Unraveling Verapamil’s Multidimensional Role in Diabetes Therapy: From β-Cell Regeneration to Cholecystokinin Induction in Zebrafish and MIN6 Cell-Line Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
2.2. Cell Survival
2.3. Cell Proliferation
2.4. Immunohistochemical Detection of Ki67
2.5. Western Blotting
2.6. Cell Cycle Analysis
2.7. Protection Assays
2.8. Glucose-Stimulated Insulin Secretion (GSIS) Assay
2.9. Metabolic Flux Analysis (Seahorse Assay)
2.10. Whole-Transcriptome Analysis
2.11. Proteome Sample Preparation
2.12. Bioinformatics Analyses
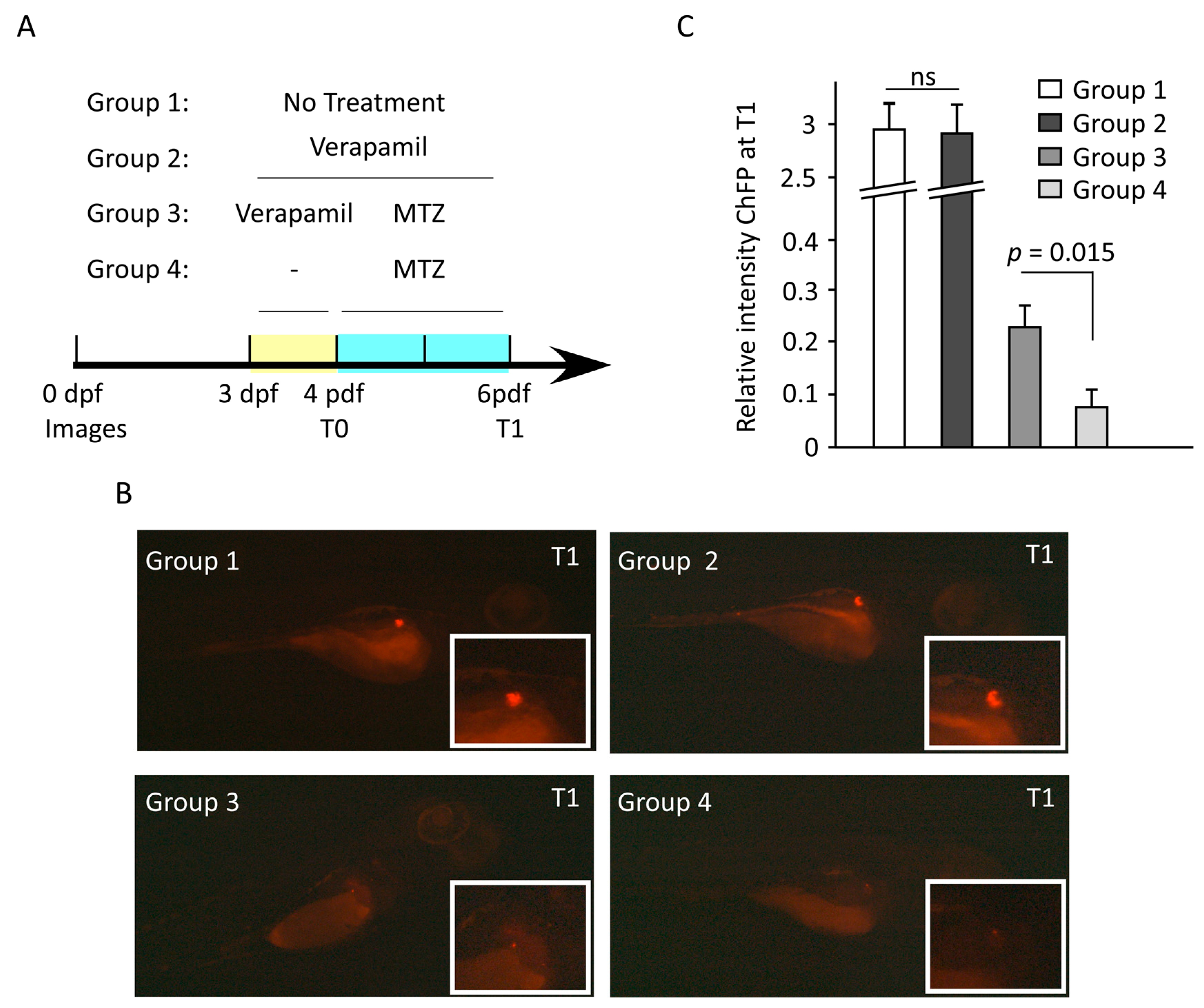
2.13. In Vivo Study Using Transgenic Zebrafish Embryos Model
2.14. Statistical Analysis
3. Results
3.1. Verapamil Induces Proliferation of MIN6 Cells
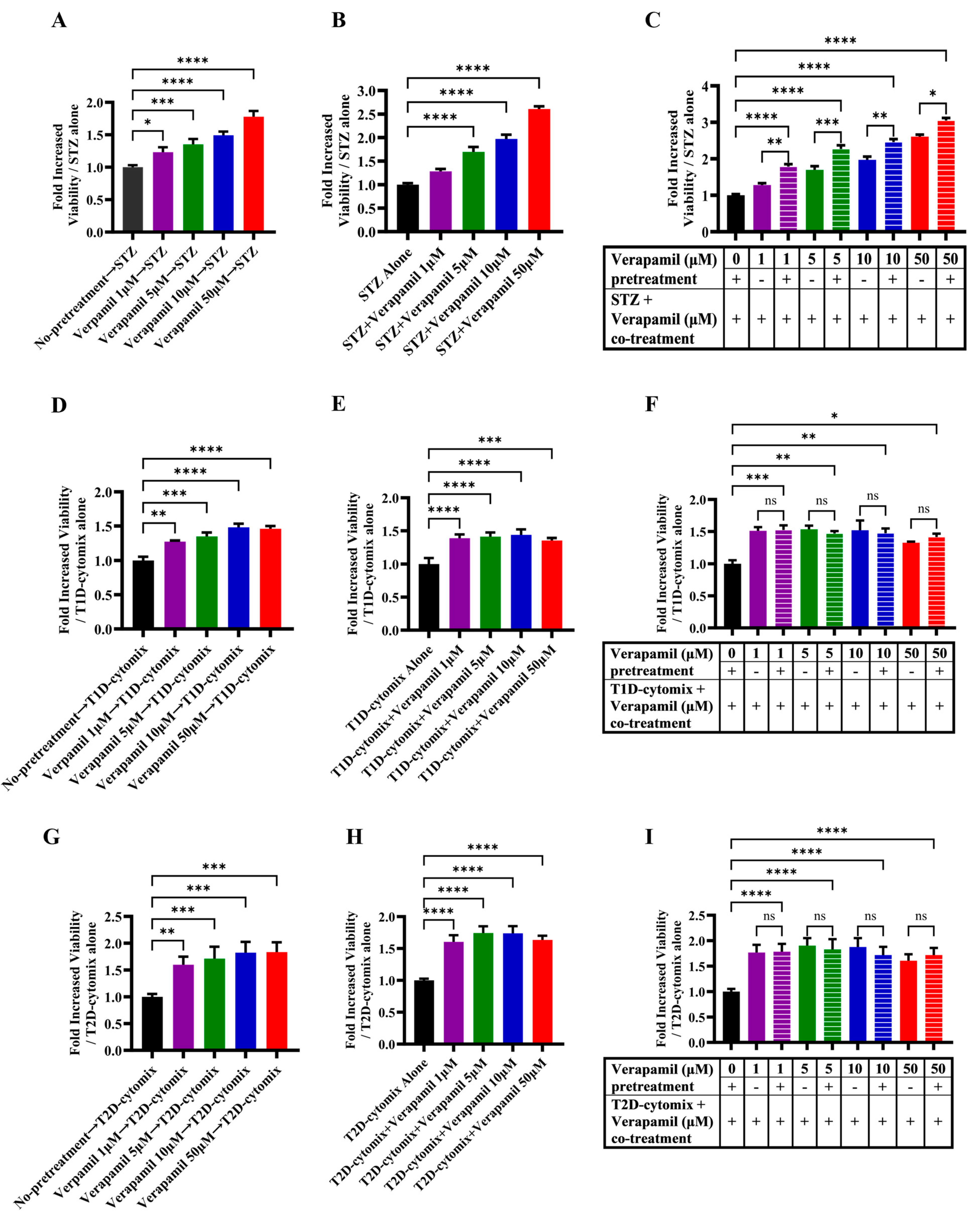
3.2. Verapamil Pretreatment Protects MIN6 β-Cells from Cytotoxic Insults Induced by STZ and T1D-/T2D-Cytomixes
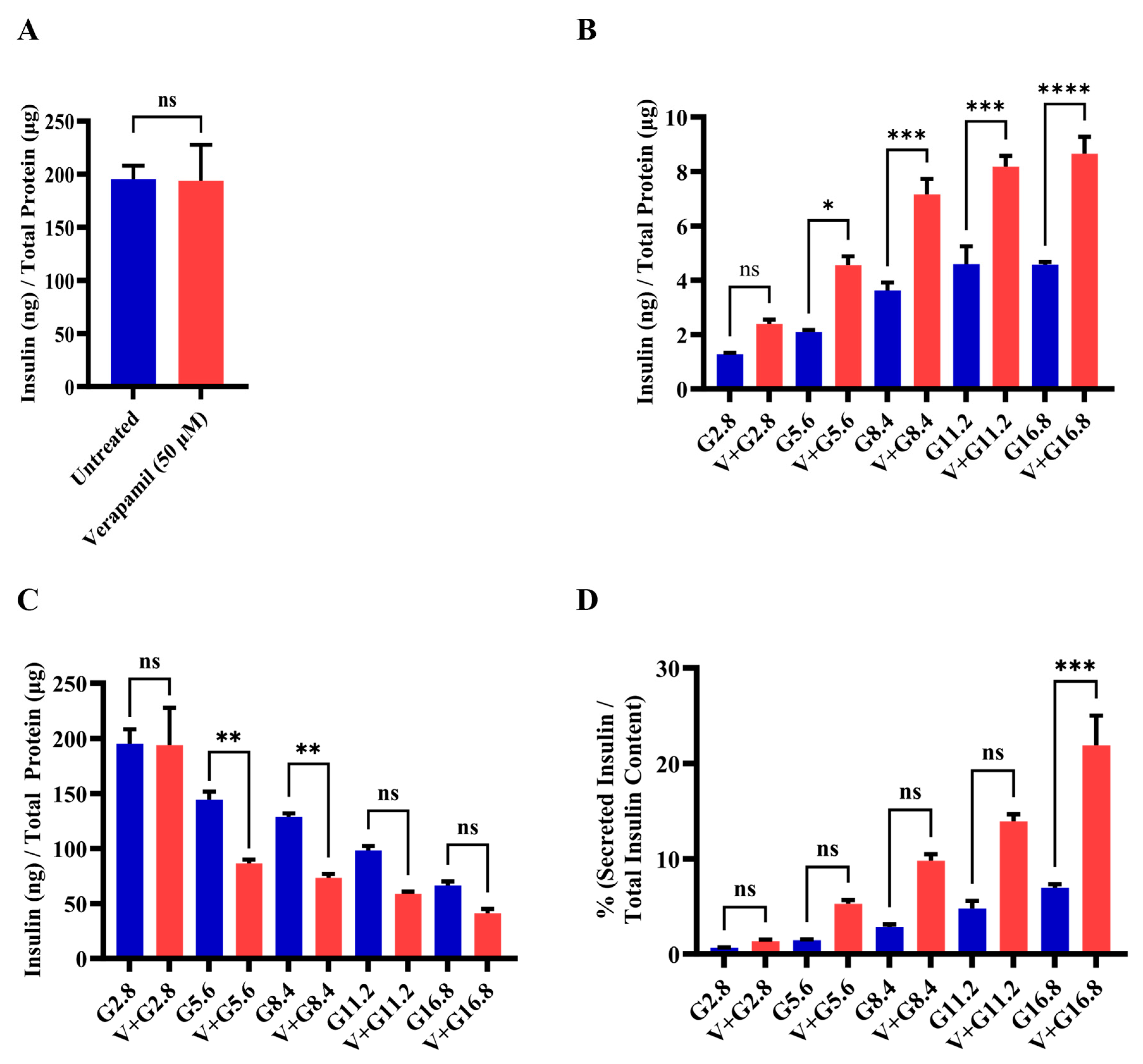
3.3. Verapamil Enhances Pancreatic β-Cell Function and Glucose Sensing in MIN6 Cells
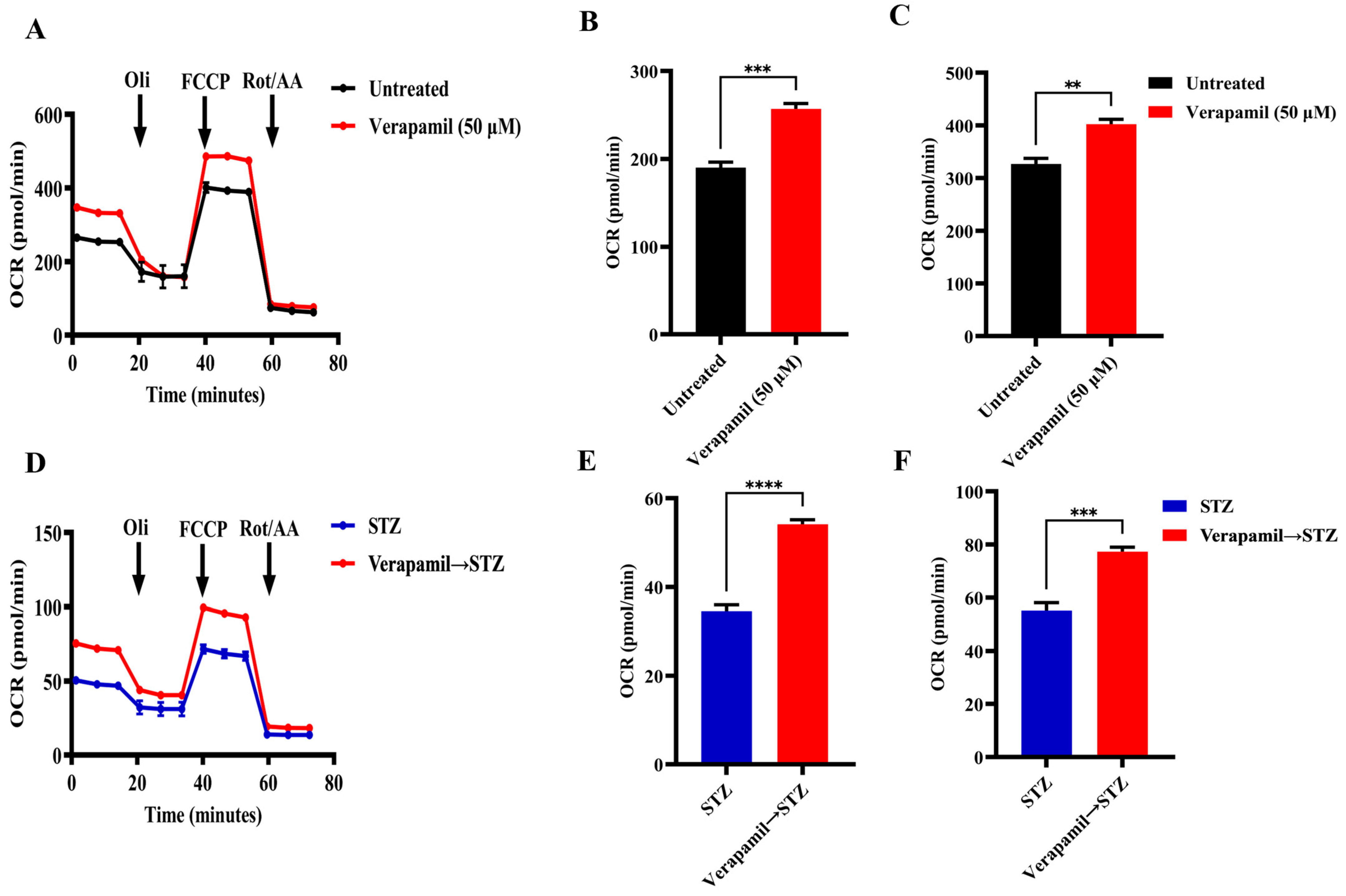
3.4. Verapamil Treatment Promotes Mitochondrial Respiration in MIN6 β-Cells
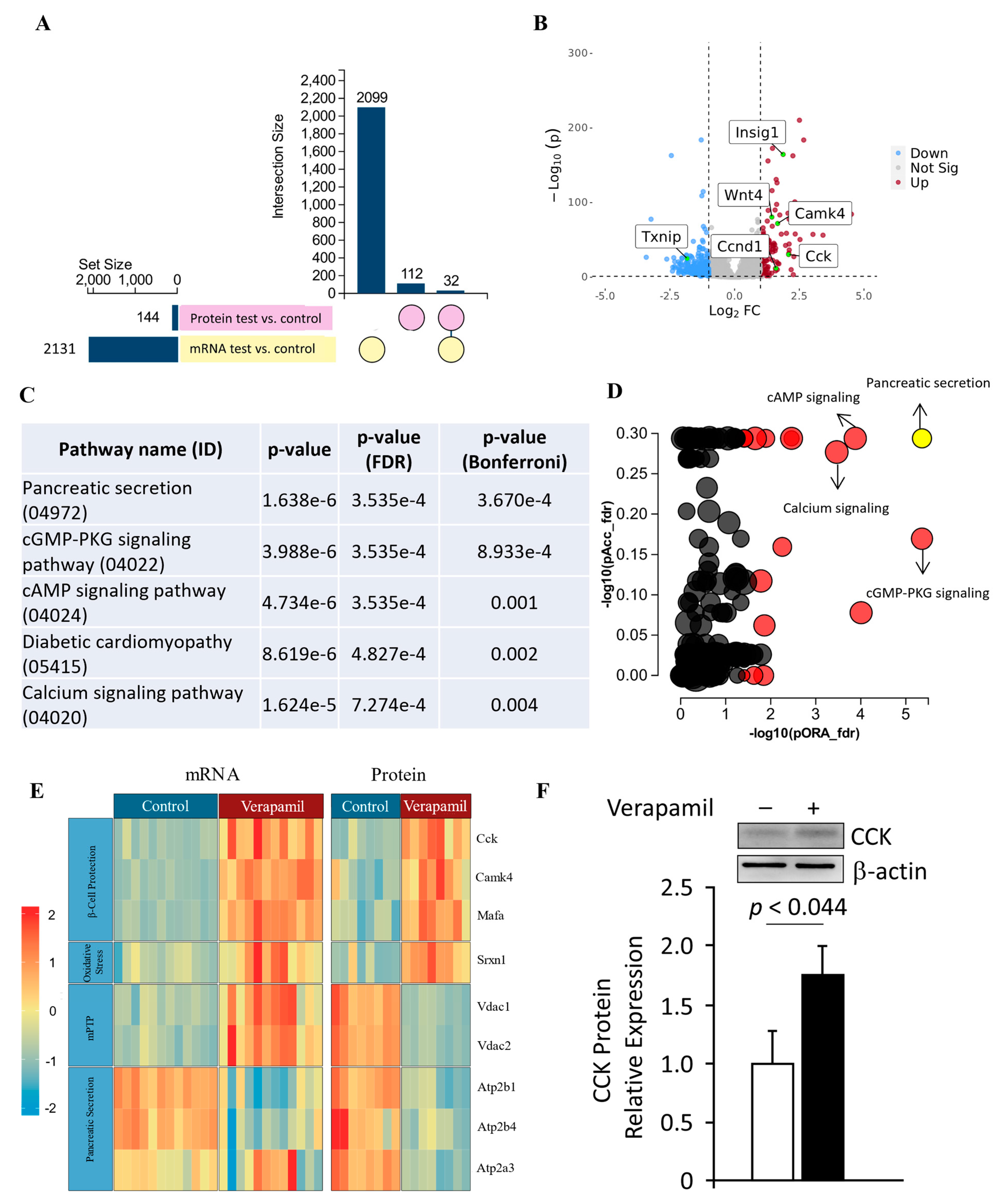
3.5. Transcriptomic and Proteomic Profiling of Verapamil-Treated MIN6 β-Cells
3.6. Verapamil Confers Protection from MTZ-Mediated Pancreatic β-Cell Damage in Transgenic Zebrafish Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.N.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef] [PubMed]
- von Scholten, B.J.; Kreiner, F.F.; Gough, S.C.L.; von Herrath, M. Current and future therapies for type 1 diabetes. Diabetologia 2021, 64, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Grimes, T.D.; Grayson, T.B.; Chen, J.; Thielen, L.A.; Tse, H.M.; Li, P.; Kanke, M.; Lin, T.T.; Schepmoes, A.A.; et al. Exploratory study reveals far reaching systemic and cellular effects of verapamil treatment in subjects with type 1 diabetes. Nat. Commun. 2022, 13, 1159. [Google Scholar] [CrossRef] [PubMed]
- Butler, A.E.; Janson, J.; Bonner-Weir, S.; Ritzel, R.; Rizza, R.A.; Butler, P.C. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes 2003, 52, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.K.; DuBose, S.N.; Haller, M.J.; Miller, K.M.; DiMeglio, L.A.; Bethin, K.E.; Goland, R.S.; Greenberg, E.M.; Liljenquist, D.R.; Ahmann, A.J.; et al. Prevalence of detectable C-Peptide according to age at diagnosis and duration of type 1 diabetes. Diabetes Care 2015, 38, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Messerli, F.H. “Cardioprotection”–Not all calcium antagonists are created equal. Am. J. Cardiol. 1990, 66, 855–856. [Google Scholar] [CrossRef] [PubMed]
- Cooper-Dehoff, R.; Cohen, J.D.; Bakris, G.L.; Messerli, F.H.; Erdine, S.; Hewkin, A.C.; Kupfer, S.; Pepine, C.J. Predictors of development of diabetes mellitus in patients with coronary artery disease taking antihypertensive medications (findings from the International Verapamil SR-Trandolapril Study [INVEST]). Am. J. Cardiol. 2006, 98, 890–894. [Google Scholar] [CrossRef] [PubMed]
- Cooper-DeHoff, R.M.; Aranda, J.M., Jr.; Gaxiola, E.; Cangiano, J.L.; Garcia-Barreto, D.; Conti, C.R.; Hewkin, A.; Pepine, C.J. Blood pressure control and cardiovascular outcomes in high-risk Hispanic patients--findings from the International Verapamil SR/Trandolapril Study (INVEST). Am. Heart J. 2006, 151, 1072–1079. [Google Scholar] [CrossRef]
- Yin, T.; Kuo, S.-C.; Chang, Y.-Y.; Chen, Y.-T.; Wang, K.-W.K. Verapamil Use Is Associated With Reduction of Newly Diagnosed Diabetes Mellitus. J. Clin. Endocrinol. Metab. 2017, 102, 2604–2610. [Google Scholar] [CrossRef]
- Khodneva, Y.; Shalev, A.; Frank, S.J.; Carson, A.P.; Safford, M.M. Calcium channel blocker use is associated with lower fasting serum glucose among adults with diabetes from the REGARDS study. Diabetes Res. Clin. Pract. 2016, 115, 115–121. [Google Scholar] [CrossRef]
- Ovalle, F.; Grimes, T.; Xu, G.; Patel, A.J.; Grayson, T.B.; Thielen, L.A.; Li, P.; Shalev, A. Verapamil and beta cell function in adults with recent-onset type 1 diabetes. Nat. Med. 2018, 24, 1108–1112. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Huang, K.C.; Lu, C.W.; Chu, C.H.; Huang, C.N.; Chen, H.S.; Lee, I.T.; Chen, J.F.; Chen, C.C.; Chen, C.S.; et al. A Randomized Controlled Trial of R-Form Verapamil Added to Ongoing Metformin Therapy in Patients with Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2022, 107, e4063–e4071. [Google Scholar] [CrossRef] [PubMed]
- Forlenza, G.P.; McVean, J.; Beck, R.W.; Bauza, C.; Bailey, R.; Buckingham, B.; DiMeglio, L.A.; Sherr, J.L.; Clements, M.; Neyman, A.; et al. Effect of Verapamil on Pancreatic Beta Cell Function in Newly Diagnosed Pediatric Type 1 Diabetes: A Randomized Clinical Trial. JAMA 2023, 329, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Malayeri, A.; Zakerkish, M.; Ramesh, F.; Galehdari, H.; Hemmati, A.A.; Angali, K.A. The Effect of Verapamil on TXNIP Gene Expression, GLP1R mRNA, FBS, HbA1c, and Lipid Profile in T2DM Patients Receiving Metformin and Sitagliptin. Diabetes Ther. Res. Treat. Educ. Diabetes Relat. Disord. 2021, 12, 2701–2713. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Chen, J.; Jing, G.; Shalev, A. Preventing β-cell loss and diabetes with calcium channel blockers. Diabetes 2012, 61, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Borowiec, A.M.; Właszczuk, A.; Olakowska, E.; Lewin-Kowalik, J. TXNIP inhibition in the treatment of diabetes. Verapamil as a novel therapeutic modality in diabetic patients. Med. Pharm. Rep. 2022, 95, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cha-Molstad, H.; Szabo, A.; Shalev, A. Diabetes induces and calcium channel blockers prevent cardiac expression of proapoptotic thioredoxin-interacting protein. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1133–E1139. [Google Scholar] [CrossRef] [PubMed]
- Carnovale, C.; Dassano, A.; Mosini, G.; Mazhar, F.; D’Addio, F.; Pozzi, M.; Radice, S.; Fiorina, P.; Clementi, E. The β-cell effect of verapamil-based treatment in patients with type 2 diabetes: A systematic review. Acta Diabetol. 2020, 57, 117–131. [Google Scholar] [CrossRef]
- Poudel, R.R.; Kafle, N.K. Verapamil in Diabetes. Indian J. Endocrinol. Metab. 2017, 21, 788–789. [Google Scholar] [CrossRef]
- Chen, J.; Hui, S.T.; Couto, F.M.; Mungrue, I.N.; Davis, D.B.; Attie, A.D.; Lusis, A.J.; Davis, R.A.; Shalev, A. Thioredoxin-interacting protein deficiency induces Akt/Bcl-xL signaling and pancreatic beta-cell mass and protects against diabetes. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2008, 22, 3581–3594. [Google Scholar] [CrossRef]
- Miyazaki, J.; Araki, K.; Yamato, E.; Ikegami, H.; Asano, T.; Shibasaki, Y.; Oka, Y.; Yamamura, K. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: Special reference to expression of glucose transporter isoforms. Endocrinology 1990, 127, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Wang, R.; Sun, B. Meteorin-Like Ameliorates β Cell Function by Inhibiting β Cell Apoptosis of and Promoting β Cell Proliferation via Activating the WNT/β-Catenin Pathway. Front. Pharmacol. 2021, 12, 627147. [Google Scholar] [CrossRef] [PubMed]
- El-Sharkawey, A. Calculate the Corrected Total Cell Fluorescence (CTCF). 2016. Available online: https://www.researchgate.net/publication/304878988_Calculate_the_Corrected_Total_Cell_Fluorescence_CTCF (accessed on 31 May 2016).
- Al Madhoun, A.; Marafie, S.K.; Haddad, D.; Melhem, M.; Abu-Farha, M.; Ali, H.; Sindhu, S.; Atari, M.; Al-Mulla, F. Comparative Proteomic Analysis Identifies EphA2 as a Specific Cell Surface Marker for Wharton’s Jelly-Derived Mesenchymal Stem Cells. Int. J. Mol. Sci. 2020, 21, 6437. [Google Scholar] [CrossRef] [PubMed]
- Al Madhoun, A.; Haddad, D.; Al Tarrah, M.; Jacob, S.; Al-Ali, W.; Nizam, R.; Miranda, L.; Al-Rashed, F.; Sindhu, S.; Ahmad, R.; et al. Microarray analysis reveals ONC201 mediated differential mechanisms of CHOP gene regulation in metastatic and nonmetastatic colorectal cancer cells. Sci. Rep. 2021, 11, 11893. [Google Scholar] [CrossRef] [PubMed]
- Pozarowski, P.; Darzynkiewicz, Z. Analysis of cell cycle by flow cytometry. In Methods in Molecular Biology (Clifton, N.J.); Humana Press: Totowa, NJ, USA, 2004; Volume 281, pp. 301–311. [Google Scholar] [CrossRef]
- Damame, H.H.; Rooge, S.B.; Patil, R.S.; Arvindekar, A.U. In vitro model using cytokine cocktail to evaluate apoptosis in Min6 pancreatic beta cells. J. Pharmacol. Toxicol. Methods 2020, 106, 106914. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Guan, Y.; Yang, J. Cytokines in the Progression of Pancreatic β-Cell Dysfunction. Int. J. Endocrinol. 2010, 2010, 515136. [Google Scholar] [CrossRef] [PubMed]
- Marafie, S.K.; Al-Shawaf, E.M.; Abubaker, J.; Arefanian, H. Palmitic acid-induced lipotoxicity promotes a novel interplay between Akt-mTOR, IRS-1, and FFAR1 signaling in pancreatic β-cells. Biol. Res. 2019, 52, 44. [Google Scholar] [CrossRef] [PubMed]
- Orliaguet, L.; Ejlalmanesh, T.; Humbert, A.; Ballaire, R.; Diedisheim, M.; Julla, J.B.; Chokr, D.; Cuenco, J.; Michieletto, J.; Charbit, J.; et al. Early macrophage response to obesity encompasses Interferon Regulatory Factor 5 regulated mitochondrial architecture remodelling. Nat. Commun. 2022, 13, 5089. [Google Scholar] [CrossRef] [PubMed]
- Abu-Farha, M.; Lambert, J.P.; Al-Madhoun, A.S.; Elisma, F.; Skerjanc, I.S.; Figeys, D. The tale of two domains: Proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol. Cell. Proteom. MCP 2008, 7, 560–572. [Google Scholar] [CrossRef]
- Rivera, C.G.; Tyler, B.M.; Murali, T.M. Sensitive detection of pathway perturbations in cancers. BMC Bioinform. 2012, 13 (Suppl. S3), S9. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Pisharath, H.; Rhee, J.M.; Swanson, M.A.; Leach, S.D.; Parsons, M.J. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech. Dev. 2007, 124, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio Rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2007. [Google Scholar]
- Wachlin, G.; Augstein, P.; Schröder, D.; Kuttler, B.; Klöting, I.; Heinke, P.; Schmidt, S. IL-1beta, IFN-gamma and TNF-alpha increase vulnerability of pancreatic beta cells to autoimmune destruction. J. Autoimmun. 2003, 20, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Banu, S.; Sur, D. Role of Macrophage in Type 2 Diabetes Mellitus: Macrophage Polarization a New Paradigm for Treatment of Type 2 Diabetes Mellitus. Endocr. Metab. Immune Disord. Drug Targets 2023, 23, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Draghici, S.; Khatri, P.; Tarca, A.L.; Amin, K.; Done, A.; Voichita, C.; Georgescu, C.; Romero, R. A systems biology approach for pathway level analysis. Genome Res. 2007, 17, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Tarca, A.L.; Draghici, S.; Khatri, P.; Hassan, S.S.; Mittal, P.; Kim, J.S.; Kim, C.J.; Kusanovic, J.P.; Romero, R. A novel signaling pathway impact analysis. Bioinformatics 2009, 25, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Khatri, P.; Voichita, C.; Kattan, K.; Ansari, N.; Khatri, A.; Georgescu, C.; Tarca, A.L.; Draghici, S. Onto-Tools: New additions and improvements in 2006. Nucleic Acids Res. 2007, 35, W206–W211. [Google Scholar] [CrossRef]
- Bo, S.; Gentile, L.; Castiglione, A.; Prandi, V.; Canil, S.; Ghigo, E.; Ciccone, G. C-peptide and the risk for incident complications and mortality in type 2 diabetic patients: A retrospective cohort study after a 14-year follow-up. Eur. J. Endocrinol. 2012, 167, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-Y.; Jung, C.-H.; Mok, J.-O.; Kang, S.-K.; Kim, C.-H. Association between serum C-peptide levels and chronic microvascular complications in Korean type 2 diabetic patients. Acta Diabetol. 2012, 49, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Kushner, J.A.; Ciemerych, M.A.; Sicinska, E.; Wartschow, L.M.; Teta, M.; Long, S.Y.; Sicinski, P.; White, M.F. Cyclins D2 and D1 are essential for postnatal pancreatic beta-cell growth. Mol. Cell. Biol. 2005, 25, 3752–3762. [Google Scholar] [CrossRef]
- Kang, J.H.; Kim, M.J.; Ko, S.H.; Jeong, I.K.; Koh, K.H.; Rhie, D.J.; Yoon, S.H.; Hahn, S.J.; Kim, M.S.; Jo, Y.H. Upregulation of rat Ccnd1 gene by exendin-4 in pancreatic beta cell line INS-1: Interaction of early growth response-1 with cis-regulatory element. Diabetologia 2006, 49, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Minn, A.H.; Pise-Masison, C.A.; Radonovich, M.; Brady, J.N.; Wang, P.; Kendziorski, C.; Shalev, A. Gene expression profiling in INS-1 cells overexpressing thioredoxin-interacting protein. Biochem. Biophys. Res. Commun. 2005, 336, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Tardivel, A.; Thorens, B.; Choi, I.; Tschopp, J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 2010, 11, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Lavine, J.A.; Kibbe, C.R.; Baan, M.; Sirinvaravong, S.; Umhoefer, H.M.; Engler, K.A.; Meske, L.M.; Sacotte, K.A.; Erhardt, D.P.; Davis, D.B. Cholecystokinin expression in the β-cell leads to increased β-cell area in aged mice and protects from streptozotocin-induced diabetes and apoptosis. Am. J. Physiol. Endocrinol. Metab. 2015, 309, E819–E828. [Google Scholar] [CrossRef] [PubMed]
- Katsumoto, K.; Yennek, S.; Chen, C.; Silva, L.F.D.; Traikov, S.; Sever, D.; Azad, A.; Shan, J.; Vainio, S.; Ninov, N.; et al. Wnt4 is heterogeneously activated in maturing β-cells to control calcium signaling, metabolism and function. Nat. Commun. 2022, 13, 6255. [Google Scholar] [CrossRef] [PubMed]
- Kurita, Y.; Ohki, T.; Soejima, E.; Yuan, X.; Kakino, S.; Wada, N.; Hashinaga, T.; Nakayama, H.; Tani, J.; Tajiri, Y.; et al. A High-Fat/High-Sucrose Diet Induces WNT4 Expression in Mouse Pancreatic β-cells. Kurume Med. J. 2019, 65, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Y.; Zhou, X.; Zhang, X.; Meng, H.; Liu, S.; Zhang, L.; He, J.; He, Q.; Geng, Y. CaMKIV limits metabolic damage through induction of hepatic autophagy by CREB in obese mice. J. Endocrinol. 2020, 244, 353–367. [Google Scholar] [CrossRef] [PubMed]
- De Marchi, U.; Fernandez-Martinez, S.; de la Fuente, S.; Wiederkehr, A.; Santo-Domingo, J. Mitochondrial ion channels in pancreatic β-cells: Novel pharmacological targets for the treatment of Type 2 diabetes. Br. J. Pharmacol. 2021, 178, 2077–2095. [Google Scholar] [CrossRef]
- Abu-Hamad, S.; Sivan, S.; Shoshan-Barmatz, V. The expression level of the voltage-dependent anion channel controls life and death of the cell. Proc. Natl. Acad. Sci. USA 2006, 103, 5787–5792. [Google Scholar] [CrossRef]
- Ahmed, M.; Muhammed, S.J.; Kessler, B.; Salehi, A. Mitochondrial proteome analysis reveals altered expression of voltage dependent anion channels in pancreatic β-cells exposed to high glucose. Islets 2010, 2, 283–292. [Google Scholar] [CrossRef]
- Zhang, E.; Mohammed Al-Amily, I.; Mohammed, S.; Luan, C.; Asplund, O.; Ahmed, M.; Ye, Y.; Ben-Hail, D.; Soni, A.; Vishnu, N.; et al. Preserving Insulin Secretion in Diabetes by Inhibiting VDAC1 Overexpression and Surface Translocation in β Cells. Cell Metab. 2019, 29, 64–77.e66. [Google Scholar] [CrossRef] [PubMed]
- Maurya, S.R.; Mahalakshmi, R. VDAC-2: Mitochondrial outer membrane regulator masquerading as a channel? FEBS J. 2016, 283, 1831–1836. [Google Scholar] [CrossRef] [PubMed]
- Persaud, S.J.; Liu, B.; Sampaio, H.B.; Jones, P.M.; Muller, D.S. Calcium/calmodulin-dependent kinase IV controls glucose-induced Irs2 expression in mouse beta cells via activation of cAMP response element-binding protein. Diabetologia 2011, 54, 1109–1120. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | logfc Protein (Mass Spec) | adjpv Protein (Mass Spec) | logfc mRNA (RNA-seq) | adjpv mRNA (RNA-seq) |
|---|---|---|---|---|
| Acss2 | 0.546956178 | 0.045430822 | 1.468439926 | 0.000001 |
| Ap3m2 | −1.258425153 | 0.004563029 | −0.631083607 | 0.00535079 |
| Atad1 | −0.878321443 | 0.003214256 | −0.69090951 | 0.001886914 |
| Atl3 | −0.775959726 | 0.01924926 | −0.705494101 | 0.000782498 |
| Atp2b1 | −0.768567592 | 0.001599313 | −0.903858193 | 0.00000179 |
| Cadm1 | 6.64385619 | 0.000001 | −1.111895426 | 0.000001 |
| Camk4 | 0.618238656 | 0.025051947 | 1.667580225 | 0.000001 |
| Cck | 1.343692069 | 0.000001 | 2.086382128 | 0.000001 |
| Fdps | 1.236339539 | 0.000001 | 2.06424327 | 0.000001 |
| Fn1 | 0.750177706 | 0.00019395 | 0.707236676 | 0.000001 |
| Gabarap | 0.779049553 | 0.000817148 | 0.608883531 | 0.000001 |
| Gc | 0.575312331 | 0.010226369 | −0.668515132 | 0.000001 |
| Gdap1 | −0.744197163 | 0.03032308 | −0.715727824 | 0.009402028 |
| Hax1 | 6.64385619 | 0.000001 | 0.708603816 | 0.000001 |
| Hectd1 | 1.205392513 | 0.000001 | −0.766523599 | 0.004572659 |
| Hmgcs1 | 1.608809243 | 0.000001 | 1.574403987 | 0.000001 |
| Idi1 | 1.136191386 | 0.000001 | 1.063478538 | 0.00444946 |
| Ins2 | −0.706041021 | 0.003576738 | −1.047050788 | 0.000001 |
| Kif23 | −0.899695094 | 0.029438363 | −1.278078727 | 0.000001 |
| Ldlr | 1.607862903 | 0.000001 | 1.459525662 | 0.000001 |
| Mafa | 0.715454127 | 0.019937366 | 0.86170978 | 0.000001 |
| Mid1ip1 | 0.82130204 | 0.000734422 | 0.902299589 | 0.000001 |
| Nsg1 | 0.568518598 | 0.029438363 | 0.821403563 | 0.000001 |
| Pam | 0.600269754 | 0.008748346 | 0.772163694 | 0.000001 |
| Ppfia2 | −2.53951953 | 0.000001 | −1.455788602 | 0.000001 |
| Prlr | 0.619178216 | 0.031249276 | −0.938951004 | 0.003238752 |
| Sdf2l1 | 0.606915942 | 0.007338519 | 1.333480725 | 0.000001 |
| Slc25a1 | −0.798366139 | 0.005415551 | 0.796013338 | 0.000001 |
| Slc9a3r1 | 0.508935662 | 0.025006868 | 0.846755275 | 0.000001 |
| Sqstm1 | 0.789103218 | 8.91352 × 10−5 | 0.605062208 | 0.000001 |
| Stc1 | 0.804053559 | 0.000223206 | 1.114392736 | 0.000001 |
| Timm8b | 0.670840336 | 0.003900441 | 0.676846573 | 0.000001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arefanian, H.; Al Madhoun, A.; Al-Rashed, F.; Alzaid, F.; Bahman, F.; Nizam, R.; Alhusayan, M.; John, S.; Jacob, S.; Williams, M.R.; et al. Unraveling Verapamil’s Multidimensional Role in Diabetes Therapy: From β-Cell Regeneration to Cholecystokinin Induction in Zebrafish and MIN6 Cell-Line Models. Cells 2024, 13, 949. https://doi.org/10.3390/cells13110949
Arefanian H, Al Madhoun A, Al-Rashed F, Alzaid F, Bahman F, Nizam R, Alhusayan M, John S, Jacob S, Williams MR, et al. Unraveling Verapamil’s Multidimensional Role in Diabetes Therapy: From β-Cell Regeneration to Cholecystokinin Induction in Zebrafish and MIN6 Cell-Line Models. Cells. 2024; 13(11):949. https://doi.org/10.3390/cells13110949
Chicago/Turabian StyleArefanian, Hossein, Ashraf Al Madhoun, Fatema Al-Rashed, Fawaz Alzaid, Fatemah Bahman, Rasheeba Nizam, Mohammed Alhusayan, Sumi John, Sindhu Jacob, Michayla R. Williams, and et al. 2024. "Unraveling Verapamil’s Multidimensional Role in Diabetes Therapy: From β-Cell Regeneration to Cholecystokinin Induction in Zebrafish and MIN6 Cell-Line Models" Cells 13, no. 11: 949. https://doi.org/10.3390/cells13110949
APA StyleArefanian, H., Al Madhoun, A., Al-Rashed, F., Alzaid, F., Bahman, F., Nizam, R., Alhusayan, M., John, S., Jacob, S., Williams, M. R., Abukhalaf, N., Shenouda, S., Joseph, S., AlSaeed, H., Kochumon, S., Mohammad, A., Koti, L., Sindhu, S., Abu-Farha, M., ... Al-Mulla, F. (2024). Unraveling Verapamil’s Multidimensional Role in Diabetes Therapy: From β-Cell Regeneration to Cholecystokinin Induction in Zebrafish and MIN6 Cell-Line Models. Cells, 13(11), 949. https://doi.org/10.3390/cells13110949











