
| Macrophages in BALF | |||
| Controls | LGALS3BP | −0.60 | 0.03 |
| BAIAP2L1 | −0.67 | 0.02 | |
| COPD | APOE | −0.49 | 0.04 |
| AQP1 | 0.51 | 0.03 | |
| Neutrophils in BALF | |||
| Controls | - | - | - |
| COPD | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartel, S.; Wolters, J.C.; Noor, H.; Rafie, K.; Fang, J.; Kirchner, B.; Nolte-′t Hoen, E.; Pfaffl, M.W.; Rutgers, S.; Timens, W.; et al. Altered Extracellular Vesicle-Derived Protein and microRNA Signatures in Bronchoalveolar Lavage Fluid from Patients with Chronic Obstructive Pulmonary Disease. Cells 2024, 13, 945. https://doi.org/10.3390/cells13110945
Bartel S, Wolters JC, Noor H, Rafie K, Fang J, Kirchner B, Nolte-′t Hoen E, Pfaffl MW, Rutgers S, Timens W, et al. Altered Extracellular Vesicle-Derived Protein and microRNA Signatures in Bronchoalveolar Lavage Fluid from Patients with Chronic Obstructive Pulmonary Disease. Cells. 2024; 13(11):945. https://doi.org/10.3390/cells13110945
Chicago/Turabian StyleBartel, Sabine, Justina C. Wolters, Hasnat Noor, Karim Rafie, Jiahua Fang, Benedikt Kirchner, Esther Nolte-′t Hoen, Michael W. Pfaffl, Steven Rutgers, Wim Timens, and et al. 2024. "Altered Extracellular Vesicle-Derived Protein and microRNA Signatures in Bronchoalveolar Lavage Fluid from Patients with Chronic Obstructive Pulmonary Disease" Cells 13, no. 11: 945. https://doi.org/10.3390/cells13110945
APA StyleBartel, S., Wolters, J. C., Noor, H., Rafie, K., Fang, J., Kirchner, B., Nolte-′t Hoen, E., Pfaffl, M. W., Rutgers, S., Timens, W., van den Berge, M., & Hylkema, M. N. (2024). Altered Extracellular Vesicle-Derived Protein and microRNA Signatures in Bronchoalveolar Lavage Fluid from Patients with Chronic Obstructive Pulmonary Disease. Cells, 13(11), 945. https://doi.org/10.3390/cells13110945







