Context-Dependent Regulation of Peripheral Nerve Abundance by the PI3K Pathway in the Tumor Microenvironment of Head and Neck Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Key Resources
2.2. Establishment of the Schwann Cell-Related Gene Set
2.3. Gene Set Variation Analysis (GSVA)
2.4. Immunohistochemical Staining and Quantification
2.5. Copy Number Alteration (CNA)
2.6. Somatic Mutations
2.7. GSVA for Regulatory Networks and Oncogenic Pathways
2.8. Oncopredict
2.9. Single-Cell RNA-Seq Analysis
2.10. Statistical Analysis
3. Results
3.1. Establishment of the Schwann Cell-Related 43-Gene Set
3.2. Association of the SC-Related 43-Gene Set with the Abundance of Peripheral Nerves in the TME
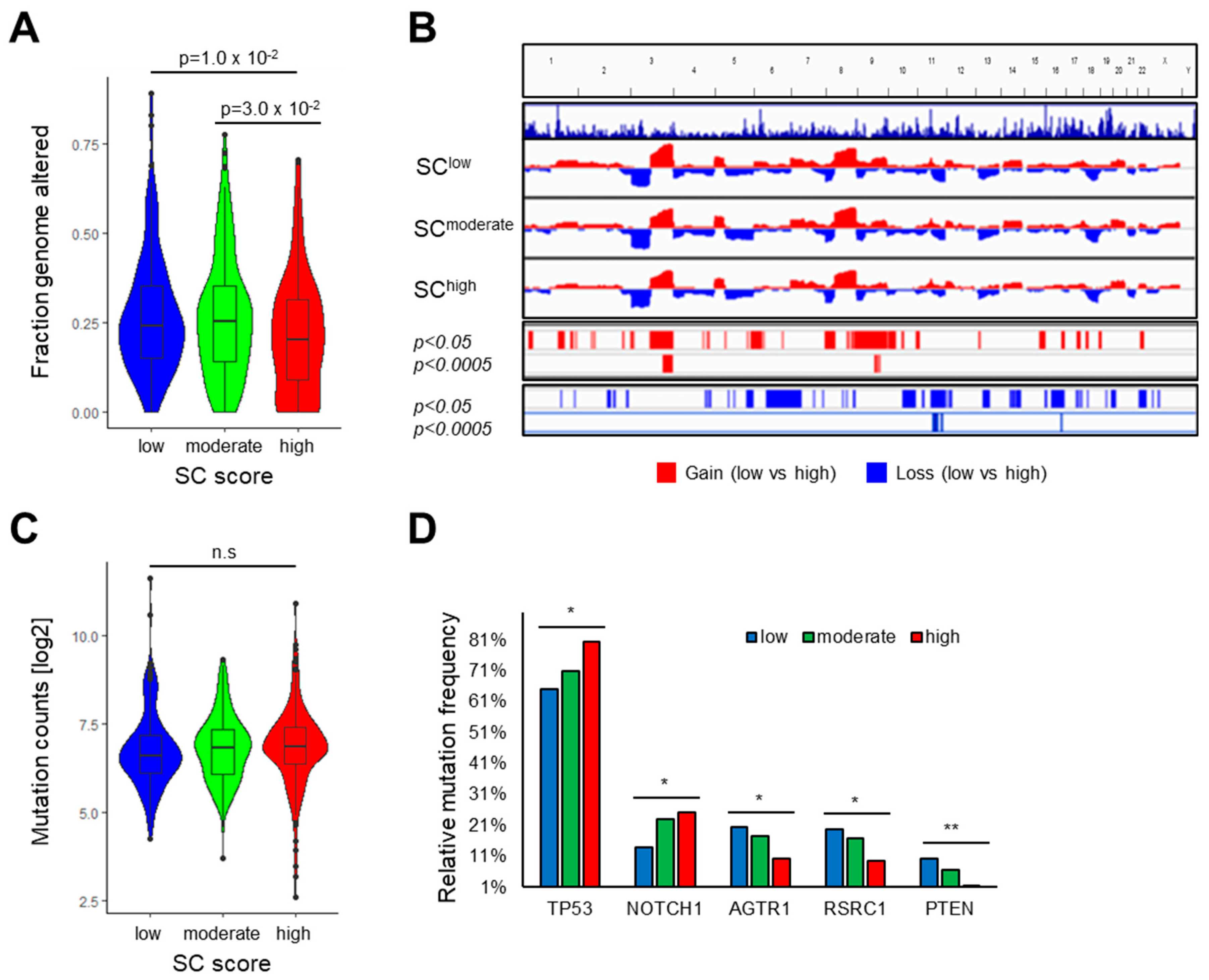
3.3. Association of the SC Score with Clinical Features and the Mutational Landscape
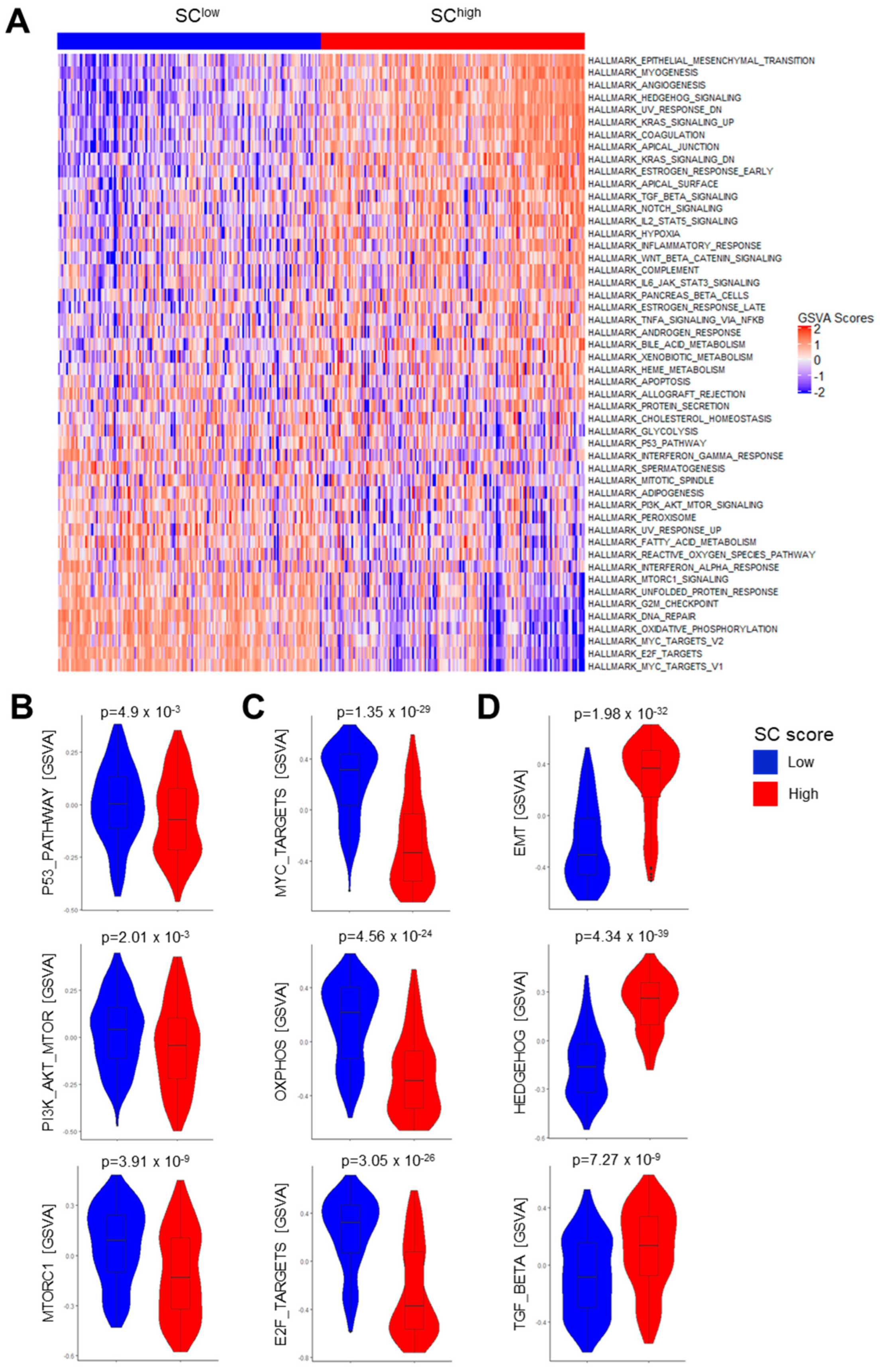
3.4. Association of the SC Score with Gene Regulatory Networks and Oncogenic Pathway Activity
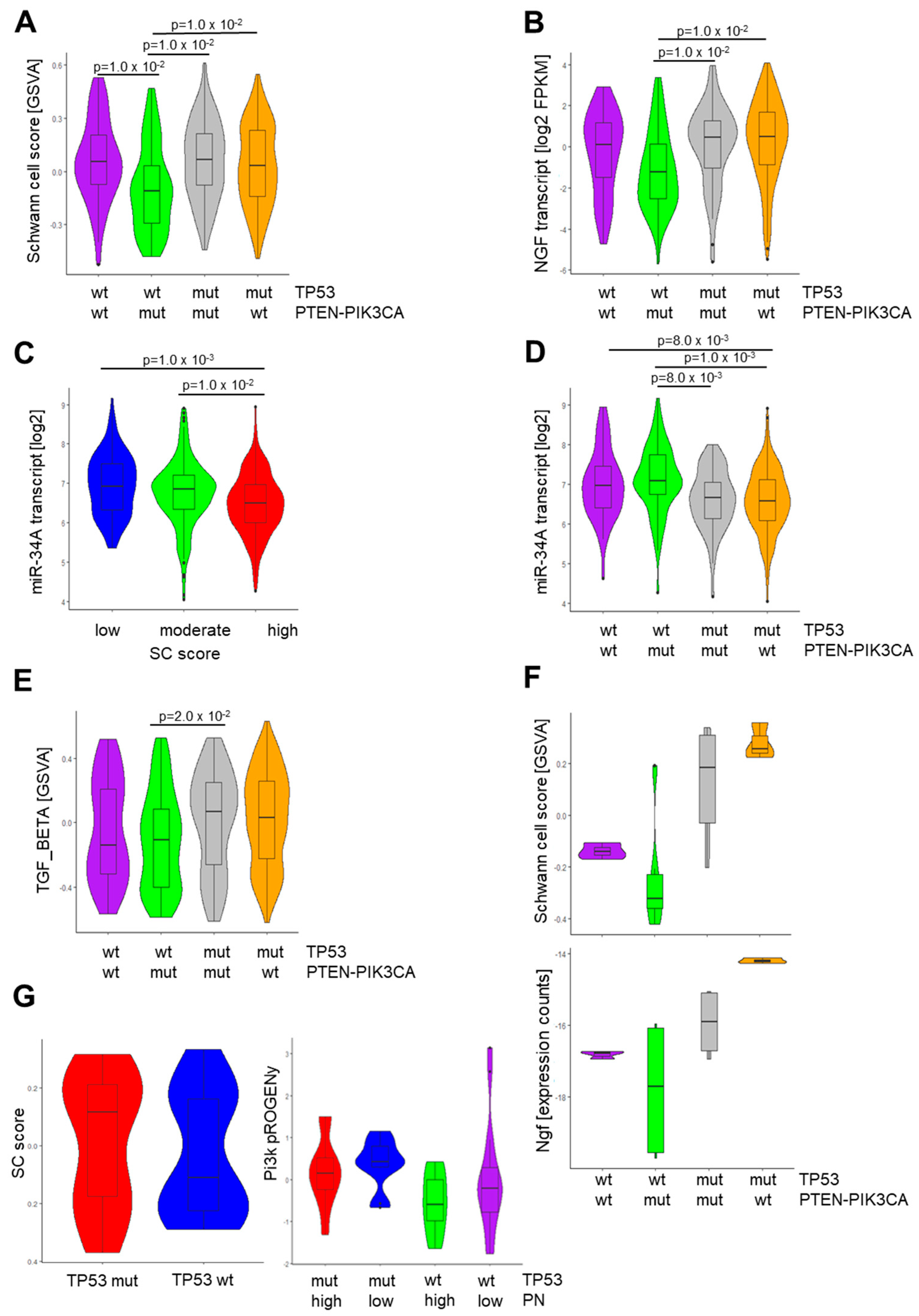
3.5. Context-Dependent Impact of PI3K Pathway Activity
3.6. Identification of Vulnerabilities by in Silico Drug Screening
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Visser, K.E.; Joyce, J.A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef] [PubMed]
- Gysler, S.M.; Drapkin, R. Tumor innervation: Peripheral nerves take control of the tumor microenvironment. J. Clin. Investig. 2021, 131, e147276. [Google Scholar] [CrossRef] [PubMed]
- Winkler, F.; Venkatesh, H.S.; Amit, M.; Batchelor, T.; Demir, I.E.; Deneen, B.; Gutmann, D.H.; Hervey-Jumper, S.; Kuner, T.; Mabbott, D.; et al. Cancer neuroscience: State of the field, emerging directions. Cell 2023, 186, 1689–1707. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Hess, J. Tumor Neurobiology in the Pathogenesis and Therapy of Head and Neck Cancer. Cells 2024, 13, 256. [Google Scholar] [CrossRef] [PubMed]
- Misztal, C.I.; Green, C.; Mei, C.; Bhatia, R.; Velez Torres, J.M.; Kamrava, B.; Moon, S.; Nicolli, E.; Weed, D.; Sargi, Z.; et al. Molecular and Cellular Mechanisms of Perineural Invasion in Oral Squamous Cell Carcinoma: Potential Targets for Therapeutic Intervention. Cancers 2021, 13, 6011. [Google Scholar] [CrossRef] [PubMed]
- Silverman, D.A.; Martinez, V.K.; Dougherty, P.M.; Myers, J.N.; Calin, G.A.; Amit, M. Cancer-Associated Neurogenesis and Nerve-Cancer Cross-talk. Cancer Res. 2021, 81, 1431–1440. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, C.; Phillips, J.A.; Djamgoz, M.B.A. Nerve input to tumours: Pathophysiological consequences of a dynamic relationship. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188411. [Google Scholar] [CrossRef]
- Schmitd, L.B.; Perez-Pacheco, C.; D’Silva, N.J. Nerve density in cancer: Less is better. FASEB Bioadv. 2021, 3, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Perez-Pacheco, C.; Schmitd, L.B.; Furgal, A.; Bellile, E.L.; Liu, M.; Fattah, A.; Gonzalez-Maldonado, L.; Unsworth, S.P.; Wong, S.Y.; Rozek, L.S.; et al. Increased Nerve Density Adversely Affects Outcome in Oral Cancer. Clin. Cancer Res. 2023, 29, 2501–2512. [Google Scholar] [CrossRef]
- Zorick, T.S.; Lemke, G. Schwann cell differentiation. Curr. Opin. Cell Biol. 1996, 8, 870–876. [Google Scholar] [CrossRef]
- Tian, Z.; Ou, G.; Su, M.; Li, R.; Pan, L.; Lin, X.; Zou, J.; Chen, S.; Li, Y.; Huang, K.; et al. TIMP1 derived from pancreatic cancer cells stimulates Schwann cells and promotes the occurrence of perineural invasion. Cancer Lett. 2022, 546, 215863. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; He, R.; Yang, W.; Zhang, Y.; Yuan, Q.; Wang, J.; Liu, Y.; Chen, S.; Zhang, S.; Zhang, W.; et al. Autophagic Schwann cells promote perineural invasion mediated by the NGF/ATG7 paracrine pathway in pancreatic cancer. J. Exp. Clin. Cancer Res. 2022, 41, 48. [Google Scholar] [CrossRef] [PubMed]
- Demir, I.E.; Boldis, A.; Pfitzinger, P.L.; Teller, S.; Brunner, E.; Klose, N.; Kehl, T.; Maak, M.; Lesina, M.; Laschinger, M.; et al. Investigation of Schwann cells at neoplastic cell sites before the onset of cancer invasion. J. Natl. Cancer Inst. 2014, 106, dju184. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fang, Y.; Jiang, W. Important Cells and Factors from Tumor Microenvironment Participated in Perineural Invasion. Cancers 2023, 15, 1360. [Google Scholar] [CrossRef] [PubMed]
- Deborde, S.; Gusain, L.; Powers, A.; Marcadis, A.; Yu, Y.; Chen, C.H.; Frants, A.; Kao, E.; Tang, L.H.; Vakiani, E.; et al. Reprogrammed Schwann Cells Organize into Dynamic Tracks that Promote Pancreatic Cancer Invasion. Cancer Discov. 2022, 12, 2454–2473. [Google Scholar] [CrossRef] [PubMed]
- Amit, M.; Na’ara, S.; Gil, Z. Mechanisms of cancer dissemination along nerves. Nat. Rev. Cancer 2016, 16, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Pascual, G.; Dominguez, D.; Elosua-Bayes, M.; Beckedorff, F.; Laudanna, C.; Bigas, C.; Douillet, D.; Greco, C.; Symeonidi, A.; Hernandez, I.; et al. Dietary palmitic acid promotes a prometastatic memory via Schwann cells. Nature 2021, 599, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Zhu, Y.; Jiang, Y.; Han, L.; Shi, M.; Su, R.; Wang, L.; Xiong, C.; Wang, C.; Wang, T.; et al. Schwann cells regulate tumor cells and cancer-associated fibroblasts in the pancreatic ductal adenocarcinoma microenvironment. Nat. Commun. 2023, 14, 4600. [Google Scholar] [CrossRef] [PubMed]
- Mody, M.D.; Rocco, J.W.; Yom, S.S.; Haddad, R.I.; Saba, N.F. Head and neck cancer. Lancet 2021, 398, 2289–2299. [Google Scholar] [CrossRef]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef] [PubMed]
- D’Silva, N.J.; Perez-Pacheco, C.; Schmitd, L.B. The 3D’s of Neural Phenotypes in Oral Cancer: Distance, Diameter, and Density. Adv. Biol. 2023, 7, e2200188. [Google Scholar] [CrossRef] [PubMed]
- Amit, M.; Takahashi, H.; Dragomir, M.P.; Lindemann, A.; Gleber-Netto, F.O.; Pickering, C.R.; Anfossi, S.; Osman, A.A.; Cai, Y.; Wang, R.; et al. Loss of p53 drives neuron reprogramming in head and neck cancer. Nature 2020, 578, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Restaino, A.C.; Walz, A.; Vermeer, S.J.; Barr, J.; Kovacs, A.; Fettig, R.R.; Vermeer, D.W.; Reavis, H.; Williamson, C.S.; Lucido, C.T.; et al. Functional neuronal circuits promote disease progression in cancer. Sci. Adv. 2023, 9, eade4443. [Google Scholar] [CrossRef] [PubMed]
- Magnon, C.; Hondermarck, H. The neural addiction of cancer. Nat. Rev. Cancer 2023, 23, 317–334. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016, baw100. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Wolbert, J.; Li, X.; Heming, M.; Mausberg, A.K.; Akkermann, D.; Frydrychowicz, C.; Fledrich, R.; Groeneweg, L.; Schulz, C.; Stettner, M.; et al. Redefining the heterogeneity of peripheral nerve cells in health and autoimmunity. Proc. Natl. Acad. Sci. USA 2020, 117, 9466–9476. [Google Scholar] [CrossRef]
- Franzen, O.; Gan, L.M.; Bjorkegren, J.L.M. PanglaoDB: A web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, 2019, baz046. [Google Scholar] [CrossRef]
- Burkart, S.; Weusthof, C.; Khorani, K.; Steen, S.; Stogbauer, F.; Unger, K.; Hess, J.; Zitzelsberger, H.; Belka, C.; Kurth, I.; et al. A Novel Subgroup of UCHL1-Related Cancers Is Associated with Genomic Instability and Sensitivity to DNA-Damaging Treatment. Cancers 2023, 15, 1655. [Google Scholar] [CrossRef] [PubMed]
- Larsen, S.J.; do Canto, L.M.; Rogatto, S.R.; Baumbach, J. CoNVaQ: A web tool for copy number variation-based association studies. BMC Genom. 2018, 19, 369. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Maeser, D.; Gruener, R.F.; Huang, R.S. oncoPredict: An R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief. Bioinform. 2021, 22, bbab260. [Google Scholar] [CrossRef] [PubMed]
- Puram, S.V.; Mints, M.; Pal, A.; Qi, Z.; Reeb, A.; Gelev, K.; Barrett, T.F.; Gerndt, S.; Liu, P.; Parikh, A.S.; et al. Cellular states are coupled to genomic and viral heterogeneity in HPV-related oropharyngeal carcinoma. Nat. Genet. 2023, 55, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Hessmann, E.; Buchholz, S.M.; Demir, I.E.; Singh, S.K.; Gress, T.M.; Ellenrieder, V.; Neesse, A. Microenvironmental Determinants of Pancreatic Cancer. Physiol. Rev. 2020, 100, 1707–1751. [Google Scholar] [CrossRef] [PubMed]
- Aran, D. Cell-Type Enrichment Analysis of Bulk Transcriptomes Using xCell. Methods Mol. Biol. 2020, 2120, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e1624. [Google Scholar] [CrossRef]
- Weusthof, C.; Burkart, S.; Semmelmayer, K.; Stogbauer, F.; Feng, B.; Khorani, K.; Bode, S.; Plinkert, P.; Plath, K.; Hess, J. Establishment of a Machine Learning Model for the Risk Assessment of Perineural Invasion in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2023, 24, 8938. [Google Scholar] [CrossRef]
- Garcia-Carracedo, D.; Cai, Y.; Qiu, W.; Saeki, K.; Friedman, R.A.; Lee, A.; Li, Y.; Goldberg, E.M.; Stratikopoulos, E.E.; Parsons, R.; et al. PIK3CA and p53 Mutations Promote 4NQO-Initated Head and Neck Tumor Progression and Metastasis in Mice. Mol. Cancer Res. 2020, 18, 822–834. [Google Scholar] [CrossRef]
- Mancusi, R.; Monje, M. The neuroscience of cancer. Nature 2023, 618, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Martyn, G.V.; Shurin, G.V.; Keskinov, A.A.; Bunimovich, Y.L.; Shurin, M.R. Schwann cells shape the neuro-immune environs and control cancer progression. Cancer Immunol. Immunother. 2019, 68, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Deborde, S.; Wong, R.J. The Role of Schwann Cells in Cancer. Adv. Biol. 2022, 6, e2200089. [Google Scholar] [CrossRef] [PubMed]
- Misso, G.; Di Martino, M.T.; De Rosa, G.; Farooqi, A.A.; Lombardi, A.; Campani, V.; Zarone, M.R.; Gulla, A.; Tagliaferri, P.; Tassone, P.; et al. Mir-34: A new weapon against cancer? Mol. Ther. Nucleic Acids 2014, 3, e194. [Google Scholar] [CrossRef]
- Jiang, C.; Cheng, Z.; Jiang, T.; Xu, Y.; Wang, B. MicroRNA-34a inhibits cell invasion and epithelial-mesenchymal transition via targeting AXL/PI3K/AKT/Snail signaling in nasopharyngeal carcinoma. Genes Genom. 2020, 42, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Batlle, E.; Massague, J. Transforming Growth Factor-beta Signaling in Immunity and Cancer. Immunity 2019, 50, 924–940. [Google Scholar] [CrossRef]
- Tauriello, D.V.F.; Sancho, E.; Batlle, E. Overcoming TGFbeta-mediated immune evasion in cancer. Nat. Rev. Cancer 2022, 22, 25–44. [Google Scholar] [CrossRef]
- Scholz, J.; Woolf, C.J. The neuropathic pain triad: Neurons, immune cells and glia. Nat. Neurosci. 2007, 10, 1361–1368. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.Y.; Wang, L.; Zhu, W.Y.; Zhang, G.; Su, Y.X. Lingual Denervation Improves the Efficacy of Anti-PD-1 Immunotherapy in Oral Squamous Cell Carcinomas by Downregulating TGFbeta Signaling. Cancer Res. Commun. 2024, 4, 418–430. [Google Scholar] [CrossRef]
- Han, S.; Lim, K.S.; Blackburn, B.J.; Yun, J.; Putnam, C.W.; Bull, D.A.; Won, Y.W. The Potential of Topoisomerase Inhibitor-Based Antibody-Drug Conjugates. Pharmaceutics 2022, 14, 1707. [Google Scholar] [CrossRef]
- Delgado, J.L.; Hsieh, C.M.; Chan, N.L.; Hiasa, H. Topoisomerases as anticancer targets. Biochem. J. 2018, 475, 373–398. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.C.; Chang, P.M.; Yang, M.H. Cisplatin/Tegafur/Uracil/Irinotecan Triple Combination Therapy for Recurrent/Metastatic Head and Neck Squamous Cell Carcinoma: A Phase I/II Clinical Study. Oncologist 2016, 21, 537–538. [Google Scholar] [CrossRef] [PubMed]
- Matias-Barrios, V.M.; Dong, X. The Implication of Topoisomerase II Inhibitors in Synthetic Lethality for Cancer Therapy. Pharmaceuticals 2023, 16, 94. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khorani, K.; Burkart, S.; Weusthof, C.; Han, R.; Liang, S.; Stögbauer, F.; Hess, J. Context-Dependent Regulation of Peripheral Nerve Abundance by the PI3K Pathway in the Tumor Microenvironment of Head and Neck Squamous Cell Carcinoma. Cells 2024, 13, 1033. https://doi.org/10.3390/cells13121033
Khorani K, Burkart S, Weusthof C, Han R, Liang S, Stögbauer F, Hess J. Context-Dependent Regulation of Peripheral Nerve Abundance by the PI3K Pathway in the Tumor Microenvironment of Head and Neck Squamous Cell Carcinoma. Cells. 2024; 13(12):1033. https://doi.org/10.3390/cells13121033
Chicago/Turabian StyleKhorani, Karam, Sebastian Burkart, Christopher Weusthof, Rui Han, Siyuan Liang, Fabian Stögbauer, and Jochen Hess. 2024. "Context-Dependent Regulation of Peripheral Nerve Abundance by the PI3K Pathway in the Tumor Microenvironment of Head and Neck Squamous Cell Carcinoma" Cells 13, no. 12: 1033. https://doi.org/10.3390/cells13121033




