Growing Role of 3D In Vitro Cell Cultures in the Study of Cellular and Molecular Mechanisms: Short Focus on Breast Cancer, Endometriosis, Liver and Infectious Diseases
Abstract
:1. Introduction
2. 3D Culture Models for Breast Cancer

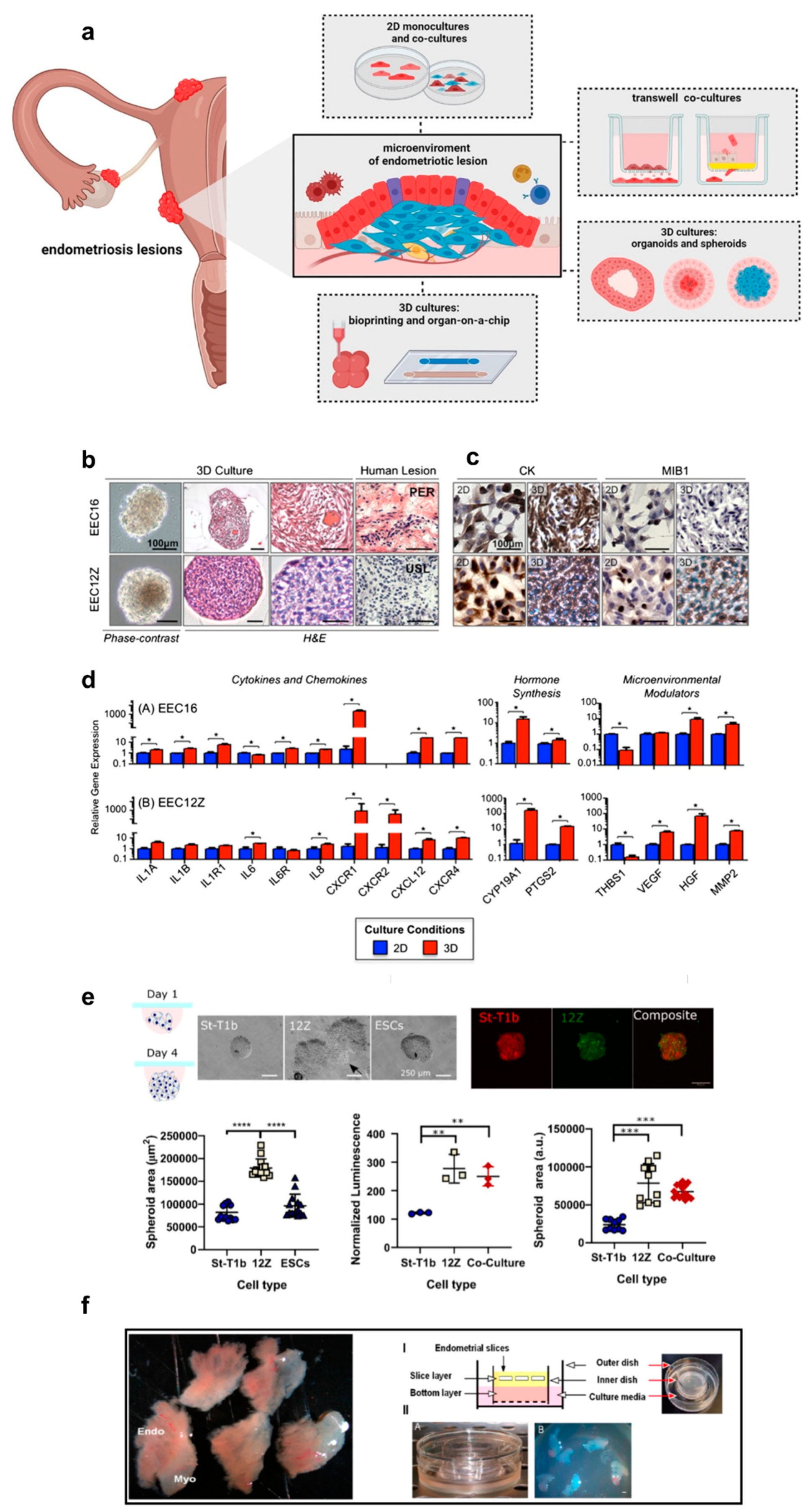
3. 3D Culture Models for Endometriosis
4. 3D Culture Models for the Liver
5. 3D Culture Models for Bacterial Infections
6. Discussion and Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pillai, S.; Munguia-Lopez, J.G.; Tran, S.D. Bioengineered Salivary Gland Microtissues—A Review of 3D Cellular Models and Their Applications. ACS Appl. Bio Mater. 2024, 7, 2620–2636. [Google Scholar] [CrossRef] [PubMed]
- Yousafzai, N.A.; El Khalki, L.; Wang, W.; Szpendyk, J.; Sossey-Alaoui, K. Advances in 3D Culture Models to Study Exosomes in Triple-Negative Breast Cancer. Cancers 2024, 16, 883. [Google Scholar] [CrossRef] [PubMed]
- Atat, O.E.; Farzaneh, Z.; Pourhamzeh, M.; Taki, F.; Abi-Habib, R.; Vosough, M.; El-Sibai, M. 3D Modeling in Cancer Studies. Hum. Cell 2022, 35, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Tofani, L.B.; Luiz, M.T.; Paes Dutra, J.A.; Abriata, J.P.; Chorilli, M. Three-Dimensional Culture Models: Emerging Platforms for Screening the Antitumoral Efficacy of Nanomedicines. Nanomedicine 2023, 18, 633–647. [Google Scholar] [CrossRef] [PubMed]
- Esparza, A.; Jimenez, N.; Borrego, E.A.; Browne, S.; Natividad-Diaz, S.L. Review: Human Stem Cell-Based 3D in Vitro Angiogenesis Models for Preclinical Drug Screening Applications. Mol. Biol. Rep. 2024, 51, 260. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Huang, S. Advances in the Application of 3D Tumor Models in Precision Oncology and Drug Screening. Front. Bioeng. Biotechnol. 2022, 10, 1021966. [Google Scholar] [CrossRef] [PubMed]
- Soeiro, J.F.; Sousa, F.L.; Monteiro, M.V.; Gaspar, V.M.; Silva, N.J.O.; Mano, J.F. Advances in Screening Hyperthermic Nanomedicines in 3D Tumor Models. Nanoscale Horiz. 2024, 9, 334–364. [Google Scholar] [CrossRef] [PubMed]
- Guller, A.; Igrunkova, A. Engineered Microenvironments for 3D Cell Culture and Regenerative Medicine: Challenges, Advances, and Trends. Bioengineering 2022, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Mulaudzi, P.E.; Abrahamse, H.; Crous, A. Insights on Three Dimensional Organoid Studies for Stem Cell Therapy in Regenerative Medicine. Stem Cell Rev. Rep. 2024, 20, 509–523. [Google Scholar] [CrossRef]
- Cacciamali, A.; Villa, R.; Dotti, S. 3D Cell Cultures: Evolution of an Ancient Tool for New Applications. Front. Physiol. 2022, 13, 836480. [Google Scholar] [CrossRef]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D Cell Cultures—A Comparison of Different Types of Cancer Cell Cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef]
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and Animal Models: Are 3D Cultures the Ideal Tool to Study Cancer-Microenvironment Interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef] [PubMed]
- d’Amora, M.; Giordani, S. The Utility of Zebrafish as a Model for Screening Developmental Neurotoxicity. Front. Neurosci. 2018, 12, 976. [Google Scholar] [CrossRef] [PubMed]
- Jubelin, C.; Muñoz-Garcia, J.; Griscom, L.; Cochonneau, D.; Ollivier, E.; Heymann, M.-F.; Vallette, F.M.; Oliver, L.; Heymann, D. Three-Dimensional in Vitro Culture Models in Oncology Research. Cell Biosci. 2022, 12, 155. [Google Scholar] [CrossRef] [PubMed]
- Guagliano, G.; Volpini, C.; Briatico-Vangosa, F.; Cornaglia, A.I.; Visai, L.; Petrini, P. Toward 3D-Bioprinted Models of the Liver to Boost Drug Development. Macromol. Biosci. 2022, 22, e2200264. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Dufour, J.M. Cell Lines: Valuable Tools or Useless Artifacts. Spermatogenesis 2012, 2, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Hawksworth, G.M. Advantages and Disadvantages of Using Human Cells for Pharmacological and Toxicological Studies. Hum. Exp. Toxicol. 1994, 13, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Ravi, M.; Paramesh, V.; Kaviya, S.R.; Anuradha, E.; Solomon, F.D.P. 3D Cell Culture Systems: Advantages and Applications. J. Cell. Physiol. 2015, 230, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Ravi, M.; Ramesh, A.; Pattabhi, A. Contributions of 3D Cell Cultures for Cancer Research. J. Cell. Physiol. 2017, 232, 2679–2697. [Google Scholar] [CrossRef]
- Gultian, K.A.; Gandhi, R.; Sarin, K.; Sladkova-Faure, M.; Zimmer, M.; de Peppo, G.M.; Vega, S.L. Human Induced Mesenchymal Stem Cells Display Increased Sensitivity to Matrix Stiffness. Sci. Rep. 2022, 12, 8483. [Google Scholar] [CrossRef]
- Septiana, W.L.; Noviantari, A.; Antarianto, R.D. Induced Pluripotent Stem Cells (Ipscs) Based Liver Organoid: The Benefits and Challenges. Cell Physiol. Biochem. 2023, 57, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Sumbalova Koledova, Z. 3D Cell Culture: Techniques for and Beyond Organoid Applications. In 3D Cell Culture: Methods and Protocols; Methods in Molecular Biology; Humana: New York, NY, USA, 2024; Volume 2764, pp. 1–12. [Google Scholar] [CrossRef]
- Nayak, P.; Bentivoglio, V.; Varani, M.; Signore, A. Three-Dimensional In Vitro Tumor Spheroid Models for Evaluation of Anticancer Therapy: Recent Updates. Cancers 2023, 15, 4846. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D Tumor Spheroids: An Overview on the Tools and Techniques Used for Their Analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Chen, X.; Dowbaj, A.M.; Sljukic, A.; Bratlie, K.; Lin, L.; Fong, E.L.S.; Balachander, G.M.; Chen, Z.; Soragni, A.; et al. Organoids. Nat. Rev. Methods Primers 2022, 2, 94. [Google Scholar] [CrossRef]
- LeSavage, B.L.; Suhar, R.A.; Broguiere, N.; Lutolf, M.P.; Heilshorn, S.C. Next-Generation Cancer Organoids. Nat. Mater. 2022, 21, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Kalyani, F.S.; Liu, L.; Cheng, T.; Chen, L. Tumor Organoids: Synergistic Applications, Current Challenges, and Future Prospects in Cancer Therapy. Cancer Commun. 2021, 41, 1331–1353. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Liu, L.; Ding, Y.; Dong, Y.; Ma, M. Advances in Biomimetic Hydrogels for Organoid Culture. Chem. Commun. 2023, 59, 9675–9686. [Google Scholar] [CrossRef] [PubMed]
- Rassomakhina, N.V.; Ryazanova, A.Y.; Likhov, A.R.; Bruskin, S.A.; Maloshenok, L.G.; Zherdeva, V.V. Tumor Organoids: The Era of Personalized Medicine. Biochemistry 2024, 89, S127–S147. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Jacob, F.; Song, M.M.; Nguyen, H.N.; Song, H.; Ming, G.-L. Generation of Human Brain Region-Specific Organoids Using a Miniaturized Spinning Bioreactor. Nat. Protoc. 2018, 13, 565–580. [Google Scholar] [CrossRef]
- Trossbach, M.; Åkerlund, E.; Langer, K.; Seashore-Ludlow, B.; Joensson, H.N. High-Throughput Cell Spheroid Production and Assembly Analysis by Microfluidics and Deep Learning. SLAS Technol. 2023, 28, 423–432. [Google Scholar] [CrossRef]
- Leung, C.M.; de Haan, P.; Ronaldson-Bouchard, K.; Kim, G.-A.; Ko, J.; Rho, H.S.; Chen, Z.; Habibovic, P.; Jeon, N.L.; Takayama, S.; et al. A Guide to the Organ-on-a-Chip. Nat. Rev. Methods Primers 2022, 2, 33. [Google Scholar] [CrossRef]
- Maharjan, S.; Ma, C.; Singh, B.; Kang, H.; Orive, G.; Yao, J.; Shrike Zhang, Y. Advanced 3D Imaging and Organoid Bioprinting for Biomedical Research and Therapeutic Applications. Adv. Drug Deliv. Rev. 2024, 208, 115237. [Google Scholar] [CrossRef] [PubMed]
- Limongi, T.; Guzzi, F.; Parrotta, E.; Candeloro, P.; Scalise, S.; Lucchino, V.; Gentile, F.; Tirinato, L.; Coluccio, M.L.; Torre, B.; et al. Microfluidics for 3D Cell and Tissue Cultures: Microfabricative and Ethical Aspects Updates. Cells 2022, 11, 1699. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lam, P.Y.; Jayaraman, A.; Han, A. Uniform Sized Cancer Spheroids Production Using Hydrogel-Based Droplet Microfluidics: A Review. Biomed. Microdevices 2024, 26, 26. [Google Scholar] [CrossRef] [PubMed]
- Yousafzai, M.S.; Hammer, J.A. Using Biosensors to Study Organoids, Spheroids and Organs-on-a-Chip: A Mechanobiology Perspective. Biosensors 2023, 13, 905. [Google Scholar] [CrossRef] [PubMed]
- Żuchowska, A.; Baranowska, P.; Flont, M.; Brzózka, Z.; Jastrzębska, E. Review: 3D Cell Models for Organ-on-a-Chip Applications. Anal. Chim. Acta 2024, 1301, 342413. [Google Scholar] [CrossRef] [PubMed]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-Dimensional Cell Culture Systems as an in Vitro Platform for Cancer and Stem Cell Modeling. World J. Stem Cells 2019, 11, 1065–1083. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, E. The Variety of 3D Breast Cancer Models for the Study of Tumor Physiology and Drug Screening. Int. J. Mol. Sci. 2023, 24, 7116. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Manduca, N.; Maccafeo, E.; De Maria, R.; Sistigu, A.; Musella, M. 3D Cancer Models: One Step Closer to in Vitro Human Studies. Front. Immunol. 2023, 14, 1175503. [Google Scholar] [CrossRef]
- Lu, X.; Lodi, A.; Konopleva, M.; Tiziani, S. Three-Dimensional Leukemia Co-Culture System for In Vitro High-Content Metabolomics Screening. SLAS Discov. 2019, 24, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Al-Kaabneh, B.; Frisch, B.; Aljitawi, O.S. The Potential Role of 3D In Vitro Acute Myeloid Leukemia Culture Models in Understanding Drug Resistance in Leukemia Stem Cells. Cancers 2022, 14, 5252. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, M.A.G.; Xavier, C.P.R.; Pereira, R.F.; Petrikaitė, V.; Vasconcelos, M.H. 3D Cell Culture Models as Recapitulators of the Tumor Microenvironment for the Screening of Anti-Cancer Drugs. Cancers 2021, 14, 190. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, D.P.; Matias, A.T.; Braga, S.; Jacinto, A.; Cabral, M.G. Establishment of a 3D Co-Culture With MDA-MB-231 Breast Cancer Cell Line and Patient-Derived Immune Cells for Application in the Development of Immunotherapies. Front. Oncol. 2020, 10, 1543. [Google Scholar] [CrossRef] [PubMed]
- Tevlek, A. The Role of Decellularized Cell Derived Extracellular Matrix in the Establishment and Culture Ofin Vitrobreast Cancer Tumor Model. Biomed. Mater. 2024, 19, 025037. [Google Scholar] [CrossRef] [PubMed]
- Kumano, K.; Nakahashi, H.; Louphrasitthiphol, P.; Kuroda, Y.; Miyazaki, Y.; Shimomura, O.; Hashimoto, S.; Akashi, Y.; Mathis, B.J.; Kim, J.; et al. Hypoxia at 3D Organoid Establishment Selects Essential Subclones within Heterogenous Pancreatic Cancer. Front. Cell Dev. Biol. 2024, 12, 1327772. [Google Scholar] [CrossRef]
- Ayuso, J.M.; Virumbrales-Munoz, M.; McMinn, P.H.; Rehman, S.; Gomez, I.; Karim, M.R.; Trusttchel, R.; Wisinski, K.B.; Beebe, D.J.; Skala, M.C. Tumor-on-a-Chip: A Microfluidic Model to Study Cell Response to Environmental Gradients. Lab Chip 2019, 19, 3461–3471. [Google Scholar] [CrossRef] [PubMed]
- Major, G.; Ahn, M.; Cho, W.-W.; Santos, M.; Wise, J.; Phillips, E.; Wise, S.G.; Jang, J.; Rnjak-Kovacina, J.; Woodfield, T.; et al. Programming Temporal Stiffness Cues within Extracellular Matrix Hydrogels for Modelling Cancer Niches. Mater. Today Bio 2024, 25, 101004. [Google Scholar] [CrossRef]
- Briem, E.; Ingthorsson, S.; Traustadottir, G.A.; Hilmarsdottir, B.; Gudjonsson, T. Application of the D492 Cell Lines to Explore Breast Morphogenesis, EMT and Cancer Progression in 3D Culture. J. Mammary Gland. Biol. Neoplasia 2019, 24, 139–147. [Google Scholar] [CrossRef]
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S.; et al. Current and Future Burden of Breast Cancer: Global Statistics for 2020 and 2040. Breast 2022, 66, 15–23. [Google Scholar] [CrossRef]
- Barzaman, K.; Karami, J.; Zarei, Z.; Hosseinzadeh, A.; Kazemi, M.H.; Moradi-Kalbolandi, S.; Safari, E.; Farahmand, L. Breast Cancer: Biology, Biomarkers, and Treatments. Int. Immunopharmacol. 2020, 84, 106535. [Google Scholar] [CrossRef] [PubMed]
- Bahcecioglu, G.; Basara, G.; Ellis, B.W.; Ren, X.; Zorlutuna, P. Breast Cancer Models: Engineering the Tumor Microenvironment. Acta Biomater. 2020, 106, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Yu, P.; Tang, J. Characterization of Triple-Negative Breast Cancer MDA-MB-231 Cell Spheroid Model. Onco Targets Ther. 2020, 13, 5395–5405. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Sang, Y.; Song, X.; Zhang, D.; Wang, L.; Zhao, W.; Liang, Y.; Zhang, N.; Yang, Q. Exosomal miR-500a-5p Derived from Cancer-Associated Fibroblasts Promotes Breast Cancer Cell Proliferation and Metastasis through Targeting USP28. Theranostics 2021, 11, 3932–3947. [Google Scholar] [CrossRef] [PubMed]
- Watzling, M.; Klaus, L.; Weidemeier, T.; Horder, H.; Ebert, R.; Blunk, T.; Bauer-Kreisel, P. Three-Dimensional Breast Cancer Model to Investigate CCL5/CCR1 Expression Mediated by Direct Contact between Breast Cancer Cells and Adipose-Derived Stromal Cells or Adipocytes. Cancers 2023, 15, 3501. [Google Scholar] [CrossRef] [PubMed]
- Jahin, I.; Phillips, T.; Marcotti, S.; Gorey, M.-A.; Cox, S.; Parsons, M. Extracellular Matrix Stiffness Activates Mechanosensitive Signals but Limits Breast Cancer Cell Spheroid Proliferation and Invasion. Front. Cell Dev. Biol. 2023, 11, 1292775. [Google Scholar] [CrossRef] [PubMed]
- Jafarpour, S.; Ahmadi, S.; Mokarian, F.; Sharifi, M.; Ghobakhloo, S.; Yazdi, M.; Nedaeinia, R.; Salehi, R. MSC-Derived Exosomes Enhance the Anticancer Activity of Drugs in 3D Spheroid of Breast Cancer Cells. J. Drug Deliv. Sci. Technol. 2024, 92, 105375. [Google Scholar] [CrossRef]
- Shibuya, N.; Kakeji, Y.; Shimono, Y. MicroRNA-93 Targets WASF3 and Functions as a Metastasis Suppressor in Breast Cancer. Cancer Sci. 2020, 111, 2093–2103. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhong, F.; Sun, S.; Ou, X.; Yuan, J.; Zhu, J.; Zeng, Z. Tamoxifen Induces Ferroptosis in MCF-7 Organoid. J. Cancer Res. Ther. 2023, 19, 1627–1635. [Google Scholar] [CrossRef]
- Engel, M.; Belfiore, L.; Aghaei, B.; Sutija, M. Enabling High Throughput Drug Discovery in 3D Cell Cultures through a Novel Bioprinting Workflow. SLAS Technol. 2022, 27, 32–38. [Google Scholar] [CrossRef]
- Jaiswal, C.; Mandal, B.B. A 3D In Vitro Triculture Hybrid Model Recapitulating Tumor Stromal Interaction of Triple-Negative Breast Cancer as a High Throughput Anticancer Drug Screening Platform. Adv. Ther. 2024, 2300450. [Google Scholar] [CrossRef]
- Brancato, V.; Kundu, B.; Oliveira, J.M.; Correlo, V.M.; Reis, R.L.; Kundu, S.C. Tumor-Stroma Interactions Alter the Sensitivity of Drug in Breast Cancer. Front. Mater. 2020, 7, 116. [Google Scholar] [CrossRef]
- Liverani, C.; De Vita, A.; Spadazzi, C.; Miserocchi, G.; Cocchi, C.; Bongiovanni, A.; De Lucia, A.; La Manna, F.; Fabbri, F.; Tebaldi, M.; et al. Lineage-Specific Mechanisms and Drivers of Breast Cancer Chemoresistance Revealed by 3D Biomimetic Culture. Mol. Oncol. 2022, 16, 921–939. [Google Scholar] [CrossRef] [PubMed]
- Itah, Z.; Chaudhry, S.; Raju Ponny, S.; Aydemir, O.; Lee, A.; Cavanagh-Kyros, J.; Tournier, C.; Muller, W.J.; Davis, R.J. HER2-Driven Breast Cancer Suppression by the JNK Signaling Pathway. Proc. Natl. Acad. Sci. USA 2023, 120, e2218373120. [Google Scholar] [CrossRef]
- Liu, C.; Li, M.; Dong, Z.-X.; Jiang, D.; Li, X.; Lin, S.; Chen, D.; Zou, X.; Zhang, X.-D.; Luker, G.D. Heterogeneous Microenvironmental Stiffness Regulates Pro-Metastatic Functions of Breast Cancer Cells. Acta Biomater. 2021, 131, 326–340. [Google Scholar] [CrossRef]
- Truong, D.D.; Kratz, A.; Park, J.G.; Barrientos, E.S.; Saini, H.; Nguyen, T.; Pockaj, B.; Mouneimne, G.; LaBaer, J.; Nikkhah, M. A Human Organotypic Microfluidic Tumor Model Permits Investigation of the Interplay between Patient-Derived Fibroblasts and Breast Cancer Cells. Cancer Res. 2019, 79, 3139–3151. [Google Scholar] [CrossRef]
- Yang, D.; Liu, J.; Qian, H.; Zhuang, Q. Cancer-Associated Fibroblasts: From Basic Science to Anticancer Therapy. Exp. Mol. Med. 2023, 55, 1322–1332. [Google Scholar] [CrossRef]
- Zheng, G.-D.; Xu, Z.-Y.; Hu, C.; Lv, H.; Xie, H.-X.; Huang, T.; Zhang, Y.-Q.; Chen, G.-P.; Fu, Y.-F.; Cheng, X.-D. Exosomal miR-590-5p in Serum as a Biomarker for the Diagnosis and Prognosis of Gastric Cancer. Front. Mol. Biosci. 2021, 8, 636566. [Google Scholar] [CrossRef]
- Manevich, L.; Okita, Y.; Okano, Y.; Sugasawa, T.; Kawanishi, K.; Poullikkas, T.; Dang Cao, L.T.L.; Zheng, L.; Nakayama, M.; Matsumoto, S.; et al. Glycoprotein NMB Promotes Tumor Formation and Malignant Progression of Laryngeal Squamous Cell Carcinoma. Cancer Sci. 2022, 113, 3244–3254. [Google Scholar] [CrossRef]
- Zhang, F.; Liu, S. Mechanistic Insights of Adipocyte Metabolism in Regulating Breast Cancer Progression. Pharmacol. Res. 2020, 155, 104741. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Takagi, K.; Narita, K.; Miki, Y.; Onodera, Y.; Miyashita, M.; Sasano, H.; Suzuki, T. Stromal CCL5 Promotes Breast Cancer Progression by Interacting with CCR3 in Tumor Cells. Int. J. Mol. Sci. 2021, 22, 1918. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, S.; Mahadik, P.; Shetty, O.; Sen, S. ECM Stiffness-Tuned Exosomes Drive Breast Cancer Motility through Thrombospondin-1. Biomaterials 2021, 279, 121185. [Google Scholar] [CrossRef] [PubMed]
- Stroka, K.M.; Wong, B.S.; Shriver, M.; Phillip, J.M.; Wirtz, D.; Kontrogianni-Konstantopoulos, A.; Konstantopoulos, K. Loss of Giant Obscurins Alters Breast Epithelial Cell Mechanosensing of Matrix Stiffness. Oncotarget 2017, 8, 54004–54020. [Google Scholar] [CrossRef] [PubMed]
- Balachander, G.M.; Kotcherlakota, R.; Nayak, B.; Kedaria, D.; Rangarajan, A.; Chatterjee, K. 3D Tumor Models for Breast Cancer: Whither We Are and What We Need. ACS Biomater. Sci. Eng. 2021, 7, 3470–3486. [Google Scholar] [CrossRef] [PubMed]
- Micalet, A.; Moeendarbary, E.; Cheema, U. 3D In Vitro Models for Investigating the Role of Stiffness in Cancer Invasion. ACS Biomater. Sci. Eng. 2021, 9, 3729–3741. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Tian, M.; Pei, Q.; Tan, F.; Pei, H. Extracellular Matrix Stiffness: New Areas Affecting Cell Metabolism. Front. Oncol. 2021, 11, 631991. [Google Scholar] [CrossRef] [PubMed]
- Ponce, I.; Garrido, N.; Tobar, N.; Melo, F.; Smith, P.C.; Martínez, J. Matrix Stiffness Modulates Metabolic Interaction between Human Stromal and Breast Cancer Cells to Stimulate Epithelial Motility. Metabolites 2021, 11, 432. [Google Scholar] [CrossRef] [PubMed]
- Nikdouz, A.; Orso, F. Emerging Roles of 3D-Culture Systems in Tackling Tumor Drug Resistance. Cancer Drug Resist. 2023, 6, 788–804. [Google Scholar] [CrossRef] [PubMed]
- Nowacka, M.; Sterzynska, K.; Andrzejewska, M.; Nowicki, M.; Januchowski, R. Drug Resistance Evaluation in Novel 3D in Vitro Model. Biomed. Pharmacother. 2021, 138, 111536. [Google Scholar] [CrossRef]
- Al-Malky, H.S.; Al Harthi, S.E.; Osman, A.-M.M. Major Obstacles to Doxorubicin Therapy: Cardiotoxicity and Drug Resistance. J. Oncol. Pharm. Pract. 2020, 26, 434–444. [Google Scholar] [CrossRef]
- Fontana, F.; Raimondi, M.; Marzagalli, M.; Sommariva, M.; Gagliano, N.; Limonta, P. Three-Dimensional Cell Cultures as an In Vitro Tool for Prostate Cancer Modeling and Drug Discovery. Int. J. Mol. Sci. 2020, 21, 6806. [Google Scholar] [CrossRef] [PubMed]
- Gołąbek-Grenda, A.; Olejnik, A. In Vitro Modeling of Endometriosis and Endometriotic Microenvironment-Challenges and Recent Advances. Cell Signal. 2022, 97, 110375. [Google Scholar] [CrossRef] [PubMed]
- Volpini, C.; Bloise, N.; Dominoni, M.; Barra, F.; Vellone, V.G.; Minzioni, P.; Gardella, B.; Ferrero, S.; Visai, L. The Nano-Revolution in the Diagnosis and Treatment of Endometriosis. Nanoscale 2023, 15, 17313–17325. [Google Scholar] [CrossRef] [PubMed]
- Laganà, A.S.; Garzon, S.; Götte, M.; Viganò, P.; Franchi, M.; Ghezzi, F.; Martin, D.C. The Pathogenesis of Endometriosis: Molecular and Cell Biology Insights. Int. J. Mol. Sci. 2019, 20, 5615. [Google Scholar] [CrossRef] [PubMed]
- Brueggmann, D.; Templeman, C.; Starzinski-Powitz, A.; Rao, N.P.; Gayther, S.A.; Lawrenson, K. Novel Three-Dimensional in Vitro Models of Ovarian Endometriosis. J. Ovarian Res. 2014, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Stejskalová, A.; Fincke, V.; Nowak, M.; Schmidt, Y.; Borrmann, K.; von Wahlde, M.-K.; Schäfer, S.D.; Kiesel, L.; Greve, B.; Götte, M. Collagen I Triggers Directional Migration, Invasion and Matrix Remodeling of Stroma Cells in a 3D Spheroid Model of Endometriosis. Sci. Rep. 2021, 11, 4115. [Google Scholar] [CrossRef] [PubMed]
- Wendel, J.R.H.; Wang, X.; Smith, L.J.; Hawkins, S.M. Three-Dimensional Biofabrication Models of Endometriosis and the Endometriotic Microenvironment. Biomedicines 2020, 8, 525. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Burns, G.W.; Joshi, N.R.; Arora, R.; Kim, J.J.; Fazleabas, A.T. Spheroids as a Model for Endometriotic Lesions. JCI Insight 2023, 8, e160815. [Google Scholar] [CrossRef] [PubMed]
- Muruganandan, S.; Fan, X.; Dhal, S.; Nayak, N.R. Development of A 3D Tissue Slice Culture Model for the Study of Human Endometrial Repair and Regeneration. Biomolecules 2020, 10, 136. [Google Scholar] [CrossRef]
- Ahn, J.; Yoon, M.-J.; Hong, S.-H.; Cha, H.; Lee, D.; Koo, H.S.; Ko, J.-E.; Lee, J.; Oh, S.; Jeon, N.L.; et al. Three-Dimensional Microengineered Vascularised Endometrium-on-a-Chip. Hum. Reprod. 2021, 36, 2720–2731. [Google Scholar] [CrossRef]
- Esfandiari, F.; Favaedi, R.; Heidari-Khoei, H.; Chitsazian, F.; Yari, S.; Piryaei, A.; Ghafari, F.; Baharvand, H.; Shahhoseini, M. Insight into Epigenetics of Human Endometriosis Organoids: DNA Methylation Analysis of HOX Genes and Their Cofactors. Fertil. Steril. 2021, 115, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Nap, A.W.; Dunselman, G.A.J.; de Goeij, A.F.P.M.; Evers, J.L.H.; Groothuis, P.G. Inhibiting MMP Activity Prevents the Development of Endometriosis in the Chicken Chorioallantoic Membrane Model. Hum. Reprod. 2004, 19, 2180–2187. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson Teague, E.M.C.; Van der Hoek, K.H.; Van der Hoek, M.B.; Perry, N.; Wagaarachchi, P.; Robertson, S.A.; Print, C.G.; Hull, L.M. MicroRNA-Regulated Pathways Associated with Endometriosis. Mol. Endocrinol. 2009, 23, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Guagliano, G.; Volpini, C.; Sardelli, L.; Bloise, N.; Briatico-Vangosa, F.; Cornaglia, A.I.; Dotti, S.; Villa, R.; Visai, L.; Petrini, P. Hep3Gel: A Shape-Shifting Extracellular Matrix-Based, Three-Dimensional Liver Model Adaptable to Different Culture Systems. ACS Biomater. Sci. Eng. 2023, 9, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.; Becattini, B.; Solinas, G. Insulin Signaling and Glucose Metabolism in Different Hepatoma Cell Lines Deviate from Hepatocyte Physiology toward a Convergent Aberrant Phenotype. Sci. Rep. 2020, 10, 12031. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Han, R.; Chang, S.; Ni, J.; Hunziker, W.; Goryachev, A.B.; Ong, S.H.; Yu, H. Improved Hepatocyte Excretory Function by Immediate Presentation of Polarity Cues. Tissue Eng. 2006, 12, 2181–2191. [Google Scholar] [CrossRef] [PubMed]
- Donato, M.T.; Tolosa, L.; Gómez-Lechón, M.J. Culture and Functional Characterization of Human Hepatoma HepG2 Cells. Methods Mol. Biol. 2015, 1250, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Khati, V.; Ramachandraiah, H.; Pati, F.; Svahn, H.A.; Gaudenzi, G.; Russom, A. 3D Bioprinting of Multi-Material Decellularized Liver Matrix Hydrogel at Physiological Temperatures. Biosensors 2022, 12, 521. [Google Scholar] [CrossRef] [PubMed]
- Khati, V.; Turkki, J.A.; Ramachandraiah, H.; Pati, F.; Gaudenzi, G.; Russom, A. Indirect 3D Bioprinting of a Robust Trilobular Hepatic Construct with Decellularized Liver Matrix Hydrogel. Bioengineering 2022, 9, 603. [Google Scholar] [CrossRef]
- Sun, L.; Yang, H.; Wang, Y.; Zhang, X.; Jin, B.; Xie, F.; Jin, Y.; Pang, Y.; Zhao, H.; Lu, X.; et al. Application of a 3D Bioprinted Hepatocellular Carcinoma Cell Model in Antitumor Drug Research. Front. Oncol. 2020, 10, 878. [Google Scholar] [CrossRef]
- Ramaiahgari, S.C.; den Braver, M.W.; Herpers, B.; Terpstra, V.; Commandeur, J.N.M.; van de Water, B.; Price, L.S. A 3D in Vitro Model of Differentiated HepG2 Cell Spheroids with Improved Liver-like Properties for Repeated Dose High-Throughput Toxicity Studies. Arch. Toxicol. 2014, 88, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Miyamoto, H.; Tokunaga, A.; Fumoto, S.; Tanaka, T.; Nishida, K. Evaluation of mRNA Expression of Drug-Metabolizing Enzymes in Acetaminophen-Induced Hepatotoxicity Using a Three-Dimensional Hepatocyte Culture System. Xenobiotica 2020, 50, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Cuvellier, M.; Ezan, F.; Oliveira, H.; Rose, S.; Fricain, J.-C.; Langouët, S.; Legagneux, V.; Baffet, G. 3D Culture of HepaRG Cells in GelMa and Its Application to Bioprinting of a Multicellular Hepatic Model. Biomaterials 2021, 269, 120611. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Hori, Y.; Yamamoto, T.; Urashima, T.; Ohara, Y.; Tanaka, H. 3D Spheroid Cultures Improve the Metabolic Gene Expression Profiles of HepaRG Cells. Biosci. Rep. 2015, 35, e00208. [Google Scholar] [CrossRef] [PubMed]
- Pihl, A.F.; Offersgaard, A.F.; Mathiesen, C.K.; Prentoe, J.; Fahnøe, U.; Krarup, H.; Bukh, J.; Gottwein, J.M. High Density Huh7.5 Cell Hollow Fiber Bioreactor Culture for High-Yield Production of Hepatitis C Virus and Studies of Antivirals. Sci. Rep. 2018, 8, 17505. [Google Scholar] [CrossRef]
- Wang, G.; Zheng, Y.; Wang, Y.; Cai, Z.; Liao, N.; Liu, J.; Zhang, W. Co-Culture System of Hepatocytes and Endothelial Cells: Two in Vitro Approaches for Enhancing Liver-Specific Functions of Hepatocytes. Cytotechnology 2018, 70, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Taymour, R.; Kilian, D.; Ahlfeld, T.; Gelinsky, M.; Lode, A. 3D Bioprinting of Hepatocytes: Core-Shell Structured Co-Cultures with Fibroblasts for Enhanced Functionality. Sci. Rep. 2021, 11, 5130. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Johnson, B.; Seirup, M.; Ardalani, H.; Duffin, B.; Barrett-Wilt, G.A.; Stewart, R.; Thomson, J.A. Co-Culture with Mouse Embryonic Fibroblasts Improves Maintenance of Metabolic Function of Human Small Hepatocyte Progenitor Cells. Curr. Res. Toxicol. 2020, 1, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Ware, B.R.; Liu, J.S.; Monckton, C.P.; Ballinger, K.R.; Khetani, S.R. Micropatterned Coculture With 3T3-J2 Fibroblasts Enhances Hepatic Functions and Drug Screening Utility of HepaRG Cells. Toxicol. Sci. 2021, 181, 90–104. [Google Scholar] [CrossRef]
- Lee, H.W.; Kook, Y.-M.; Lee, H.J.; Park, H.; Koh, W.-G. A Three-Dimensional Co-Culture of HepG2 Spheroids and Fibroblasts Using Double-Layered Fibrous Scaffolds Incorporated with Hydrogel Micropatterns. RSC Adv. 2014, 4, 61005–61011. [Google Scholar] [CrossRef]
- Kiamehr, M.; Heiskanen, L.; Laufer, T.; Düsterloh, A.; Kahraman, M.; Käkelä, R.; Laaksonen, R.; Aalto-Setälä, K. Dedifferentiation of Primary Hepatocytes Is Accompanied with Reorganization of Lipid Metabolism Indicated by Altered Molecular Lipid and miRNA Profiles. Int. J. Mol. Sci. 2019, 20, 2910. [Google Scholar] [CrossRef] [PubMed]
- Handin, N.; Mickols, E.; Ölander, M.; Rudfeldt, J.; Blom, K.; Nyberg, F.; Senkowski, W.; Urdzik, J.; Maturi, V.; Fryknäs, M.; et al. Conditions for Maintenance of Hepatocyte Differentiation and Function in 3D Cultures. iScience 2021, 24, 103235. [Google Scholar] [CrossRef]
- Nagata, S.; Ozawa, F.; Nie, M.; Takeuchi, S. 3D Culture of Functional Human iPSC-Derived Hepatocytes Using a Core-Shell Microfiber. PLoS ONE 2020, 15, e0234441. [Google Scholar] [CrossRef] [PubMed]
- Fattahi, P.; de Hoyos-Vega, J.M.; Choi, J.H.; Duffy, C.D.; Gonzalez-Suarez, A.M.; Ishida, Y.; Nguyen, K.M.; Gwon, K.; Peterson, Q.P.; Saito, T.; et al. Guiding Hepatic Differentiation of Pluripotent Stem Cells Using 3D Microfluidic Co-Cultures with Human Hepatocytes. Cells 2023, 12, 1982. [Google Scholar] [CrossRef] [PubMed]
- Meier, F.; Freyer, N.; Brzeszczynska, J.; Knöspel, F.; Armstrong, L.; Lako, M.; Greuel, S.; Damm, G.; Ludwig-Schwellinger, E.; Deschl, U.; et al. Hepatic Differentiation of Human iPSCs in Different 3D Models: A Comparative Study. Int. J. Mol. Med. 2017, 40, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Pettinato, G.; Ramanathan, R.; Fisher, R.A.; Mangino, M.J.; Zhang, N.; Wen, X. Scalable Differentiation of Human iPSCs in a Multicellular Spheroid-Based 3D Culture into Hepatocyte-like Cells through Direct Wnt/β-Catenin Pathway Inhibition. Sci. Rep. 2016, 6, 32888. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Qi, K.; Wang, L.; Yu, X.; Zhang, Q.; Yu, L.; Wang, L.; Yang, C.; Fan, L. E3 Ubiquitin Ligase CHIP Interacts with Transferrin Receptor 1 for Degradation and Promotes Cell Proliferation through Inhibiting Ferroptosis in Hepatocellular Carcinoma. Cell Signal. 2024, 118, 111148. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Ding, C.; Yu, T.; Liu, B.; Tang, W.; Wang, Z.; Tang, X.; Liang, G.; Peng, J.; Zhang, X.; et al. SIRT7 Promotes Hippo/YAP Activation and Cancer Cell Proliferation in Hepatocellular Carcinoma via Suppressing MST1. Cancer Sci. 2024, 115, 1209–1223. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhao, Z.; Zhao, Y.; Zhao, J.; Xia, L.; Xia, Q. Macroscopic Inhibition of DNA Damage Repair Pathways by Targeting AP-2α with LEI110 Eradicates Hepatocellular Carcinoma. Commun. Biol. 2024, 7, 342. [Google Scholar] [CrossRef]
- Lan, T.; Li, P.; Zhang, S.-J.; Liu, S.-Y.; Zeng, X.-X.; Chai, F.; Tong, Y.-H.; Mao, Z.-J.; Wang, S.-W. Paeoniflorin Promotes PPARγ Expression to Suppress HSCs Activation by Inhibiting EZH2-Mediated Histone H3K27 Trimethylation. Phytomedicine 2024, 128, 155477. [Google Scholar] [CrossRef]
- Henneke, P.; Golenbock, D.T. Phagocytosis, Innate Immunity, and Host-Pathogen Specificity. J. Exp. Med. 2004, 199, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Fasciano, A.C.; Mecsas, J.; Isberg, R.R. New Age Strategies to Reconstruct Mucosal Tissue Colonization and Growth in Cell Culture Systems. Microbiol. Spectr. 2019, 7, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.D.; Raghani, N.R.; Chorawala, M.R.; Singh, S.; Prajapati, B.G. Harnessing Three-Dimensional (3D) Cell Culture Models for Pulmonary Infections: State of the Art and Future Directions. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2023, 396, 2861–2880. [Google Scholar] [CrossRef] [PubMed]
- Rajan, D.; Gaston, K.A.; McCracken, C.E.; Erdman, D.D.; Anderson, L.J. Response to Rhinovirus Infection by Human Airway Epithelial Cells and Peripheral Blood Mononuclear Cells in an in Vitro Two-Chamber Tissue Culture System. PLoS ONE 2013, 8, e66600. [Google Scholar] [CrossRef] [PubMed]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Liu, T.; Liu, Q.; Lian, L.; Tang, G.; Mille, L.S.; García, F.R.; Engstrand, L.; Zhang, Y.S.; Du, J. A 3D Bioprinted Gut Anaerobic Model for Studying Bacteria-Host Interactions. Research 2023, 6, 0058. [Google Scholar] [CrossRef] [PubMed]
- Łaniewski, P.; Herbst-Kralovetz, M.M. Bacterial Vaginosis and Health-Associated Bacteria Modulate the Immunometabolic Landscape in 3D Model of Human Cervix. NPJ Biofilms Microbiomes 2021, 7, 88. [Google Scholar] [CrossRef] [PubMed]
- Calatayud, M.; Dezutter, O.; Hernandez-Sanabria, E.; Hidalgo-Martinez, S.; Meysman, F.J.R.; Van De Wiele, T. Development of a Host-Microbiome Model of the Small Intestine. FASEB J. 2019, 33, 3985–3996. [Google Scholar] [CrossRef] [PubMed]
- Roodsant, T.; Navis, M.; Aknouch, I.; Renes, I.B.; van Elburg, R.M.; Pajkrt, D.; Wolthers, K.C.; Schultsz, C.; van der Ark, K.C.H.; Sridhar, A.; et al. A Human 2D Primary Organoid-Derived Epithelial Monolayer Model to Study Host-Pathogen Interaction in the Small Intestine. Front. Cell. Infect. Microbiol. 2020, 10, 272. [Google Scholar] [CrossRef]
- Koestler, B.J.; Ward, C.M.; Fisher, C.R.; Rajan, A.; Maresso, A.W.; Payne, S.M. Human Intestinal Enteroids as a Model System of Shigella Pathogenesis. Infect. Immun. 2019, 87, e00733-18. [Google Scholar] [CrossRef]
- Barrila, J.; Yang, J.; Franco Meléndez, K.P.; Yang, S.; Buss, K.; Davis, T.J.; Aronow, B.J.; Bean, H.D.; Davis, R.R.; Forsyth, R.J.; et al. Spaceflight Analogue Culture Enhances the Host-Pathogen Interaction Between Salmonella and a 3-D Biomimetic Intestinal Co-Culture Model. Front. Cell. Infect. Microbiol. 2022, 12, 705647. [Google Scholar] [CrossRef] [PubMed]
- Barua, N.; Huang, L.; Li, C.; Yang, Y.; Luo, M.; Wei, W.I.; Wong, K.T.; Lo, N.W.S.; Kwok, K.O.; Ip, M. Comparative Study of Two-Dimensional (2D) vs. Three-Dimensional (3D) Organotypic Kertatinocyte-Fibroblast Skin Models for Staphylococcus Aureus (MRSA) Infection. Int. J. Mol. Sci. 2022, 23, 299. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, Invasion and Evasion: The Many Functions of the Surface Proteins of Staphylococcus Aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Schwarz-Linek, U.; Höök, M.; Potts, J.R. Fibronectin-Binding Proteins of Gram-Positive Cocci. Microbes Infect. 2006, 8, 2291–2298. [Google Scholar] [CrossRef] [PubMed]
- Anifantaki, F.; Boutas, I.; Kalampokas, T.; Kalampokas, E.; Sofoudis, C.; Salakos, N. Association of Endometriosis and Breast Cancer: Mini Review of the Literature. Arch. Gynecol. Obstet. 2016, 293, 5–10. [Google Scholar] [CrossRef] [PubMed]
- van Elsland, D.; Neefjes, J. Bacterial Infections and Cancer. EMBO Rep. 2018, 19, e46632. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, K.; Sampath, V.; Umar, S. Bacterial Infections and Cancer: Exploring This Association and Its Implications for Cancer Patients. Int. J. Mol. Sci. 2023, 24, 3110. [Google Scholar] [CrossRef]
- Peneda Pacheco, D.; Bertoglio, F.; Butnarasu, C.; Suarez Vargas, N.; Guagliano, G.; Ziccarelli, A.; Briatico-Vangosa, F.; Ruzzi, V.; Buzzaccaro, S.; Piazza, R.; et al. Heterogeneity Governs 3D-Cultures of Clinically Relevant Microbial Communities. Adv. Funct. Mater. 2023, 33, 2306116. [Google Scholar] [CrossRef]
- Temple, J.; Velliou, E.; Shehata, M.; Lévy, R.; Gupta, P. Current Strategies with Implementation of Three-Dimensional Cell Culture: The Challenge of Quantification. Interface Focus 2022, 12, 20220019. [Google Scholar] [CrossRef]

| Type of Platform | Technical Features | Cells | End-Point | Ref. |
|---|---|---|---|---|
| Spheroid | Agarose-based | MDA-MB-231 | The cell invasion capacity increased compared to that of the 2D model; the E-cadherin expression was down-regulated while N-cadherin was up-regulated | [54] |
| Spheroid | Matrigel | MDA-MB-231; MCF-7; CAFs; NFs | miR-500a-5p was highly expressed in breast cancer cell lines. CAFs-derived exosomes promoted breast cancer progression and metastasis via miR-500a-5p by binding USP28 | [55] |
| Spheroid | Agarose molds, which were cast in MicroTissues®3D Petri Dishes® | ASCs; adipocytes; MDA-MB-231; MCF-7 | The model showed a direct interaction between breast cancer cells, adipocytes and ASCs. Gene expression revealed a remarkable up-regulation of CCL5 and its receptor CCR1. The CCL5/CCR1 axis promoted tumor progression when the cells were in close contact in 3D TME | [56] |
| Spheroid | 3D collagen gels with increasing stiffness | MCF-7; HCC1954 | The breast cancer spheroids model evidenced how ECM stiffness influenced cell invasion capacity. In the lower stiffness, ERK activity was increased and operated upstream of the YAP signaling, determining ECM remodeling through MMPs | [57] |
| Spheroid | Agarose | MDA-MB-231; HFFF2; BT474 | Exosomes secreted by ADMSCs were able to deliver anti-cancer drugs, using a low concentration of Cis and PTX. Exosomes loaded with drugs increased the chemotherapy response by reducing cell viability and activating the apoptosis pathway | [58] |
| Organoid | Matrigel | MDA-MB-231; T-47D; MCF7 | miR-93 was able to affect cell viability. The constitutive up-regulation of miR-93 suppressed invasion and the metastasis process reducing WASF3 expression | [59] |
| Organoid | Resuspension with matrix glue in 48-well plates | MCF-7 | The tamoxifen anticancer drug inhibited the growth of MCF-7 organoids, inducing ferropstosis. Erastin, a ferrosptosis activator, enhanced the sensitivity of TAMR cells to the anticancer drug | [60] |
| Bioprinted Hydrogel | Small Plug Cell Model with PEG bionk formulation | MDA-MB-231; MCF-7; NHDF | DOX treatment determined cell death by entering in the 3D hydrogel and altering ERK1-ERK2 phosphorylation. Pharmacological treatment increased ATP production. GSKβ3 phosphorylation was decreased due to DOX-induced cell stress. | [61] |
| Hydrogel | 3D silk scaffold encapsulated in a GelMA hydrogel-based hybrid system | h-ADMSCs; HUVEC; MDA-MB-231 | TC-TBNC recapitulated, in a realistic way, TME. DOX and Cis treatments determined cell death through an apoptosis process: Bcl-2 was down-regulated while Bax was up-regulated. The model showed an increase in ABCC1 expression in the Cis-treated hydrogel compared to the DOX -treatment. Cis was more potent in causing a cytotoxicity effect. | [62] |
| Scaffold-based | Freeze-dried Silk Fibroin-Scaffold | HMF; MDA-MB-231; MCF-7 | 3D scaffolds were cultured in two different manners with and without fibroblasts. Cell growth was monitored, and it was higher in the co-culture model. Gene expression, related to TME, revealed that Col-I, FN, MMP1, MMP2 and MMP3 expression was higher in the heterotypic tumor culture compared to that in the homotypic one | [63] |
| Scaffold-based | Biomimetic collagen scaffolds | MDA-MB-231; MCF-7 | MCF-7 DOX-resistant cells were characterized by an overexpression of TP53I3 and TAP1 correlated with multidrug resistance phenomena. Cells showed an enhanced expression of the GADD45 family, correlated with a reduced DNA damage response MDA-MB-231 cells reduced drug accumulation by down-regulating the endocytic pathway and activating the lysosomal pathway | [64] |
| Acini | GFR Matrigel® | MCF-10A.B2 | The JNK signaling pathway was involved in HER 2+ breast cancer tumor progression: its deficiency promoted an acceleration of cell proliferation | [65] |
| Hybrid hydrogel system | Tumor spheroids surrounded by a CAF-embedded collagen -hydrogel | MDA-MB-231; MCF-7; CAFs | 3D complex models were characterized by different stiffnesses. The higher expression of matrix genes RNA sequencing revealed that cells in a soft environment up-regulated YAP1, while cells in a stiffer matrix up-regulated proangiogenic proteins (FN1 and MMP9). BC cells shift from glycolysis to OXPHOS and FA metabolism, responding to a stiff microenvironment | [66] |
| Co-culture microfluidic tumor model | Hydrogel-based matrices injected into tumor microfluidic chips | SUM-159; MDA-MB-231; MCF-7; CAFs | The 3D co-culture model revealed that CAFs enhanced breast cancer proliferation. The model showed the central role of GPNMB in tumor progression: the knockdown of this protein was able to reduce the effects of CAFs on cancer invasion | [67] |
| Type of Platform | Technical Features | Cells | End-Point | Ref. |
|---|---|---|---|---|
| Spheroid | polyHEMA coated multiwell | EEC16, EEC16 | Up-regulation of the gene involved in the immune response and hormonal signaling | [86] |
| Spheroid | hanging-drop method | St-T1b | Up-regulation of the gene involved in invasion and EMT | [87] |
| Spheroid | Kenzan method | 12Z | Up-regulation of the gene involved in the inflammatory response and hormonal signaling | [88] |
| Spheroid | micro-molded agarose well plates | 12Z, iEc-ESCs, iHUFs | Up-regulation of the gene involved in the immune, inflammation and invasion process | [89] |
| Tissue slices | 3D air–liquid interface cultures-collagen type I | Primary endometrial cells | Up-regulation of the gene involved in cell proliferation and hormonal signaling | [90] |
| Organ on a chip | microfluidic 3D tri-culture model | HUVECs; Ishikawa; ESFs: CRL-4003 cells | Up-regulation of the gene involved in angiogenesis | [91] |
| Organoid | liquefied growth factor reduced Matrigel | Primary endometrial cells | Epigenetic modification for the HOX genes and their cofactors | [92] |
| Type of Platform | Technical Features | Cells | End-Point | Ref. |
|---|---|---|---|---|
| Spheroid | Matrigel-embedded spheroids | HepG2 | Enhanced secretion of albumin and urea. Up-regulation of genes involved in drug metabolism (CYP450 enzymes) | [102] |
| Spheroid | Fabricated with a commercial system based on the hanging drop principle (GravityPLUS, Insphero) | Monoculteres of either HepaRG or HepG2 | Enhanced secretion of albumin and urea. Up-regulation of genes related to drug metabolism (CYP1A2, CYP2B6, CYP3A4), gluconeogenesis (G6Pase, PEPCK2), glycolysis (L-PK), energetic lipid synthesis (SREBF1, SCD1, DGAT2), bile acid metabolism (CYP7A1, CYP8B1, ABCB11) and lipoprotein metabolism (ApoE, ApoA-1) | [105] |
| Bioprinted cell-laden scaffold | 3D structure printed with a GelMa-based bioink | HepaRG | Enhanced albumin and urea secretion. Up-regulation of genes related to phenotypical stability. Up-regulation of phase-1 drug metabolizing enzymes (CYP1A2, CYP2B6, CYP3A4) | [104] |
| Bioprinted cell-laden scaffold | Pinewood structure bioprinted with a custom alginate-based bioink | HepG2 | Up-regulation of genes involved in hepatic functionality (ALB), lipid metabolism (ApoA4, ApoC3) and cell proliferation (VTN) | [101] |
| Bioprinted cell-laden scaffold | Hexagonal structures bioprinted with an ECM-based custom bioink | HepG2 | Enhanced secretion of albumin and urea | [100] |
| Hollow fiber bioreactor | Commercial device (C2011, FiberCell Systems) | HUH7.5 | Enhanced metabolic activity | [106] |
| Bioprinted cell-laden scaffold | Core-shell structures printed with a custom ink based on methacrylated alginate and Matrigel. The coaxial structure enabled the segregated coculture of different cell types | HepG2 NIH 3T3 | The coculture with fibroblasts enhanced albumin and urea secretion and proliferation and promoted the aggregation of HepG2 | [108] |
| Spheroid | Self-assembly in ultra-low-adhesion U-bottom plates | HepaRG 3T3-J2 | The coculture enhanced albumin and urea secretion and incremented the metabolic activity of HepaRG | [110] |
| Mixed | This study exploited self-assembled spheroids in U-bottom, agarose-coated wells and cells seeded in a 3D hollow fiber bioreactor | iPSCs-derived hepatocytes. | Enhanced differentiation performances in both 3D cultures (down-regulation of pluripotency markers, up-regulation of hepatic markers ALB, CYP34A and HNF4A, metabolic shift toward oxidative phosphorylation). Bioreactor-based cultures outperformed spheroids | [116] |
| Spheroid | The formation of spheroids was guided and achieved in a custom microfluidic device | Primary hepatocytes derived from humanized chimeric mice and murine embryonic fibroblasts (H9 SOX17-mCHERRY) | Long-term (33 days) 3D cultures of functional and polarized primary hepatocytes with enhanced albumin, urea and bile acids synthesis compared to that of 2D cultures. Promoted the differentiation of murine embryonic fibroblasts when cocultured with primary hepatocytes in the microfluidic device | [115] |
| Spheroid | Cell aggregation into spheroids was achieved by using custom-made agarose micro-wells arrays | iPSCs derived from iPS(foreskin)-3 fibroblasts | Successful differentiation into functional hepatocytes (albumin and urea synthesis, expression of active CYP450 enzymes) | [117] |
| Type of Platform | Technical Features | Pathogens | End-Point | Ref. |
|---|---|---|---|---|
| 3D gut epithelium | Microchannel-embedded hydrogel | co-cultures of Salmonella enterica and Lactobacillus reuteri | Overexpression of NF-κB and TNF signaling pathways; Rap1 signaling; homologous recombination in the Caco-2 cells co-cultured with pathogens | [127] |
| 3D human endocervical model | RWV bioreactor technology | L. crispatus, A. vaginae, G. vaginalis, P. bivia, S. amnii | Exhibition of the pro-inflammatory potential through the induction of specific cytokines (e.g., IL-6, IL-8, INF-ϒ-induced protein-10, monocyte chemotactic protein 10), iNOS and oxidative stress-associated compounds | [128] |
| Model of the small intestine | Transwell | E. faecalis, E. coli, S. salivarius, S. mitis, L. plantarum, V. parvula, V. atypica and P. intermedia | Secretion of IL-6, IL-8, TNF-α, and CXCL16; Protein expression (Actin); Gene expression (TLR-2 and -4 and DUOX2) | [129] |
| Gut organoid | Transwell | Listeria monocytogenes | Bacterial translocation study (increase in the pro-inflammatory response: IL-8, TNF-α) | [130] |
| Enteroids | Human intestinal enteroids (HIE) | Shigella flexneri | Increased HIE proinflammatory signals and the amino acid transporter SLC7A5 | [131] |
| Hollow fiber bioreactor | 3D model of human colonic epithelial cells (HT-29) cultured on bioreactors (with/without macrophages) in microgravity | Salmonella enterica (serovar Typhimurium) | Increased bacterial virulence in an spaceflight analogue culture (expression of adherence, invasion, motility and chemotaxis genes); Increased expression of host inflammatory genes (IL-8) | [132] |
| 3D organotypic skin model | HaCaT keratinocyte-fibroblast co-cultured on transwell | Staphylococcus aureus MRSA strains | Assessment of MRSA pathogenicity and HaCaT/fibroblasts DNA damage detection | [133] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bloise, N.; Giannaccari, M.; Guagliano, G.; Peluso, E.; Restivo, E.; Strada, S.; Volpini, C.; Petrini, P.; Visai, L. Growing Role of 3D In Vitro Cell Cultures in the Study of Cellular and Molecular Mechanisms: Short Focus on Breast Cancer, Endometriosis, Liver and Infectious Diseases. Cells 2024, 13, 1054. https://doi.org/10.3390/cells13121054
Bloise N, Giannaccari M, Guagliano G, Peluso E, Restivo E, Strada S, Volpini C, Petrini P, Visai L. Growing Role of 3D In Vitro Cell Cultures in the Study of Cellular and Molecular Mechanisms: Short Focus on Breast Cancer, Endometriosis, Liver and Infectious Diseases. Cells. 2024; 13(12):1054. https://doi.org/10.3390/cells13121054
Chicago/Turabian StyleBloise, Nora, Marialaura Giannaccari, Giuseppe Guagliano, Emanuela Peluso, Elisa Restivo, Silvia Strada, Cristina Volpini, Paola Petrini, and Livia Visai. 2024. "Growing Role of 3D In Vitro Cell Cultures in the Study of Cellular and Molecular Mechanisms: Short Focus on Breast Cancer, Endometriosis, Liver and Infectious Diseases" Cells 13, no. 12: 1054. https://doi.org/10.3390/cells13121054







