Exploring the Role of Circular RNA in Bone Biology: A Comprehensive Review
Abstract
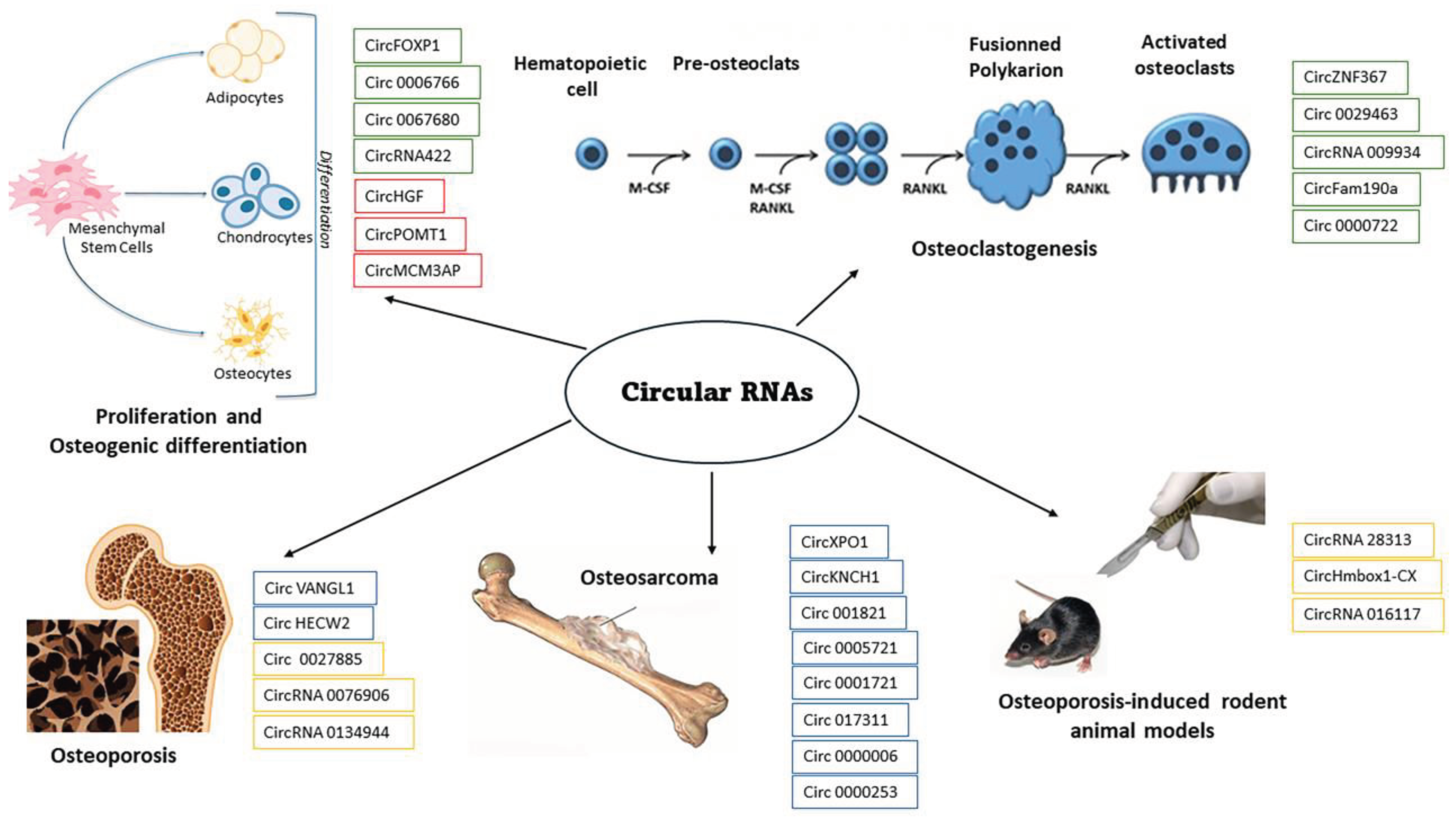
:1. Introduction
2. Circular RNAs (circRNAs)
2.1. circRNAs in Bone Development and Maintenance
2.2. circRNAs in Bone Disorders
3. Animal Models
4. Therapeutic Strategies Targeting circRNAs in Bone Disorders
5. Perspective
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bombieri, C.; Corsi, A.; Trabetti, E.; Ruggiero, A.; Marchetto, G.; Vattemi, G.; Valenti, M.T.; Zipeto, D.; Romanelli, M.G. Advanced Cellular Models for Rare Disease Study: Exploring Neural, Muscle and Skeletal Organoids. Int. J. Mol. Sci. 2024, 25, 1014. [Google Scholar] [CrossRef]
- Valenti, M.T.; Dalle Carbonare, L.; Mottes, M. Osteogenic differentiation in healthy and pathological conditions. Int. J. Mol. Sci. 2016, 18, 41. [Google Scholar] [CrossRef]
- Li, D.; Yang, C.; Tian, Y.; Chen, Z.; Qian, A.; Yin, C. Roles and Mechanism of Long Noncoding RNAs in Bone Diseases. In Noncoding RNAs Bone; Springer: Singapore, 2021; pp. 95–128. [Google Scholar]
- Haque, S.; Harries, L.W. Circular RNAs (circRNAs) in health and disease. Genes 2017, 8, 353. [Google Scholar] [CrossRef]
- Kirgiafini, D.; Kyrgiafini, M.-A.; Gournaris, T.; Mamuris, Z. Understanding Circular RNAs in Health, Welfare, and Productive Traits of Cattle, Goats, and Sheep. Animals 2024, 14, 733. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, C.; Xu, L.; Li, X. Roles of circular RNAs in osteogenic/osteoclastogenic differentiation. Biofactors 2024, 50, 6–15. [Google Scholar] [CrossRef]
- Chen, G.; Wang, S.; Wei, R.; Liu, Y.; Xu, T.; Liu, Z.; Tan, Z.; Xie, Y.; Yang, D.; Liang, Z. Circular RNA circ-3626 promotes bone formation by modulating the miR-338-3p/Runx2 axis. Jt. Bone Spine 2024, 91, 105669. [Google Scholar] [CrossRef]
- Mazziotta, C.; Badiale, G.; Cervellera, C.F.; Tognon, M.; Martini, F.; Rotondo, J.C. Regulatory mechanisms of circular RNAs during human mesenchymal stem cell osteogenic differentiation. Theranostics 2024, 14, 143. [Google Scholar] [CrossRef]
- Li, F.; Zhao, X.; Zhang, Y.; Zhuang, Q.; Wang, S.; Fang, X.; Xu, T.; Li, X.; Chen, G. Exosomal circFAM63Bsuppresses bone regeneration of postmenopausal osteoporosis via regulating miR-578/HMGA2 axis. J. Orthop. Res. 2024, 42, 1244–1253. [Google Scholar] [CrossRef]
- Tang, L.; Yuan, L.; Yan, J.; Ge, J.; Lian, Z.; Li, Z. circ_0029463 promotes osteoclast differentiation by mediating miR-134-5p/Rab27a axis. J. Orthop. Surg. Res. 2024, 19, 128. [Google Scholar] [CrossRef]
- Geng, Y.; Shao, R.; Xu, T.; Zhang, L. Identification of a potential signature to predict the risk of postmenopausal osteoporosis. Gene 2024, 894, 147942. [Google Scholar] [CrossRef]
- Himič, V.; Syrmos, N.; Ligarotti, G.K.; Kato, S.; Fehlings, M.G.; Ganau, M. The role of genetic and epigenetic factors in determining the risk of spinal fragility fractures: New insights in the management of spinal osteoporosis. Quant. Imaging Med. Surg. 2023, 13, 7632. [Google Scholar] [CrossRef]
- Wang, L.; Sheng, Z.; Yao, T. Association between circHIPK3/miR-378a-3p/HDAC4 axis and osteoporotic fractures: A comprehensive investigation. J. Orthop. Surg. 2023, 31, 10225536231219637. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, K.; Wang, X.; Yu, L.; Xu, F.; Ding, H.; Ma, H. Circular RNAs are involved in the pathogenesis of osteoarthritis through intracellular mechanisms. Chin. J. Tissue Eng. Res. 2024, 28, 5716. [Google Scholar]
- Zhang, Z.; Yu, P.; Bai, L. Hsa_circular RNA_0045474 Facilitates Osteoarthritis Via Modulating microRNA-485-3p and Augmenting Transcription Factor 4. Mol. Biotechnol. 2024, 66, 1174–1187. [Google Scholar] [CrossRef]
- Chen, H.; Qu, Z.; Shi, T.; Zhao, H.; Huang, S.; Ma, C. Circular RNA CircACAP2 regulates temporomandibular joint osteoarthritis via miR-21-5p/PLAG1 axis. Oral Dis. 2024; online ahead of print. [Google Scholar]
- Yan, B.; Li, Z.; Su, H.; Xue, H.; Qiu, D.; Xu, Z.; Tan, G. Regulatory mechanisms of autophagy-related ncRNAs in bone metabolic diseases. Front. Pharmacol. 2023, 14, 1178310. [Google Scholar] [CrossRef]
- Zhou, F.; Wu, H.; Yin, Z.; Zhou, W.; Chen, W. Circular RNA circ_0096041 promotes osteosarcoma cell proliferation and migration via sponging miR-556-5p and regulating LIN28A expression. Cell. Mol. Biol. 2024, 70, 113–119. [Google Scholar] [CrossRef]
- Zeng, L.; Liu, L.; Ni, W.-J.; Xie, F.; Leng, X.-M. Circular RNAs in osteosarcoma: An update of recent studies. Int. J. Oncol. 2023, 63, 123. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Li, Z. Advancements in Understanding the Role of Circular RNA in Osteosarcoma. Mol. Biotechnol. 2023; online ahead of print. [Google Scholar]
- Xu, Y.; Zhang, S.; Liao, X.; Li, M.; Chen, S.; Li, X.; Wu, X.; Yang, M.; Tang, M.; Hu, Y. Circular RNA circIKBKB promotes breast cancer bone metastasis through sustaining NF-κB/bone remodeling factors signaling. Mol. Cancer 2021, 20, 98. [Google Scholar] [CrossRef]
- Fu, M.; Fang, L.; Xiang, X.; Fan, X.; Wu, J.; Wang, J. Microarray analysis of circRNAs sequencing profile in exosomes derived from bone marrow mesenchymal stem cells in postmenopausal osteoporosis patients. J. Clin. Lab. Anal. 2022, 36, e23916. [Google Scholar] [CrossRef]
- Wu, J.; Chen, H.; Xie, D.-H.; Zhang, H.-Y.; Zhao, C.; Zou, Y.-C. Analysis of CircRNA expression profile of pathological bone formation in ankylosing spondylitis. Int. J. Rheum. Dis. 2021, 26, 1403–1406. [Google Scholar]
- Li, H.; Yang, H.; Sun, Z.; Tang, H.; Min, J. Whole-transcriptome sequencing of knee joint cartilage from osteoarthritis patients. Bone Jt. Res. 2019, 8, 290–303. [Google Scholar] [CrossRef]
- Greene, J.; Baird, A.-M.; Brady, L.; Lim, M.; Gray, S.G.; McDermott, R.; Finn, S.P. Circular RNAs: Biogenesis, function and role in human diseases. Front. Mol. Biosci. 2017, 4, 38. [Google Scholar] [CrossRef]
- Chen, L.-L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Feng, X.-Y.; Zhu, S.-X.; Pu, K.-J.; Huang, H.-J.; Chen, Y.-Q.; Wang, W.-T. New insight into circRNAs: Characterization, strategies, and biomedical applications. Exp. Hematol. Oncol. 2023, 12, 91. [Google Scholar] [CrossRef]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Verduci, L.; Tarcitano, E.; Strano, S.; Yarden, Y.; Blandino, G. CircRNAs: Role in human diseases and potential use as biomarkers. Cell Death Dis. 2021, 12, 468. [Google Scholar] [CrossRef]
- Jung, Y.-S.; Park, J.-I. Wnt signaling in cancer: Therapeutic targeting of Wnt signaling beyond β-catenin and the destruction complex. Exp. Mol. Med. 2020, 52, 183–191. [Google Scholar] [CrossRef]
- Stoll, L.; Rodríguez-Trejo, A.; Guay, C.; Brozzi, F.; Bayazit, M.B.; Gattesco, S.; Menoud, V.; Sobel, J.; Marques, A.C.; Venø, M.T. A circular RNA generated from an intron of the insulin gene controls insulin secretion. Nat. Commun. 2020, 11, 5611. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Liu, C.-X.; Li, X.; Nan, F.; Jiang, S.; Gao, X.; Guo, S.-K.; Xue, W.; Cui, Y.; Dong, K.; Ding, H. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 2019, 177, 865–880.e821. [Google Scholar] [CrossRef]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol. Cell 2019, 74, 494–507.e498. [Google Scholar] [CrossRef]
- Singh, S.; Shyamal, S.; Das, A.; Panda, A.C. Global identification of mRNA-interacting circular RNAs by CLiPPR-Seq. Nucleic Acids Res. 2024, 52, e29. [Google Scholar] [CrossRef]
- Zhao, W.; Cheng, Y.; Zhang, C.; You, Q.; Shen, X.; Guo, W.; Jiao, Y. Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci. Rep. 2017, 7, 5636. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, N.; Wu, X.; Wu, P.; Song, N.; Ma, J. Identification of differentially expressed circular RNAs in keloid and normal skin tissue by high-throughput sequencing. Dermatol. Ther. 2021, 34, e14745. [Google Scholar] [CrossRef]
- Dong, R.; Ma, X.-K.; Chen, L.-L.; Yang, L. Increased complexity of circRNA expression during species evolution. RNA Biol. 2017, 14, 1064–1074. [Google Scholar] [CrossRef]
- Panda, A.C.; Grammatikakis, I.; Munk, R.; Gorospe, M.; Abdelmohsen, K. Emerging roles and context of circular RNAs. Wiley Interdiscip. Rev. RNA 2017, 8, e1386. [Google Scholar] [CrossRef]
- Huang, S.; Yang, B.; Chen, B.; Bliim, N.; Ueberham, U.; Arendt, T.; Janitz, M. The emerging role of circular RNAs in transcriptome regulation. Genomics 2017, 109, 401–407. [Google Scholar] [CrossRef]
- Liu, C.-X.; Chen, L.-L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef]
- Shen, W.; Sun, B.; Zhou, C.; Ming, W.; Zhang, S.; Wu, X. CircFOXP1/FOXP1 promotes osteogenic differentiation in adipose-derived mesenchymal stem cells and bone regeneration in osteoporosis via miR-33a-5p. J. Cell. Mol. Med. 2020, 24, 12513–12524. [Google Scholar] [CrossRef]
- Guo, Z.; Xie, M.; Zou, Y.; Liang, Q.; Liu, F.; Su, J.; He, Z.; Cai, X.; Chen, Z.; Zhao, Q. Circular RNA Hsa_circ_0006766 targets microRNA miR-4739 to regulate osteogenic differentiation of human bone marrow mesenchymal stem cells. Bioengineered 2021, 12, 5679–5687. [Google Scholar] [CrossRef]
- Huang, Y.; Wan, S.; Yang, M. Circ_0067680 expedites the osteogenic differentiation of human bone marrow-derived mesenchymal stem cells through miR-4429/CTNNB1/Wnt/β-catenin pathway. Biol. Direct 2021, 16, 16. [Google Scholar] [CrossRef]
- Della Bella, E.; Menzel, U.; Basoli, V.; Tourbier, C.; Alini, M.; Stoddart, M.J. Differential regulation of circRNA, miRNA, and piRNA during early osteogenic and chondrogenic differentiation of human mesenchymal stromal cells. Cells 2020, 9, 398. [Google Scholar] [CrossRef]
- Cherubini, A.; Barilani, M.; Rossi, R.L.; Jalal, M.M.K.; Rusconi, F.; Buono, G.; Ragni, E.; Cantarella, G.; Simpson, H.A.R.W.; Péault, B. FOXP1 circular RNA sustains mesenchymal stem cell identity via microRNA inhibition. Nucleic Acids Res. 2019, 47, 5325–5340. [Google Scholar] [CrossRef]
- Xin, W.; Yuan, S.; Wang, B.; Qian, Q.; Chen, Y. Hsa_circ_0066523 promotes the proliferation and osteogenic differentiation of bone mesenchymal stem cells by repressing PTEN. Bone Jt. Res. 2021, 10, 526–535. [Google Scholar] [CrossRef]
- Yu, K.; Jiang, Z.; Miao, X.; Yu, Z.; Du, X.; Lai, K.; Wang, Y.; Yang, G. circRNA422 enhanced osteogenic differentiation of bone marrow mesenchymal stem cells during early osseointegration through the SP7/LRP5 axis. Mol. Ther. 2022, 30, 3226–3240. [Google Scholar] [CrossRef]
- Feng, X.; Xiang, Q.; Jia, J.; Guo, T.; Liao, Z.; Yang, S.; Cai, X.; Liu, X. CircHGF suppressed cell proliferation and osteogenic differentiation of BMSCs in ONFH via inhibiting miR-25-3p binding to SMAD7. Mol. Ther.-Nucleic Acids 2022, 28, 99–113. [Google Scholar] [CrossRef]
- Huang, X.-Q.; Cen, X.; Sun, W.-T.; Xia, K.; Yu, L.-Y.; Liu, J.; Zhao, Z.-H. CircPOMT1 and circMCM3AP inhibit osteogenic differentiation of human adipose-derived stem cells by targeting miR-6881-3p. Am. J. Transl. Res. 2019, 11, 4776. [Google Scholar]
- Chia, W.; Liu, J.; Huang, Y.-G.; Zhang, C. A circular RNA derived from DAB1 promotes cell proliferation and osteogenic differentiation of BMSCs via RBPJ/DAB1 axis. Cell Death Dis. 2020, 11, 372. [Google Scholar] [CrossRef]
- Wen, J.; Guan, Z.; Yu, B.; Guo, J.; Shi, Y.; Hu, L. Circular RNA hsa_circ_0076906 competes with OGN for miR-1305 biding site to alleviate the progression of osteoporosis. Int. J. Biochem. Cell Biol. 2020, 122, 105719. [Google Scholar] [CrossRef]
- Li, M.; Li, C.; Zheng, H.; Zhou, Z.; Yang, W.; Gong, Y.; Wu, X.; Li, L. CircRNA_0001795 sponges miRNA-339-5p to regulate yes-associated protein 1 expression and attenuate osteoporosis progression. Bioengineered 2022, 13, 2803–2815. [Google Scholar] [CrossRef]
- Huang, Y.; Xiao, D.; Huang, S.; Zhuang, J.; Zheng, X.; Chang, Y.; Yin, D. Circular RNA YAP1 attenuates osteoporosis through up-regulation of YAP1 and activation of Wnt/β-catenin pathway. Biomed. Pharmacother. 2020, 129, 110365. [Google Scholar] [CrossRef]
- Yin, P.; Xue, Y. CircRNA hsa_circ_0006859 inhibits the osteogenic differentiation of BMSCs and aggravates osteoporosis by targeting miR-642b-5p/miR-483-3p and upregulating EFNA2/DOCK3. Int. Immunopharmacol. 2023, 116, 109844. [Google Scholar] [CrossRef]
- Gao, J.-W.; Song, M.-K.; Wu, D.-Z.; Yan, T.; Zhao, K.; Huang, Y.-S.; Chen, X.-Y.; Tu, C.; Deng, G.-X.; Chen, Z.-S. CircRBM23 regulates the switch between osteogenesis and adipogenesis of mesenchymal stem cells via sponging miR-338-3p. Clin. Sci. 2023, 137, 495–510. [Google Scholar] [CrossRef]
- Shen, Y.; Jiang, B.; Luo, B.; Jiang, X.; Zhang, Y.; Wang, Q. Circular RNA-FK501 binding protein 51 boosts bone marrow mesenchymal stem cell proliferation and osteogenic differentiation via modulating microRNA-205-5p/Runt-associated transcription factor 2 axis. J. Orthop. Surg. Res. 2023, 18, 782. [Google Scholar] [CrossRef]
- Huang, W.; Wu, Y.; Zhao, Y.; Gan, X.; Zhang, B.; Cen, X.; Huang, X.; Zhao, Z. Down-regulation of hsa-circ-0107593 promotes osteogenic differentiation of hADSCs via miR-20a-5p/SMAD6 signaling. Oral Dis. 2023, 29, 3447–3459. [Google Scholar] [CrossRef]
- Pang, J.; Wu, Y.; Ji, Y.; Si, Y.; Liang, F. Circ_0006873 suppresses the osteogenic differentiation of human-derived mesenchymal stem cells through mediating miR-20a/SMURF2 axis in vitro. APMIS 2023, 131, 313–324. [Google Scholar] [CrossRef]
- Li, H.; Wang, C.; Yao, J.; Jin, Y.; Song, X.; Meng, Q.; Wu, J.; Liu, Q.; Liu, M.; Sun, H. Circ_0114581 promotes osteogenic differentiation of BMSCs via the MiR-155-5p/HNRNPA3 axis. Life Sci. 2023, 333, 122127. [Google Scholar] [CrossRef]
- Zheng, C.; Ding, L.; Xiang, Z.; Feng, M.; Zhao, F.; Zhou, Z.; She, C. Circ_0001825 promotes osteogenic differentiation in human-derived mesenchymal stem cells via miR-1270/SMAD5 axis. J. Orthop. Surg. Res. 2023, 18, 663. [Google Scholar] [CrossRef]
- Zhou, J.; Sui, M.; Ji, F.; Shen, S.; Lin, Y.; Jin, M.; Tao, J. Hsa_circ_0036872 has an important promotional effect in enhancing osteogenesis of dental pulp stem cells by regulating the miR-143-3p/IGF2 axis. Int. Immunopharmacol. 2024, 130, 111744. [Google Scholar] [CrossRef]
- Liu, G.; Luo, J.; Wang, Z.; Zhou, Y.; Li, Y. CircZNF367 suppresses osteogenic differentiation of human bone marrow mesenchymal stromal/stem cells via reducing HuR-mediated mRNA stability of LRP5. Hum. Cell 2023, 36, 146–162. [Google Scholar] [CrossRef]
- Zhang, Z.-M.; Huang, C.-X.; Huo, J.-Z. Circ_C4orf36 Promotes the Proliferation and Osteogenic Differentiation of BMSCs by Regulating VEGFA. Biochem. Genet. 2023, 61, 931–944. [Google Scholar] [CrossRef]
- Li, Y.; Nie, J.; Wu, Q.; Yang, X.; Jiang, P. Circ-Sirt1 promotes osteoblast differentiation by activating Sirt1 and Wnt/β-catenin pathway. Acta Biochim. Pol. 2023, 70, 51–57. [Google Scholar] [CrossRef]
- Zhi, F.; Ding, Y.; Wang, R.; Yang, Y.; Luo, K.; Hua, F. Exosomal hsa_circ_0006859 is a potential biomarker for postmenopausal osteoporosis and enhances adipogenic versus osteogenic differentiation in human bone marrow mesenchymal stem cells by sponging miR-431-5p. Stem Cell Res. Ther. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Chen, G.; Wang, Q.; Li, Z.; Yang, Q.; Liu, Y.; Du, Z.; Zhang, G.; Song, Y. Circular RNA CDR1as promotes adipogenic and suppresses osteogenic differentiation of BMSCs in steroid-induced osteonecrosis of the femoral head. Bone 2020, 133, 115258. [Google Scholar] [CrossRef]
- Mohamed, A.M. An overview of bone cells and their regulating factors of differentiation. Malays. J. Med. Sci. MJMS 2008, 15, 4. [Google Scholar]
- Deng, M.; Wang, Z.; Luo, J.; Cao, H.; Li, Y.; Chen, L.; Liu, G. CircZNF367 promotes osteoclast differentiation and osteoporosis by interacting with FUS to maintain CRY2 mRNA stability. J. Orthop. Surg. Res. 2023, 18, 492. [Google Scholar] [CrossRef]
- Miao, F.; Yin, B.-H.; Zhang, X.; Xue, D.-D.; Ma, C. CircRNA_009934 induces osteoclast bone resorption via silencing miR-5107. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7580–7588. [Google Scholar]
- Chen, K.; Chen, X.; Lang, C.; Yuan, X.; Huang, J.; Li, Z.; Xu, M.; Wu, K.; Zhou, C.; Li, Q. CircFam190a: A critical positive regulator of osteoclast differentiation via enhancement of the AKT1/HSP90β complex. Exp. Mol. Med. 2023, 55, 2051–2066. [Google Scholar] [CrossRef]
- Xie, L.; Ren, X.; Yang, Z.; Zhou, T.; Zhang, M.; An, W.; Guan, Z. Exosomal circ_0000722 derived from periodontal ligament stem cells undergoing osteogenic differentiation promotes osteoclastogenesis. Int. Immunopharmacol. 2024, 128, 111520. [Google Scholar] [CrossRef]
- Yang, L.; Zeng, Z.; Kang, N.; Yang, J.-C.; Wei, X.; Hai, Y. Circ-VANGL1 promotes the progression of osteoporosis by absorbing miRNA-217 to regulate RUNX2 expression. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 949–957. [Google Scholar]
- Fang, S.; Cao, D.; Wu, Z.; Chen, J.; Huang, Y.; Shen, Y.; Gao, Z. Circ_0027885 sponges miR-203-3p to regulate RUNX2 expression and alleviates osteoporosis progression. BMC Musculoskelet. Disord. 2024, 25, 5. [Google Scholar] [CrossRef]
- Huang, Y.; Xie, J.; Li, E. Comprehensive circular RNA profiling reveals circ_0002060 as a potential diagnostic biomarkers for osteoporosis. J. Cell. Biochem. 2019, 120, 15688–15694. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, W.; Li, F.; Xu, N.; Sun, P. Dysregulation of circRNA-0076906 and circRNA-0134944 is Correlated with Susceptibility to Osteoporosis and Osteoporotic Fracture in Postmenopausal Females from the Chinese Han Population. Pharmacogenomics Pers. Med. 2023, 16, 183–194. [Google Scholar] [CrossRef]
- Moura, S.R.; Fernandes, M.J.; Santos, S.G.; Almeida, M.I. Circular RNAs: Promising targets in osteoporosis. Curr. Osteoporos. Rep. 2023, 21, 289–302. [Google Scholar] [CrossRef]
- Yang, J.; Wu, J. Discovery of potential biomarkers for osteoporosis diagnosis by individual omics and multi-omics technologies. Expert Rev. Mol. Diagn. 2023, 23, 505–520. [Google Scholar] [CrossRef]
- Zhao, K.; Zhao, Q.; Guo, Z.; Chen, Z.; Hu, Y.; Su, J.; Chen, L.; He, Z.; Cai, X.; Chen, M. Hsa_Circ_0001275: A potential novel diagnostic biomarker for postmenopausal osteoporosis. Cell. Physiol. Biochem. 2018, 46, 2508–2516. [Google Scholar] [CrossRef]
- Zhang, C.; Li, Q.; Ye, Z.; Wang, X.; Zhao, H.; Wang, Y.; Zheng, X. Mechanism of Circ_HECW2 regulating osteoblast apoptosis in osteoporosis by attenuating the maturation of miR-1224-5p. J. Orthop. Surg. Res. 2024, 19, 40. [Google Scholar] [CrossRef]
- Mirabello, L.; Troisi, R.J.; Savage, S.A. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int. J. Cancer 2009, 125, 229–234. [Google Scholar] [CrossRef]
- Wang, X.; Qin, G.; Liang, X.; Wang, W.; Wang, Z.; Liao, D.; Zhong, L.; Zhang, R.; Zeng, Y.-X.; Wu, Y. Targeting the CK1α/CBX4 axis for metastasis in osteosarcoma. Nat. Commun. 2020, 11, 1141. [Google Scholar] [CrossRef]
- Berner, K.; Johannesen, T.B.; Berner, A.; Haugland, H.K.; Bjerkehagen, B.; Bøhler, P.J.; Bruland, Ø.S. Time-trends on incidence and survival in a nationwide and unselected cohort of patients with skeletal osteosarcoma. Acta Oncol. 2015, 54, 25–33. [Google Scholar] [CrossRef]
- de Azevedo, J.W.V.; Fernandes, T.A.A.d.M.; Fernandes, J.V.; de Azevedo, J.C.V.; Lanza, D.C.F.; Bezerra, C.M.; Andrade, V.S.; de Araújo, J.M.G. Biology and pathogenesis of human osteosarcoma. Oncol. Lett. 2020, 19, 1099–1116. [Google Scholar]
- Zhou, C.; Balmer, L.; Song, M.; Mahara, G.; Wu, K.; Wang, W.; Wang, H. Identification of CircRNA biomarkers in osteosarcoma: An updated systematic review and meta-analysis. Non-Coding RNA Res. 2024, 9, 341–349. [Google Scholar] [CrossRef]
- Jiang, Y.; Hou, J.; Zhang, X.; Xu, G.; Wang, Y.; Shen, L.; Wu, Y.; Li, Y.; Yao, L. Circ-XPO1 upregulates XPO1 expression by sponging multiple miRNAs to facilitate osteosarcoma cell progression. Exp. Mol. Pathol. 2020, 117, 104553. [Google Scholar] [CrossRef]
- Li, Z.; Fu, Y.; Ouyang, W.; He, M.; Wang, Y.; Wang, X.; Tan, W. Circ_0016347 promotes osteosarcoma progression by regulating miR-1225-3p/KCNH1 axis. Cancer Biother. Radiopharm. 2023, 38, 619–631. [Google Scholar] [CrossRef]
- Liu, Y.-P.; Wan, J.; Long, F.; Tian, J.; Zhang, C. circPVT1 facilitates invasion and metastasis by regulating miR-205-5p/c-FLIP axis in osteosarcoma. Cancer Manag. Res. 2020, 12, 1229–1240. [Google Scholar] [CrossRef]
- Huang, S.-X.; Mei, H.-B.; Liu, K.; Tang, J.; Wu, J.-Y.; Zhu, G.-H.; Ye, W.-H. CircPVT1 promotes the tumorigenesis and metastasis of osteosarcoma via mediation of miR-26b-5p/CCNB1 axis. J. Bone Miner. Metab. 2022, 40, 581–593. [Google Scholar] [CrossRef]
- Yan, M.; Gao, H.; Lv, Z.; Liu, Y.; Zhao, S.; Gong, W.; Liu, W. Circular RNA PVT1 promotes metastasis via regulating of miR-526b/FOXC2 signals in OS cells. J. Cell. Mol. Med. 2020, 24, 5593–5604. [Google Scholar] [CrossRef]
- Xiang, L.; Zhang, W. Expression level of circular RNA hsa_circ_0005721 in tissues and serum of patients with osteosarcoma and its clinical significance. J. Clin. Pathol. Res. 2020, 40, 1136–1143. [Google Scholar]
- Xu, M.; Sun, X.; Liu, Y.; Chang, L.; Wang, H.; Wang, S. hsa_circ_0005721 triggers proliferation, migration and invasion of osteosarcoma by upregulating the linear transcript TEP1. J. BUON Off. J. Balk. Union Oncol. 2021, 26, 1588–1594. [Google Scholar]
- Gao, Y.; Ma, H.; Gao, Y.; Tao, K.; Fu, L.; Ren, R.; Hu, X.; Kou, M.; Chen, B.; Shi, J. CircRNA Circ_0001721 promotes the progression of osteosarcoma through miR-372-3p/MAPK7 axis. Cancer Manag. Res. 2020, 12, 8287–8302. [Google Scholar] [CrossRef]
- Hu, R.; Chen, S.; Yan, J. Blocking circ-CNST suppresses malignant behaviors of osteosarcoma cells and inhibits glycolysis through circ-CNST-miR-578-LDHA/PDK1 ceRNA networks. J. Orthop. Surg. Res. 2021, 16, 300. [Google Scholar] [CrossRef]
- Ji, X.; Shan, L.; Shen, P.; He, M. Circular RNA circ_001621 promotes osteosarcoma cells proliferation and migration by sponging miR-578 and regulating VEGF expression. Cell Death Dis. 2020, 11, 18. [Google Scholar] [CrossRef]
- Shen, S.; Yao, T.; Xu, Y.; Zhang, D.; Fan, S.; Ma, J. CircECE1 activates energy metabolism in osteosarcoma by stabilizing c-Myc. Mol. Cancer 2020, 19, 151. [Google Scholar] [CrossRef]
- Kornak, U.; Mundlos, S. Genetic disorders of the skeleton: A developmental approach. Am. J. Hum. Genet. 2003, 73, 447–474. [Google Scholar] [CrossRef]
- Spranger, J.W.; Superti-Furga, A.; Unger, S. Bone Dysplasias: An Atlas of Genetic Disorders of Skeletal Development; Oxford University Press: Cary, NC, USA, 2018. [Google Scholar]
- Arnold, W.V.; Fertala, A. Skeletal diseases caused by mutations that affect collagen structure and function. Int. J. Biochem. Cell Biol. 2013, 45, 1556–1567. [Google Scholar] [CrossRef]
- Yip, R.K.; Chan, D.; Cheah, K.S. Mechanistic insights into skeletal development gained from genetic disorders. Curr. Top. Dev. Biol. 2019, 133, 343–385. [Google Scholar]
- Zhai, N.; Lu, Y.; Wang, Y.; Ren, X.; Han, J. Circular RNAs and hereditary bone diseases. Intractable Rare Dis. Res. 2018, 7, 1–6. [Google Scholar] [CrossRef]
- McCauley, L.K. Transgenic mouse models of metabolic bone disease. Curr. Opin. Rheumatol. 2001, 13, 316–325. [Google Scholar] [CrossRef]
- Pearce, A.; Richards, R.; Milz, S.; Schneider, E.; Pearce, S. Animal models for implant biomaterial research in bone: A review. Eur. Cell Mater. 2007, 13, 1–10. [Google Scholar] [CrossRef]
- Dalle Carbonare, L.; Bertacco, J.; Marchetto, G.; Cheri, S.; Deiana, M.; Minoia, A.; Tiso, N.; Mottes, M.; Valenti, M.T. Methylsulfonylmethane enhances MSC chondrogenic commitment and promotes pre-osteoblasts formation. Stem Cell Res. Ther. 2021, 12, 326. [Google Scholar] [CrossRef]
- Valenti, M.T.; Marchetto, G.; Mottes, M.; Dalle Carbonare, L. Zebrafish: A suitable tool for the study of cell signaling in bone. Cells 2020, 9, 1911. [Google Scholar] [CrossRef]
- Chen, X.; Ouyang, Z.; Shen, Y.; Liu, B.; Zhang, Q.; Wan, L.; Yin, Z.; Zhu, W.; Li, S.; Peng, D. CircRNA_28313/miR-195a/CSF1 axis modulates osteoclast differentiation to affect OVX-induced bone absorption in mice. RNA Biol. 2019, 16, 1249–1262. [Google Scholar] [CrossRef]
- Patil, S.; Dang, K.; Zhao, X.; Gao, Y.; Qian, A. Role of LncRNAs and CircRNAs in bone metabolism and osteoporosis. Front. Genet. 2020, 11, 584118. [Google Scholar] [CrossRef]
- Zhang, Z.; Yue, L.; Wang, Y.; Jiang, Y.; Xiang, L.; Cheng, Y.; Ju, D.; Chen, Y. A circRNA-miRNA-mRNA network plays a role in the protective effect of diosgenin on alveolar bone loss in ovariectomized rats. BMC Complement. Med. Ther. 2020, 20, 220. [Google Scholar] [CrossRef]
- Ping, J.; Li, L.; Dong, Y.; Wu, X.; Huang, X.; Sun, B.; Zeng, B.; Xu, F.; Liang, W. The role of long non-coding RNAs and circular RNAs in bone regeneration: Modulating miRNAs function. J. Tissue Eng. Regen. Med. 2022, 16, 227–243. [Google Scholar] [CrossRef]
- Wen, X.; Zhang, J.; Yang, W.; Nie, X.; Gui, R.; Shan, D.; Huang, R.; Deng, H. CircRNA-016901 silencing attenuates irradiation-induced injury in bone mesenchymal stem cells via regulating the miR-1249-5p/HIPK2 axis. Exp. Ther. Med. 2021, 21, 355. [Google Scholar] [CrossRef]
- Liu, Z.; Li, C.; Huang, P.; Hu, F.; Jiang, M.; Xu, X.; Li, B.; Deng, L.; Ye, T.; Guo, L. CircHmbox1 targeting miRNA-1247-5p is involved in the regulation of bone metabolism by TNF-α in postmenopausal osteoporosis. Front. Cell Dev. Biol. 2020, 8, 594785. [Google Scholar] [CrossRef]
- Wang, Z.; Yi, X.; Liu, Y.; Liu, Q.; Li, Z.; Yu, A. Differential expression profiles and functional prediction of circRNA in mice with traumatic heterotopic ossification. Front. Immunol. 2023, 13, 1090529. [Google Scholar] [CrossRef]
- Lin, C.; Zhong, W.; Yan, W.; Yang, J.; Zheng, W.; Wu, Q. Circ-SLC8A1 regulates osteoporosis through blocking the inhibitory effect of miR-516b-5p on AKAP2 expression. J. Gene Med. 2020, 22, e3263. [Google Scholar] [CrossRef]
- Dong, Y.; Shan, S.; Liu, J.; Han, X.; He, H. Circular RNA mmu_circ_0001775 knockdown improves the osteogenic ability of mouse bone marrow mesenchymal stem cells. Chin. J. Tissue Eng. Res. 2022, 26, 4767. [Google Scholar]
- Chen, Z.; Lin, W.; Zhao, S.; Mo, X.; Wen, Z.; Cheung, W.H.; Fu, D.; Chen, B. Identification of circRNA expression profiles in BMSCs from glucocorticoid-induced osteoporosis model. Stem Cells Int. 2022, 2022, 3249737. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Xing, Z.; Tang, S.; Chen, H.; Zhang, Z.; Li, J.; Li, Y. circStrn3 is involved in bone cancer pain regulation in a rat model. Acta Biochim. Biophys. Sin. 2020, 52, 495–505. [Google Scholar] [CrossRef]
- Zhang, M.; He, Y.; Zhang, X.; Gan, S.; Xie, X.; Zheng, Z.; Liao, J.; Chen, W. Engineered cell-overexpression of circular RNA hybrid hydrogels promotes healing of calvarial defects. Biomater. Sci. 2023, 11, 1665–1676. [Google Scholar] [CrossRef]
- He, A.T.; Liu, J.; Li, F.; Yang, B.B. Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct. Target. Ther. 2021, 6, 185. [Google Scholar] [CrossRef]
- Ouyang, Z.; Tan, T.; Zhang, X.; Wan, J.; Zhou, Y.; Jiang, G.; Yang, D.; Guo, X.; Liu, T. CircRNA hsa_circ_0074834 promotes the osteogenesis-angiogenesis coupling process in bone mesenchymal stem cells (BMSCs) by acting as a ceRNA for miR-942-5p. Cell Death Dis. 2019, 10, 932. [Google Scholar] [CrossRef]
| circRNAs | Function | References |
|---|---|---|
| circ-DAB1 | Promotes cell proliferation and osteogenic differentiation via the NOTCH/RBPJ axis | [52] |
| circ_0076906 | Induces osteogenic differentiation via miR-1305/osteoglycin pathway | [53] |
| circ_0001795 | Promotes the osteogenic differentiation via circ_0001795/miR-339-5p/YAP1 axis | [54] |
| circRNA_0024097 | Promotes osteogenesis through miRNA-376b-3p/YAP1 axis | [55] |
| circ_0006859 | Inhibits osteogenic differentiation by targeting miR-642b-5p/miR-483-3p | [56] |
| circRBM23 | Regulates the switch between osteogenesis and adipogenesis by sponging miR-338-3p | [57] |
| circ-3626 | Promotes osteogenesis by via miR-338-3p/Runx2 axis | [7] |
| Circ-FK501 | Promotes osteogenic differentiation via microRNA-205-5p/RUNX2 axis | [58] |
| hsa-circ-0107593 | Inhibits osteogenic differentiation via miR-20a-5p/SMAD6 signaling | [59] |
| Circ_0006873 | Inhibits osteogenic differentiation via miR-20a-/SMURF2 signaling | [60] |
| Circ_0114581 | Promotes osteogenic differentiation via the MiR-155-5p/HNRNPA3 axis | [61] |
| Circ_0001825 | Promotes osteogenesis via miR-1270/SMAD5 axis | [62] |
| Circ_0036872 | Promotes osteogenesis via miR-143-3p/IGF2 axis | [63] |
| CircZNF367 | Inhibits osteogenic differentiation via reducing HuR-mediated mRNA stability of LRP5 | [64] |
| Circ_C4orf36 | Promotes osteogenesis by regulating VEGFA | [65] |
| Circ-Sirt1 | Promotes osteogenesis by activating Sirt1 and Wnt/β-catenin pathway | [66] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valenti, M.T.; Zerlotin, R.; Cominacini, M.; Bolognin, S.; Grano, M.; Dalle Carbonare, L. Exploring the Role of Circular RNA in Bone Biology: A Comprehensive Review. Cells 2024, 13, 999. https://doi.org/10.3390/cells13120999
Valenti MT, Zerlotin R, Cominacini M, Bolognin S, Grano M, Dalle Carbonare L. Exploring the Role of Circular RNA in Bone Biology: A Comprehensive Review. Cells. 2024; 13(12):999. https://doi.org/10.3390/cells13120999
Chicago/Turabian StyleValenti, Maria Teresa, Roberta Zerlotin, Mattia Cominacini, Silvia Bolognin, Maria Grano, and Luca Dalle Carbonare. 2024. "Exploring the Role of Circular RNA in Bone Biology: A Comprehensive Review" Cells 13, no. 12: 999. https://doi.org/10.3390/cells13120999







