Abstract
Autism spectrum disorders (ASDs) are complex neurodevelopmental conditions characterized by deficits in social interaction and communication, as well as repetitive behaviors. Although the etiology of ASD is multifactorial, with both genetic and environmental factors contributing to its development, a strong genetic basis is widely recognized. Recent research has identified numerous genetic mutations and genomic rearrangements associated with ASD-characterizing genes involved in brain development. Alterations in developmental programs are particularly harmful during critical periods of brain development. Notably, studies have indicated that genetic disruptions occurring during the second trimester of pregnancy affect cortical development, while disturbances in the perinatal and early postnatal period affect cerebellar development. The developmental defects must be viewed in the context of the role of the cerebellum in cognitive processes, which is now well established. The present review emphasizes the genetic complexity and neuropathological mechanisms underlying ASD and aims to provide insights into the cerebellar involvement in the disorder, focusing on recent advances in the molecular landscape governing its development in humans. Furthermore, we highlight when and in which cerebellar neurons the ASD-associated genes may play a role in the development of cortico–cerebellar circuits. Finally, we discuss improvements in protocols for generating cerebellar organoids to recapitulate the long period of development and maturation of this organ. These models, if generated from patient-induced pluripotent stem cells (iPSC), could provide a valuable approach to elucidate the contribution of defective genes to ASD pathology and inform diagnostic and therapeutic strategies.
1. Introduction
Autism spectrum disorders (ASDs) are clinically heterogeneous neurodevelopmental disorders that predominantly occur in infancy or early childhood. They comprise two symptom domains: the social domain, which includes difficulties with social interactions and communication, the non-social domain, which is characterized by repetitive behaviors, and sensory perception abnormalities (as indicated in the Diagnostic and Statistical Manual of Mental Disorders 5th Edn DSM-5 [1] and in the International Classification of Diseases, 11th Revision, of World Health Organization 2019, available online: https://icd.who.int/en code 6A02). The etiology of ASD is multifaceted, with a significant genetic component that is exacerbated by environmental factors, including prenatal exposures such as valproic acid, which are known to increase ASD risk [2,3].
In particular, the genetic contributions to ASD are substantial, with heritability estimates of around 80% from twin and family studies [2,4,5], involving numerous mutations and genomic rearrangements. These genetic perturbations significantly impact neurodevelopmental pathways, particularly in the cortex during the second trimester of pregnancy and the cerebellum during the perinatal and early/postnatal period [6,7]. During the second trimester, the cortex, a complex neural network critical for cognition and social interactions, undergoes significant developmental processes such as neuronal proliferation, migration, and cortical layer formation. Genetic abnormalities in these processes are strongly associated with ASD symptoms that affect social communication and cognitive functioning. Conversely, the cerebellum, beyond its traditional motor functions, has emerged as a critical player in the development of behavioral circuitry.
During the perinatal and early/postnatal periods, the cerebellum undergoes significant growth and structural refinement, involving granule cell (GC) migration, Purkinje cell (PC) dendritic arborization, and synaptogenesis.
Genetic perturbations during this critical period can profoundly affect cerebellar circuitry and function, contributing to the neurodevelopmental alterations observed in individuals with ASD [7,8].
This review aims to explore the recent advancements in understanding the role of the cerebellum in ASD, focusing on the impact of ASD-associated genetic alterations on cerebellar development and their contribution to ASD pathogenesis. By examining experimental models that have elucidated the effects of these genetic factors, this review will shed light on the potential for future therapeutic interventions that target the intricate relationship between genetic influences and cerebellar development in ASD.
2. Syndromic and Non-Syndromic ASD
ASDs are categorized into syndromic and non-syndromic (idiopathic) types based on clinical criteria. These terms have recently been extended over to annotate genes (as in “syndromic” and “non-syndromic” genes for ASD), but like all classifications, it is simplistic [9,10].
Syndromic ASD, which accounts for less than 10% of clinical cases of autism [10,11], includes disorders characterized by both genetic anomalies and physical manifestations, such as Fragile X (FXS), MECP2 duplication, Phelan–McDermid, and tuberous sclerosis complex (TSC) syndromes.
These disorders are often characterized by significant genetic etiologies including chromosomal abnormalities like the 15q11-q13 duplication, 6p11.2 microdeletion, and 22q11.2 deletion syndrome, copy number variations (CNVs), or mutations in a single gene resulting in complex clinical profiles. The genetic underpinnings of syndromic ASD involve disruptions in transcriptional regulation and synaptic homeostasis, affecting the development of cognitive and behavioral circuits [11]. Notable genes involved include the SHANK3 gene, mapped to chromosome 22, which encodes a postsynaptic membrane scaffolding protein necessary for the development, maturation, and plasticity of excitatory glutamatergic synapses [12]. SHANK3 is deleted in Phelan–McDermid syndrome, which is characterized by severe intellectual disability, language delay, and autism [13]. This gene, along with SHANK1 and SHANK2, is considered a high confidence gene associated with ASD in SFARI, the database with the most up-to-date information on all known human genes associated with ASD (https://sfari.org/).
Studies in Shank3-deficient mice, which express this gene predominantly in the granule cell layer (GCL) of the cerebellum, highlight its critical role in sensory learning and its association with ASD [14]. These mice have normal cerebellar anatomy but show a significant reduction in the number of dendritic spines of PCs, suggesting an involvement of SHANK3 in the development of mature neuronal connections [14].
Another significant gene, the X-linked MECP2 gene, associated with MECP2 duplication syndrome in boys [15] and Rett syndrome in girls [16], regulates gene expression through interactions with methylated DNA and plays a critical role in proper brain development from infancy through childhood [17,18]. Studies in mouse models have demonstrated that mutations in the MECP2 gene result in reduced cerebellar volume and impaired motor and associative behaviors. These findings underscore the critical role of MECP2 in cerebellar function and of the alteration of this brain area associated with ASD pathophysiology [19].
Non-syndromic ASD, typically referred to as classic autism, first described by Kanner in 1943 [20], is characterized by cognitive and behavioral symptoms only. The etiology of these forms of autism is unknown, with both rare and common variants contributing to their heritability [21,22]. Although individually these variants have a small effect, cumulatively they influence the risk of autism [23].
3. Genomic Complexity in ASD
The complexity of the gene network associated with ASD has become apparent with the advances in molecular genetics and technologies for analyzing large genomic datasets. This complexity is evident in the diversity of genetic alterations, including copy number variants (CNVs), the primary form of inherited gene risk mutations for ASD, and de novo mutations, which play a significant role in ASD pathology.
CNVs, which involve duplications or deletions of a portion of a chromosomal region, are the primary form of inherited gene risk mutations for ASD, although the consequences of these cytogenetic abnormalities are variable. They characterize most syndromic ASD, such as the Angelman syndrome (AS) [24], which is caused by a 15q11.13 deletion and accompanied by a deficit in the maternally inherited UBE3A gene. UBE3A is critical for protein degradation by the proteasome, and its deficiency at the embryonic ages leads to an accumulation of UBE3A targets with the disruption of specific brain functions. The gene is expressed preferentially in distinct brain areas, including PCs in the cerebellum, and many dysregulated pathways have been described in the cerebellar mouse model of the syndrome [25]. Specifically, an imbalance of mTORC1/mTORC2 signaling and oxidative stress in PCs, in addition to an abnormal tonic inhibition in GCs linked to reduced degradation and activity of the GABA transporter 1 (GAT1), have been associated with gait dysfunction and ataxia in AS mice [26,27].
In addition to common and rare CNVs, ASD patients frequently exhibit de novo structural genomic mutations that differ significantly from those of neurotypical individuals.
These mutations primarily affect chromosomal regions linked to ASD-related genes, as detailed in a review by Vicari et al. [28].
The largest genome-wide association study (GWAS), involving 18,000 ASD patients and 28,000 controls from the Danish population, demonstrated the importance of single nucleotide polymorphisms (SNPs) in five ASD-specific-loci, predominantly located in regulatory regions crucial for human cortical development [29]. To expand the sample sizes of GWAS, the SPARK dataset (https://sparkforautism.org/) was utilized, resulting in the identification of two loci with novel variants [30]. The majority of the identified variants were found to be enriched in enhancers [31], with a particular focus on cerebellar enhancers at different developmental stages [32].
Among the predicted target genes of these enhancers, PAX6, TCF4 and ZMIZ1 have been annotated in the SFARI database. PAX6 encodes a transcription factor that is crucial for cerebellar development, and its deletion in mice leads to aberrant growth of the glutamatergic neurons as GCs [33].
Over the past decade, genetic laboratories and consortia, using whole exome sequencing (WES), have identified rare inherited and de novo variants within the coding regions of the genome that are associated with ASD risk. These variants, predominantly loss of function (LoF) or insertion/deletion mutations, have a significant impact on the encoded proteins compared to common variants [34,35].
Using validated statistical approaches, such as TADA [36], susceptibility genes associated with ASD have been identified with high confidence (false discovery rate (FDR) threshold of <10%) [21,23,37]. The number of such genes increases with the number of samples sequenced.
Data from a WES analysis of approximately 12,000 ASD patients from the Autism Sequencing Consortium (ASC) [38] revealed coding mutations in 102 autosomal genes [39], including 30 novel genes [21,34,39]. By increasing the number of individuals studied from the SPARK Consortium and SFARI Simons Simplex Collection (SSC), additional new genes have been identified [40,41].
A study by Trost et al. [42], which involved more than 7000 individuals with autism and 13,000 family members, identified 134 genes through whole genome sequencing (WGS). Of these, 67 were novel genes beyond complex DNA rearrangements such as tandem repeat expansions.
A functional networks analysis of ASD risk genes reveals their involvement in specific biological processes such as gene expression regulation, neuronal communication, migration, and proliferation, all of which are important for neuronal development [21,34,39,40,41,42].
The WGS approach has shifted attention to the regulatory regions of genes and to non-coding genes such as lncRNA or miRNA [43]. The importance of these genomic studies is illustrated by the hyperexpansion of a CGG repeat in the 5′ untranslated region (UTR) of the FMR1 gene of the FXS [44]. The hypermethylation of the repeats in the promoter region results in the silencing of the gene and the absence or greatly reduced synthesis of FMRP. FMRP is critical for normal brain function, particularly during specific periods of embryonic and early postnatal development [45,46]. It is responsible for the localization, stabilization, and translation of many brain mRNAs, including those encoding synaptic and ion channel proteins.
In FXS, the most apparent dysfunction relates to motor behaviors like tremor and ataxia [47]. Alterations in the structure and function of the cerebellum have been observed in FXS patients. Studies in a mouse model have demonstrated altered synaptic plasticity and reduced spontaneous activity and excitability of PCs [48]. Interestingly, global Fmr1KO mice showed that in restoring Fmr1 expression solely in PCs, an improvement in behavioral deficits was obtained, underscoring the pivotal role of the cerebellum in ASD behaviors [49].
All genetic approaches relevant to the identification of candidate gene variants associated with ASD require well-defined statistical applications to generate a significant gene list that can be refined and extended by gene ontology (GO) enrichment analysis to categorize the genes and by the establishment of a protein–protein interaction (PPI) network. One challenge lies in the retrieval of gene lists from different studies and databases, which often exhibit discrepancies. This is partly due to the heterogeneity of the samples analyzed and the statistical approaches employed. To consolidate evidences from different sources, we combined the 232 genes classified as high-confidence ratings for ASD (score of 1) in the SFARI gene database (https://gene.sfari.org/database/human-gene/ accessed on 5 July 2024), the 162-gene list from the SPARK Committee (http://sparkforautism.org/portal/page/spark-gene-list/ accessed on 5 July 2024), the 102 genes identified by Satterstrom and colleagues [39] starting from ASC, the 185 genes identified from four independent cohorts [41], and the 135 genes identified by the Autism Speaks Genomics Program (MSSNG, https://research.mss.ng) [42].
By overlaying the different sources, a core set of 43 genes was identified that consistently appeared across the datasets (Figure 1A). These genes are listed in Table 1. Many of the genes did not yield the highest SFARI gene score for ASD association. Further analysis through GO functional enrichment highlighted their involvement in crucial neural functions, including gene expression regulation and synaptic communication (Figure 1B). This suggests that they may have potential translational medical applications.
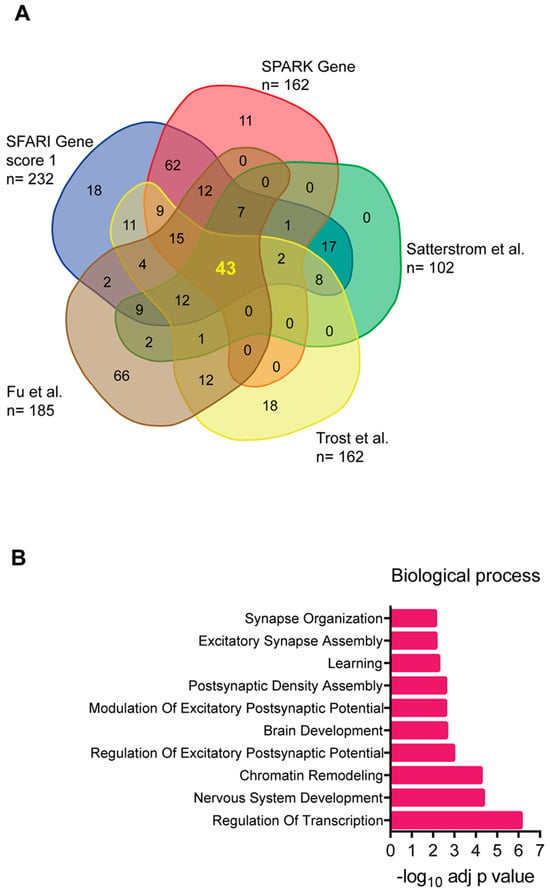
Figure 1.
(A) Venn diagram showing the 43 ASD-associated genes obtained by overlapping the gene list from the different genomic databases and the author’s studies mentioned in the text [39,41,42]. (B) Gene ontology biological process of the 43 ASD-associated genes. Each term is expressed as −log10 of the adjusted p-value (Benjamini and Hochberg).

Table 1.
List of 43 genes derived by overlap analysis shown in the Figure 1.
4. Neuropathological Mechanism of ASD
The identification of predisposing genes does not immediately translate into a mechanistic understanding of ASD. The critical questions of where, when, and in which cell types neurophysiology is altered for the development of this brain disorder remain unanswered.
A comprehensive analysis of RNA-seq data from the Genotype-Tissue Expression (GTEx) resource (https://gtexportal.org) revealed that the strongest expression was observed in the developing cortex and cerebellar hemisphere [39,50].
Furthermore, the transcriptomic landscapes of ASD have been extensively studied through analyses of post-mortem brain tissue. The differential gene expression (DEG) between ASD samples and controls reveals a significant downregulation of genes related to synaptic signaling and plasticity. Additionally, there is a notable upregulation of microglial, astrocyte, and neural immune genes, which suggests a potential link between ASD and neuroimmune dysfunction [43,51,52]. Genetic studies have demonstrated that immunogenic factors involved in autoimmunity and allergic responses may have a regulatory function in both the mature and the developing brain and that these factors are correlated to autistic-like traits [53,54]. The most significant unanswered question arising from these studies is whether the immune dysfunction is a primary cause or a consequence of the underlying disorder.
Although advances in genomic and transcriptomic analysis have increased our knowledge of the biological basis of ASD risk, few studies have investigated the proteomic network in this pathology. Several biochemical approaches have been employed to elucidate the ASD proteome as reviewed by Murtaza et al. [55]. An early proteomics analysis of post-mortem ASD prefrontal cortex and cerebellum samples revealed differential protein expression. Specifically, those associated with myelination, synaptic function, and energy metabolism were decreased in the cortex, while their expression was increased in the cerebellum. This suggests a perturbation in functional connectivity in both regions [56]. Recent studies have demonstrated significant proteomic differences in the occipital cortex and cerebellum between individuals with autism ASD and those without the condition. These differences have been observed in various biological processes, including alterations in synaptic scaffolding, glutamatergic transmission, calcium signaling, and axonal cytoskeletal proteins [57]. These findings align with genetic and transcriptomic studies.
In addition to differential protein expression analysis, the identification of biologically relevant PPI networks and signaling pathways is crucial for elucidating pathogenic processes underlying ASD. However, the PPI networks built from ASD risk proteins through public databases may not accurately represent the active networks in the brain. Efforts to construct more specific neural “interactomes” using cultured neurons and hiPSC-derived neuronal models have begun to reveal the functional roles of ASD-related proteins in the frontal cortex and cerebellar hemispheres [55,58].
5. Cerebellar Involvement in ASD
Over the past decade, a few key characteristics of ASD have been identified. These include the involvement of specific brain regions, collectively referred to as the “social brain” and those closely associated with it. These include the prefrontal and occipital cortex, amygdala, hippocampus, limbic system, and dopaminergic areas. The cerebellum, which is traditionally associated with the modulation of motor functions, has been demonstrated to play a pivotal role in cognitive circuits and complex behaviors [59,60,61]. The “cognitive” functions of the cerebellum depend on its reciprocal connections with the cortex, which are established during development, and on the processing of the signals exchanged between different cortical regions. For a comprehensive review of the cerebellar connectivity within the “social brain” network, see Stoodley and Tsai (2021) [62] and Mapelli et al. (2022) [63].
New research indicates that insults to the cerebellum during a specific developmental window, particularly in the perinatal and early postnatal period, may lead to alterations in cognitive processes that manifest later in life and underlie the clinical outcomes of ASD [7,8].
The developmental alterations associated with ASD in the cerebellum, particularly in the cytoarchitecture of this brain area, have been studied mainly in mouse models that mimic both syndromic and idiopathic ASD pathology. For a detailed description, see Mapelli et al., 2022 [63].
5.1. Morphological and Functional Cerebellar Alterations in ASD
Neuroimaging studies, histopathological and molecular analysis of post-mortem brains (reviewed in Fetit 2021 [64]), and functional analysis of cerebellar connectivity have revealed abnormalities associated with neurodevelopmental disorders such as ASD.
5.1.1. Cerebellar Anatomy and Neuronal Specification
The cerebellum consists of a central part, the vermis, and two lateral extensions, the cerebellar hemispheres (Figure 2A). The folia divide the cerebellum anterior–posteriorly into three lobes, the anterior lobe (corresponding to the sensorimotor cerebellum), which is separated from the posterior lobe by the primary fissure, and the flocculonodular lobe below the posterolateral fissure (Figure 2B). The deep cerebellar nuclei (DCN: dentate, interpositus, and fastigial) consist of excitatory neurons that make polysynaptic cerebellar–cerebral projections through the thalamus and inhibitory neurons that project to the inferior olive (Figure 2C).
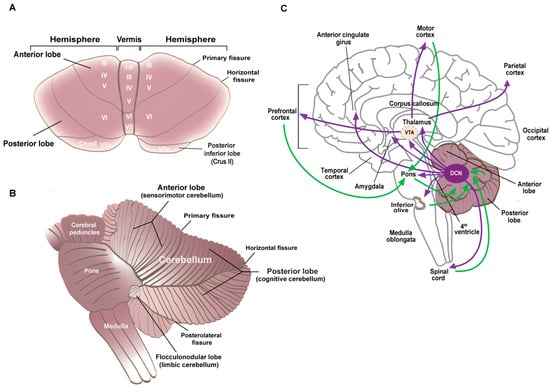
Figure 2.
(A) Superior view of the isolated cerebellum. The lobes and the fissures that separate them are indicated. Roman numerals indicate the lobules. (B) Left lateral view of the cerebellum and brainstem. The functional divisions of the lobes are indicated with posterior lobules VI and VII in the vermis and the hemispheric extensions Crus I and Crus II forming the cognitive cerebellum. (C) Medial view of the brain with the efferent cerebellar connections indicated in purple and the afferents to the cerebellum in green. The deep cerebellar nuclei (DCN) project to the thalamus and midbrain, including the ventral tegmental area (VTA), amygdala, and anterior cingulate cortex. These regions then synapse with different cortical regions to form the cortico–cerebellar–cortical loops.
The cerebellar neurons are organized into apparently simple circuits whose diversity and dynamics have recently been highlighted [65,66]. In the cerebellar cortex, a monolayer of inhibitory GABAergic PCs (Purkinje cell layer, PCL) represents the only output neurons projecting to the DCN, which in turn project to the brainstem, spinal cord, ventral tegmental area (VTA), and thalamus, and from there to the cortex. The DCN–thalamic projections cross the midline to the contralateral side in a manner analogous to the pontine–cerebellar afferents. PCs integrate direct excitatory afferent signals from the climbing fibers originating in the inferior olive and the mossy fibers from various nuclei (in the midbrain, pons, and spinal cord), which indirectly influence the PCs via the parallel fibers of the GCs. These axons establish excitatory synapses with the distal dendrites of PCs in the outer molecular layer (ML) of the cerebellar cortex (Figure 3). The refinement of the output signal is regulated by several populations of inhibitory interneurons, including Golgi cells in the GCL, molecular layer interneurons I and II (MLI, MLII), and Bergmann glial cells in the PCL, all of which play a crucial role in the proper development of the cerebellar cortex.
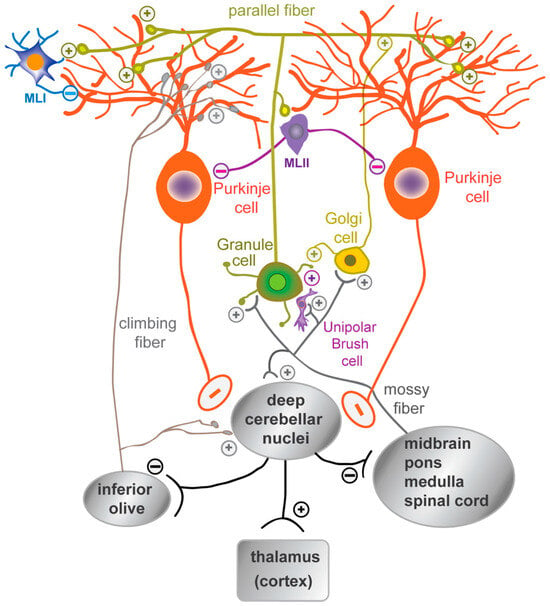
Figure 3.
A schematic representation of the organization of mature cerebellar cells. The synaptic connections between the dendritic tree of the Purkinje cells and the parallel fibers of granule cells, as well as with the inhibitory molecular layer interneurons (MLI, MLII), are shown. Granule cell activity is modulated by Golgi cells and unipolar brush cells. The mossy fibers, originating from the brainstem and spinal cord nuclei, and the climbing fibers, originating from the inferior olive, are the excitatory afferent fibers that reach the cerebellar cortex. The mossy fibers regulate granule cell activity, while the climbing fibers synapse on the proximal dendrites of Purkinje cells.
It is noteworthy that the development of the human cerebellum remains relatively understudied. This process extends from 30 days post conception (dpc) to the second postnatal year [67]. The prolonged period of its development renders it susceptible to insults, including genetic and environmental factors.
Cerebellar neurons originate from radial glial progenitors in two primary zones of neurogenesis: the ventricular zone (VZ) and the rhombic lip (RL). Recently, the molecular characteristics and heterogeneity of progenitors in the two niches have been elucidated [68,69].
The VZ is responsible for the generation of all GABAergic populations during the embryonic period, including PCs. These neurons begin to differentiate at an early stage and can be identified as early as 47 dpc in a region that, similarly in the developing cortex, constitutes the sub-ventricular zone (SVZ) containing the mitotic progenitors [70,71].
During the fetal period, PCs migrate towards the outer surface of the cerebellar anlage, initially aggregating in multiple layers. The PCL, formed by a monolayer of PCs, is completed in the third trimester. After this, PCs undergo axon extension and dendritic tree expansion. In the meantime, the RL, which expands after VZ neurogenesis ceases, gives rise to all cerebellar glutamatergic neurons, including GCs. RL-derived progenitors migrate initially to give rise to neurons of the cerebellar nuclei and, subsequently, tangentially to the surface of the anlage, to form the external granule layer (EGL), which is populated by the immature GCs. In humans, the RL persists throughout gestation, undergoes internalization, and remains proliferative postnatally, contributing GC progenitors to the posterior vermis (Figure 4).
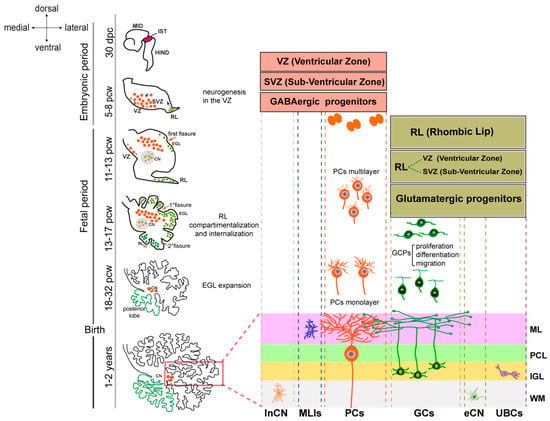
Figure 4.
Human cerebellar development. The timeline of cerebellar development is shown on the left. The drawings show a 30 day post conception (dpc) embryo MID—midbrain, IST—isthmus, HIND—hindbrain, and cerebellar morphology at different stages of development. In the embryonic period, the cerebellar anlage is evident at 30 dcp. The progenitor zone of GABAergic neurons, including Purkinje cells (PCs), inhibitory neurons of the cerebellar nuclei (inCN), and molecular layer interneurons (MLIs), corresponds to a ventricular zone (VZ) and a sub-ventricular zone (SVZ). These regions expand until 8 postconceptional weeks (pcws) before regressing to a single cell layer. The glutamatergic progenitors of the excitatory neurons of the cerebellar nuclei (eCN), the granule cells (GCs), and the unipolar brush cells (UBCs) are born in the rhombic lip (RL), which begins its expansion in the fetal period. The external granular layer (EGL) appears at the end of the embryonic period and grows extensively throughout the fetal period due to the proliferation of granule cell precursors (GCPs). Between 13 and 14 pcw, the elongated RL divides into an RL-SVZ and an RL-VZ, which internalize and give rise to the neurons of the posterior lobe. The cortical layers in the mature cerebellum are marked (boxed area) and labeled on the right as follows: ML—molecular layer, PCL—Purkinje cell layer, IGL—internal granule layer, and WM—white matter.
During the third trimester, the cerebellum undergoes extensive growth due to the proliferation of the GCs, which migrate to become granule neurons of the internal granule layer (IGL). The migration of GCs is completed when the EGL disappears at the end of the first postnatal year.
During the postnatal period, cerebellar neurons undergo a maturation process that is characterized by the acquisition of intrinsic spontaneous activity, synaptic refinement, and pruning due to experience-induced plasticity and the activation of different signaling pathways.
Advances in prenatal diagnosis have enabled the monitoring of cerebellar development during fetal life, thereby facilitating the early detection of malformations that were previously diagnosed in adolescents or adult patients.
5.1.2. Cerebellar Morphology and Connectivity Abnormalities in ASD
Although there is conflicting evidence between different approaches, the cerebellum consistently emerges as one of the most affected brain areas in ASD, exhibiting abnormalities that also correlate with the severity of symptoms.
The first description of hypoplasia of the posterior vermis was in a 21-year-old man diagnosed with autism [72]. This was subsequently confirmed by neuroimaging studies in children and adolescents [73,74]. A recent study employing magnetic resonance imaging (MRI) analysis revealed a statistically significant increase in lobular volume in several cerebellar regions, including the vermis, left and right lobule I–V, right Crus II, and right lobules VIIb and VIIIb, in children with ASD aged between 15 and 36 months when compared with both siblings and healthy controls [75].
Moreover, correlations were identified between morphological changes in the cerebellum and clinical scores based on cognitive, motor, and language functions. These studies confirm the previous observations [76] that altered cerebellar topography is associated with clinical deficits observed in children with ASD. Nevertheless, a more recent, larger study [77] did not yield compelling evidence for volumetric alterations in the cerebellum and its regions. This highlights the limitations of MRI-based methodologies, particularly when applied to small sample sizes, and the difficulties inherent in classifying lobules using disparate atlases as references. To accurately assess structural changes in the cerebellum in ASD, more sophisticated measures, such as local surface area and thickness of the cerebellar cortex, are required.
The involvement of the cerebellum in ASD is also evidenced by the consequences of deficits that occur in this brain area, particularly at early stages of its development. This is exemplified by studies on preterm infants with isolated cerebellar injury. Limperopoulos et al. [78,79] have demonstrated that such injuries not only result in long-term motor deficits but also lead to the impairment of regional growth in the contralateral cerebral hemisphere. Additionally, they have shown that these injuries cause deficits in the development of cognition, communication, and social interaction. These deficits are indicative of contralateral cerebro-cerebellar projections, whereby the left cerebellar hemisphere forms circuits with right cerebral cortical regions that are involved in spatial processing, and the right cerebellar hemisphere interconnects with left cerebral language regions.
It has been demonstrated that perinatal damage to the cerebellum is associated with a high predicted risk of ASD and may result in the development of long-term ASD symptoms, including behavioral problems, deficits in social interactions, and in affect, attention, and other cognitive functions [8,78]. Wang et al. [7] proposed that the impact of early cerebellar damage on the neuropathology of ASD was due to the influence of this area on the development of cerebral cortical regions to which the cerebellum projects.
The morphological alterations of the cerebellum are not exclusive to autism. This implies that the differences between neurodevelopmental disorders are likely to be found at the cellular and molecular levels. Post-mortem studies offer the opportunity to highlight the cellular changes in the autistic cerebellum. Among the most observed findings are a reduced number of PCs and alterations in their shape, particularly in the lateral hemispheres at Crus I and II [80,81]. This is accompanied by a reduction in GAD65 and GAD67, which catalyze the synthesis of GABA [80], and a reduction in the GABA receptor subunits, GABBR1 and 2 [82]. The reduction of GABAergic inputs from the cerebellum, in conjunction with alterations in GABA signaling, may contribute significantly to the hyperexcitability of cerebellar connections, particularly with the cortex.
Cerebellar alterations have also been identified in animal models of idiopathic and monogenic ASD. For instance, rodents exposed to VPA during fetal development exhibit autistic features postnatally [83]. These mice exhibited morphological alterations in the posterior cerebellar lobules and a reduction in the number of PCs in these regions [84,85]. In addition, the BTBR inbred mouse line, which is known to display robust and consistent autism-relevant behaviors [86], exhibits delayed eyeblink conditioning accompanied by an expansion of the vermis and aberrant foliation in both the early postnatal and adult cerebellum [87,88]. BTBR mice exhibit a slight but significant reduction in PC density and a marked reduction in dendritic spines, the loss of which in ASD remains an open question.
Cerebellar pathology has also been observed in individuals with FXS, characterized by atrophy of the vermis and reduced numbers of PCs [89]. A mouse model lacking Fmr1 has been observed to exhibit alterations in the structure and plasticity of synapses on PCs [48,90].
Furthermore, atypical PC structure and function have been documented in the TSC disorder caused by mutations in TSC1 and TSC2 proteins, which negatively regulate the mTOR pathway [91]. A TSC1 mouse model of autism also exhibited similar cerebellar abnormalities [92].
In addition to the presence or absence of structural abnormalities, the cerebellum of individuals with ASD exhibit differences in functional connectivity. These differences frequently manifest as a reduction in the lateralization of the contralateral cerebellar-cerebral communications that are characteristic of motor, sensory, language, and cognitive functioning [93]. Deficits in sensorimotor behaviors, which emerge early in development, are a common core symptom in ASD patients. In a functional MRI (fMRI) study by Mostofsky and colleagues [94], a cohort of children were observed performing a finger motor task. It was observed that there was hyperactivation of premotor cortical regions, accompanied by decreased cerebellar activation in ASD patients. A reduction in the connectivity between the left anterior intraparietal lobule and the right Crus I has been observed in individuals with ASD during a visuomotor task. This alteration is more pronounced in children than in adolescents or adults, and it is associated with an increased severity of ASD symptoms [95]. Furthermore, atypical fMRI activation of cerebro-cerebellar circuitries has been observed in children with ASD during tasks involving motion perception or social interaction [96,97].
In contrast, Oldehinkel and colleagues [98] analyzed 20 well-characterized resting-state networks and demonstrated increased cerebellar connectivity between visual association, somatosensory, and motor networks in ASD subjects compared to typically developing subjects. This finding is consistent with those of other research groups [99,100]. It is of note that adolescents with ASD and infants at high familial risk for ASD exhibit hypoconnectivity between the right Crus I and language-related regions [101,102]. The findings on functional cerebellar connectivity alterations in ASD are dependent on the approach used (resting-state fMRI vs. task-based fMRI), the age of the subjects analyzed, and, most importantly, the inter-individual variance in cerebellar functional networks [103].
The findings collectively indicate a correlation between the morphological and functional cerebellar anomalies and the sensorimotor behaviors. Nevertheless, several questions remain unanswered. One such question is whether the sensorimotor deficit causes the reduced development of the cerebellum and thus a reduced number of functional cells, or whether the reverse is true.
5.2. ASD Susceptibility Genes and Cerebellar Dysregulation
The complex interactions between the cerebellum and the brain areas involved in ASD, which undergo development and maturation during the first years of life, offers insights into why insults to the cerebellum during this developmental period can result in more severe behavioral and cognitive impairments than those observed in children and adults.
Nevertheless, the molecular mechanisms underlying the link between the cerebellum and ASD remain unclear. To gain further insight, the use of mouse models has been invaluable.
The transcriptional program governing mouse cerebellar development is highly dynamic and includes modulation of ASD-associated genes as evidenced by the significant overlap with genes dysregulated in human ASD brains and present in the SFARI and Autism Gene databases (AutDB, http://www.mindspec.org/autdb.html accessed on 5 July 2024) [84].
During the maturation of the mouse cerebellum, 162 ASD-associated genes are expressed, which significantly fall into functional processes that are frequently dysregulated in ASD. The most significant categories were related to neurogenesis, behavior, synapse maturation, and function, with a specific enrichment observed in genes related to the synapse involving the parallel fibers of GCs and PC dendrites [84]. Furthermore, an analysis of the transcriptome of mouse PCs, laser-captured at different developmental stages, confirmed the expression of ASD-associated genes in these neurons. This finding underscores the vulnerability of PCs during this early developmental stage [104]. It is of note that the gene expression profile of PCs differed from that observed in the cortex during the same developmental period, thus highlighting the unique molecular features of these cerebellar neurons [104].
In contrast to mice, the transcriptional landscape of the human cerebellum is underrepresented in transcriptomic studies of the developing brain. This limitation diminishes the statistical power of ASD gene enrichment analysis. Moreover, the gene expression patterns observed in the human cerebellum diverge from those observed in the mouse during development [69,105].
A molecular program that characterizes progenitors and differentiating cells in the human cerebellum, particularly in the fetal period, has been recently highlighted using single-cell and spatial transcriptomic approaches [68,69,105]. These approaches have revealed the existence of a complex molecular landscape underlying the differentiation of cerebellar cells. These studies demonstrated that cerebellar neurons generated during early development exhibit spatial and molecular heterogeneity, which will lead to the formation of distinct neural projections and connections in the adult organ.
By employing single-cell chromatin accessibility methods, researchers have elucidated the cis-regulatory elements and transcription factors that are instrumental in determining the fate of distinct cerebellar cells [69,105]. This approach has revealed that PCs and GCs exhibit spatial heterogeneity and are determined in a hierarchical manner, each with a distinct molecular signature. For instance, at post-conception week (pcw) 10, all progenitor cells in the ventricular zone (VZ) express RORA, a type of retinoic acid-related orphan receptor. However, by postnatal day 14, only a subset of RORA-positive PCs that migrated to the outer edge of the cerebellar cortex expressed RORB [69].
Interestingly, RORB, which plays a role in regulating PC differentiation and maturation, is among the 43 ASD-associated genes (Figure 1, Table 1) and is characterized by rare single mutations [34,39]. A consensus sequence analysis of the 286 potential target genes expressed by PCs at the beginning of differentiation [69] identified 13 genes, including RORB, that were highly significant for ASD in the SFARI database (Table 2). These genes, including SCN2A, SHANK2, GRIN1, and NRXN1, and 3, play a critical role in axonogenesis, synaptic assembly, and signaling (Table 2). Although this in silico analysis requires further experimental confirmation, it suggests a possible involvement of RORB in ASD neuropathology.

Table 2.
RORB-target genes expressed by PCs and included in the SFARI database.
The importance of accurate molecular and regional PC differentiation in establishing the correct connections within the “cognitive brain” is underscored by the observation that ASD-associated genes are prominently expressed in prenatal PCs [68,69,105]. It is not surprising that these genes are classified into functional categories that mirror those observed in the cortex: chromatin organization and neuronal development [68,106]. Furthermore, it has been observed that PCs display an enrichment of SNPs in both coding and non-coding regions of genes associated with ASD, providing additional confirmation of their involvement in the pathogenesis of the disorders [69].
During the transcriptionally mapped fetal period, when ASD-associated genes are highly expressed, neurogenesis takes place in the cerebellar nuclei, and PCs emerge as the most predominant neuronal population. These cells migrate and initiate axonogenesis before organizing into a monolayer. Meanwhile, GCPs migrate to the cerebellar cortex to form the EGL before initiating their expansion phase, which characterizes the postnatal period. The lack of functional and molecular changes in highly proliferative and therefore vulnerable GCPs and GCs is intriguing. GCs play a crucial role in the proper maturation of PCs and receive mitogenic signals from them, such as SHH, and are driven by PCs in their migration from the EGL to the IGL. It can be hypothesized that ASD-mutated genes may indirectly influence the differentiation of GCs due to the close functional relationship between PCs and GCs.
By examining the gene expression profiles of ASD-associated genes during cerebellar development, it became clear that they are expressed at higher levels during prenatal development compared to postnatal development [105]. However, the cerebellar maturation process during the neonatal and early postnatal periods is a complex one. The number of inputs undergoes a rapid change, and synaptogenesis occurs, particularly the formation of synapses between parallel fibers and PCs. Furthermore, the activity-dependent maturation of climbing fibers involves the pruning and relocation of these fibers from the soma to the dendrites of PCs.
Brandenburg and colleagues have demonstrated transcriptomic differences in PCs that were laser-microdissected from post-mortem cerebella of young ASD patients (mean age 19 years) compared to normal tissue [107]. Of the 427 differentially expressed genes, the majority were downregulated compared to controls. These genes produced protein–protein interaction clusters related to neurogenesis, cell adhesion and migration, and cell–cell communication. This finding confirms an altered developmental trajectory of PCs in ASD pathology.
Ament and colleagues recently proposed a single-cell transcriptomic atlas of the human cerebellum during childhood (one to five years), which highlights the heterogeneity of PCs [108].
These neurons are typically identified by the expression of ALDOC/ZEBRIN2, and each of the two differentiated populations expresses specific genes, thus indicating functional differences between stripes and representing specific developmental trajectories. While the distribution of spatial stripes and the function of the two main subtypes of PCs are known in rodents, the heterogeneity of PCs in humans is still being elucidated. Ament and colleagues confirmed that risk genes for ASD are enriched in prenatal PCs and decline during early childhood. The authors observed that PCs, other than Golgi interneurons, are particularly susceptible to inflammation during childhood, which is a well-known risk factor for ASD [109].
In post-mortem cerebella exhibiting inflammatory conditions, synaptic genes associated with ASD, such as SHANK1, are downregulated in PCs. This suggests a premature defective in their maturation, which could result in the altered connectivity typical of ASDs. The transcriptomic alterations associated with early-life inflammation appear to rescue the GCs, even though they are the most abundant and morphologically simple neuronal population in the cerebellum. These findings corroborate the hypothesis that genetic and environmental factors converge to influence the development of cerebellar neurons, potentially contributing to the etiology of ASD. However, it is not clear how the ASD risk genes affect the development and functionality of PCs. Mouse models have been used to induce PC-specific deletion of a number of these genes, which has revealed the effects on specific behaviors indicative of social impairment. A variety of behavioral tests are employed to assess social, cognitive, and repetitive processing in mice. Most of the tests can be conducted exclusively on adult animals, apart from ultrasonic vocalization, which can be evaluated in postnatal young animals. Most behavioral paradigms are affected in PC-conditional knockout (KO) animals when the deleted genes are involved in synaptic structure or plasticity. For example, Shank2KO mice do not exhibit motor impairment, but they display deficits in motor learning, as well as in social interaction in the three-chamber test, and in specific repetitive behaviors [110,111].
6. ASD Models to Highlight Pathology in the Cerebellum
The study of the cerebellar development is crucial for the understanding of its role in neurodevelopmental disorders, but access to human cerebellar tissue is challenging. In addition to animal models, in vitro models and protocols can be used to differentiate cerebellar neurons and generate a 3D self-organizing organoid from hiPSCs.
The differentiation of hiPSCs into cerebellar neurons is intended to reproduce the neurogenesis of the cerebellum, which is a vulnerable period for the risk of ASD. Previous studies have primarily focused on the generation of immature PCs in hiPSC cultures, which can develop a low-branched dendritic tree after 90 days when co-cultured with mouse cerebellar cells [112].
Furthermore, excitatory neurons derived from RL have been differentiated using a chemically defined medium, resulting in the generation of GCs after 48 days in vitro [113]. A model of ASD using hiPSC-differentiated PCs was obtained from individuals diagnosed with TSC and ASD, carrying a mutation in the TCS2 gene that causes hyperactivation of the mTORC1 signal [114]. TSC2-deficient hiPSCs exhibit differentiation deficits, resulting in PCs with increased soma size, synaptic alterations, and reduced excitability. Additionally, the neurons exhibited indications of autophagy activation and oxidative stress, which ultimately resulted in cell death [114]. Treatment with mTORC1 inhibitors, such as rapamycin and Torin1, has been shown to rescue the synaptic alterations observed in neurons [114], thus highlighting the utility of hiPSCs as a model for pathology and as a platform for therapeutic screening.
Organoid models offer a more accurate representation of the complex ontogenesis of the cerebellum, which undergoes a lengthy developmental period. In a pioneering study published in 2015, Muguruma and colleagues developed a protocol for generating cerebellar neurons through 3D cultures of hiPSCs [115]. The cells aggregated and formed a neuroepithelial structure in the presence of specific growth factors and inhibitors. Upon aggregation with mouse EGLs, the neural precursors differentiated into neurons that exhibited morphological and electrophysiological characteristics similar to those of human PCs. GCs, interneurons, and DN neurons were identified in the 3D aggregates, albeit with a low efficiency [115]. The maturation of the neurons obtained through mouse co-culture exhibited characteristics consistent with those observed during the first trimester of pregnancy. In 2021, Nayler and colleagues [116] developed a long-term culture system that allowed neural networks to form and lobular morphogenesis to occur without the need for mouse cells. This was achieved by adapting the organoid protocol using hiPSCs, which introduced Matrigel, a solubilized basement membrane preparation, to induce differentiation within a defined microenvironment.
Over a 90-day culture period, all the major cerebellar developmental neuronal cell types were identified, including RL cells, GCPs, and immature PCs. Notably, these cells exhibited low levels of ALDOC or RORA marker genes, which are indicative of mid to late embryonic cerebellar development [116]. It is noteworthy that certain genes associated with ASD were found to be enriched in the glutamatergic DCN, suggesting that the organoid model may offer insights into the pathogenesis of the disease [116].
Recent advances have improved the cerebellar organoid models, allowing the recapitulation of more advanced developmental stages, in particular the establishment of functional neural circuits between GCs and PCs. Atamian and colleagues [117] successfully generated organoids that, after 60 days of culture, exhibited spatial segregation of stem niches reminiscent of the VZ and the RL, including the human-specific RLVZ and RLSVZ. Furthermore, Bergmann glial cells, immature PCs, and GCPs, which are characteristic of fetal cerebellar development, were also observed in these organoids. The addition of SDF1, a chemoattractant factor that drives the migration of GCPs, resulted in the identification of organoid regions that resembled the EGL and PCL. At the six-month time point, lamination of GCs and PCs was evident, with both excitatory and inhibitory neurons exhibiting functional spontaneous activity at 60 days and more coordinated firing at six months [117]. Importantly, for the first time, the in vitro differentiated PCs closely resembled late midgestational in vivo neurons, suggesting the potential utility of cerebellar organoids to explore ASD neurophysiology. Furthermore, expression analysis revealed a significant enrichment of ASD risk genes in different neuronal cell types within the organoids. These included progenitor and mature molecular layer interneurons, immature and mature inhibitory neurons of the cerebellar nuclei, and especially immature and differentiated PCs.
The advancement of cerebellar organoid models has yielded invaluable insights into the development and functionality of the human cerebellum, while also elucidating the alterations in these processes that predispose individuals to ASD. Furthermore, the use of organoids developed from patient-derived hiPSCs could potentially be useful in elucidating the specific events that initiate the pathology, as opposed to those that are merely associated with it. This would represent a significant advancement in the development of precision therapy.
It is important to note that these models have limitations that need to be overcome in the future. Most notably, there is variability and reproducibility in the protocols used (Figure 5). Moreover, the profiling of non-neuronal cell types, including astrocytes, Bergmann glial cells, microglia, and vascular endothelial cells, in cerebellar organoids must be enhanced. This is because most of ASD-associated genes are expressed in these cells. The resolution of these critical issues will constitute the inaugural stage in the advancement of cerebellar assembloids, the subsequent generation of brain organoids that will permit the combination of multiple brain regions and/or cell lineages in three-dimensional culture [118].
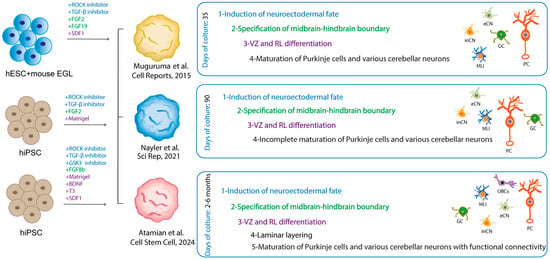
Figure 5.
Comparative representation of the cerebellar organoids. The figure presents a comparative analysis of cerebellar organoids derived from different protocols, as illustrated in the indicated studies. The cultured cells and the factors added to the medium to obtain the self-assembling organoid are indicated. The use of fonts with the same colors was employed to indicate the different stages identified in the organoid, along with the factors responsible for them [115,116,117]. The neurons differentiated in the organoids are schematized in the manner indicated in Figure 3.
7. Conclusions
This review highlights the pivotal role of the cerebellum in the etiology and pathophysiology of ASD, a role that has been historically underappreciated compared to cerebral functions. The involvement of this brain area in ASD has led to the proposal of a clinical trial using non-invasive transcranial magnetic or direct current stimulation as a possible treatment for some of the symptoms of ASD in children (such as repetitive behaviors and hyperactivity) (https://clinicaltrials.gov/ NCT04446442). In recent years, significant progress has been made in identifying ASD-associated genes with hundreds of genetic variants implicated in the disorder. These genes modulate essential processes at the cytoplasmic, nuclear, and synaptic levels, thereby influencing the overall functionality and connectivity of the neurons. Figure 6 illustrates the pivotal role of Purkinje cells in the pathophysiology of the disorder, since ASD-associated genes are expressed in distinct compartments of these neurons, particularly during their maturation. Since ASD is a neurodevelopmental disorder, it is essential to know how the affected areas develop. The knowledge of the mechanisms underlying the determination and maturation of the human cerebellum is still at an early stage compared to what is known about other brain areas, particularly the cerebral cortex. An important aspect is understanding why genetic and environmental insults during a critical period of cerebellar development can have broad effects on cognition and behavior. It is clear that the interplay between GCs and PCs is fundamental for the proper refinement of cerebellar circuitry, and it is intriguing that only PCs appear to be susceptible to ASD development, as evidenced by their morphological, functional, and molecular alterations. It is likely that the expression of ASD-associated genes enriched in PCs during the prenatal period [68,69,105] also characterizes other cell types such as glutamatergic and GABAergic DCN or MLIs, which may be affected in the disorder. In particular, MLIs, which are important for modulating PC activity through their inhibitory input, may contribute to changes in excitatory/inhibitory network connectivity driven by altered GABA signaling of ASD-altered PCs.
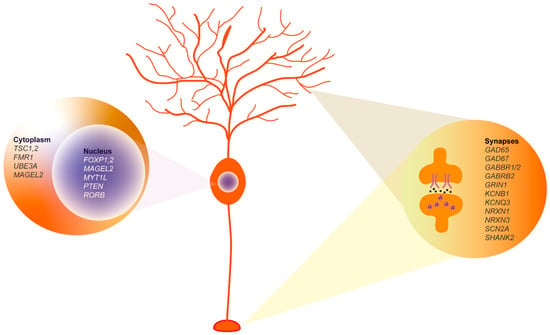
Figure 6.
Purkinje cell molecular profile highlighting key ASD-associated genes. The genes shown, derived from extensive database analyses and synthesized conclusions of our review, represent key elements in ASD pathophysiology. Nucleus: Genes known to influence transcriptional regulation and neuronal differentiation are shown, including FOXP1-2, MAGEL2, MYT1L, PTEN, and RORB. Cytoplasm: The TSC1, 2, and FMR1 proteins involved in signaling pathways critical for neuronal communication and synaptic function are shown. Synaptic region: Includes synaptic proteins such as GAD65, GAD67, GABBR1/2, and SHANK2, among others, which are essential for synaptic transmission and plasticity.
The elucidation of the genomic, transcriptomic, and proteomic landscape that characterizes ASD has been greatly accelerated by the ability to acquire and analyze a vast amount of data. At the same time, this large body of data must be subjected to rigorous and unambiguous interpretation in order to ensure convergence between basic and clinical research. For instance, it is evident that there are discrepancies between the various databases that collect data on autism-associated genes, which makes it challenging to identify which are the most promising candidates for translational research. In this regard, an interesting pipeline has been recently proposed by Schaaf and colleagues [119].
Recent studies and reviews have highlighted the potential of artificial intelligence (AI) to revolutionize our understanding of ASD. This technology offers insights into the integration and the analysis of vast amounts of genomic, transcriptomic, and proteomic data, as well as the identification of patterns and prediction of disease phenotypes with unprecedented precision [120,121]. The integration of comprehensive genetic data obtained through AI with functional analysis provided by organoids, particularly those derived from patient-specific hiPSCs, will enable researchers to elucidate the multifaceted nature of ASD more effectively and facilitate the development of more efficacious treatments.
Basically, this review highlights the critical role of the cerebellum in ASD and emphasizes the importance of integrating genetic, neurobiological, and modelling approaches to elucidate the complex mechanisms underlying the disorder. Continued research efforts aimed at overcoming current challenges and translating findings into clinical applications have the potential to advance personalized therapies for individuals with ASD.
Author Contributions
M.G., V.M., G.L.S. and D.F. contributed to the writing and editing of this review. All authors approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
We thank Massimo De Felici for critical reading of the manuscript.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Glossary
GWAS genome-wide association study: Observational study of a genome-wide set of genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases (commercial genome-wide SNP arrays). This type of study asks if the allele of a genetic variant is found more often than expected in individuals with the phenotype of interest. A key step in most GWA studies is the imputation of genotypes in SNPs not on the genotype chip used in the study.
WES: Whole-exome sequencing. A technique for sequencing the protein-coding regions of the genome (exome) unlike the WGS, which provides whole genome sequencing. WES has the advantage of lower costs thus allowing the sequencing of a large number of samples.
Enhancers: Short sequences of DNA recognized by transcription factors that, by binding upstream or downstream from the start site of a gene, increases its expression, stimulating its transcription.
Eyeblink conditioning: A form of associative learning that involves the cerebellum and its circuits. It consists of the ability to associate two sensations that arrive within a very short time of each other. To study this kind of learning, a bright light is shone at the mice, followed a split second later by a puff of air that always causes the mice to blink. After this happens dozens of times, the mice begin to blink earlier, as soon as the light appears, in anticipation of the puff of air.
TADA method: A statistical framework based on a hierarchical Bayesian method to narrow the pool of genetic variants generated by high-throughput genome sequencing methods and to establish their relevance to the disease of interest. To identify risk genes, the method includes de novo and rare variants transmitted in family trios (father, mother, and an affected child) and data obtained by case-control studies.
Protein–protein interaction (PPI) network: Refers to the dataset based on the physical associations between two or more proteins in a cell or organism. In the network, a node is a protein and an edge is an interaction between two proteins. The hub proteins in the network are those with higher connectivity and therefore more important biological role. With the correlation approach, the PPI network provides functional information about unannotated proteins.
Induced human pluripotent stem cells (hiPSCs): Adult somatic cells reprogrammed into stem cells with a self-renewal capacity that can be induced to differentiate into multiple cell lineages in vitro. These cells maintain the genetic background of the healthy and patient donors.
Organoid: Self-organized 3D tissue derived from either pluripotent or tissue-resident stem cells, or progenitor or differentiated cells from healthy or diseased tissues. This in vitro model aims to mimic the key functional, structural, and biological complexity of an organ.
Assembloid: Culture model resulting from the integration of two or more organoids or an organoid and cells or tissue fragments. Brain assembloids permit the integration of different regions to model cell migration and neuronal projection and connectivity integration. Different cell lineages cultured with organoids permit, for example, the formation of the vasculature in the 3D models.
Resting state networks (RSN): In the functional magnetic resonance imaging (fMRI), sets of brain areas that undergo correlated spontaneous slow fluctuations in the blood oxygenation level dependent (BOLD) signal when no tasks are performed. These areas are functionally connected and correspond to the default mode network (DMN) composed of the medial prefrontal cortex, the posterior cingulate cortex, the inferior parietal lobule, the lateral temporal cortex, and the hippocampal formation.
Anterior intraparietal lobule: In the posterior parietal cortex, corresponds to the area anterior to the intraparietal sulcus. It contains neurons that respond primarily to the hand postures used to grasp an object.
References
- First, M.B. Diagnostic and statistical manual of mental disorders, 5th edition, and clinical utility. J. Nerv. Ment. Dis. 2013, 201, 727–729. [Google Scholar] [CrossRef] [PubMed]
- Sandin, S.; Lichtenstein, P.; Kuja-Halkola, R.; Hultman, C.; Larsson, H.; Reichenberg, A. The Heritability of Autism Spectrum Disorder. JAMA 2017, 318, 1182–1184. [Google Scholar] [CrossRef]
- Christensen, J.; Gronborg, T.K.; Sorensen, M.J.; Schendel, D.; Parner, E.T.; Pedersen, L.H.; Vestergaard, M. Prenatal valproate exposure and risk of autism spectrum disorders and childhood autism. JAMA 2013, 309, 1696–1703. [Google Scholar] [CrossRef]
- Castelbaum, L.; Sylvester, C.M.; Zhang, Y.; Yu, Q.; Constantino, J.N. On the Nature of Monozygotic Twin Concordance and Discordance for Autistic Trait Severity: A Quantitative Analysis. Behav. Genet. 2020, 50, 263–272. [Google Scholar] [CrossRef]
- Dworzynski, K.; Happé, F.; Bolton, P.; Ronald, A. Relationship between Symptom Domains in Autism Spectrum Disorders: A Population Based Twin Study. J. Autism Dev. Disord. 2009, 39, 1197–1210. [Google Scholar] [CrossRef] [PubMed]
- Willsey, A.J.; Sanders, S.J.; Li, M.; Dong, S.; Tebbenkamp, A.T.; Muhle, R.A.; Reilly, S.K.; Lin, L.; Fertuzinhos, S.; Miller, J.A.; et al. Coexpression Networks Implicate Human Midfetal Deep Cortical Projection Neurons in the Pathogenesis of Autism. Cell 2013, 155, 997–1007. [Google Scholar] [CrossRef]
- Wang, S.S.; Kloth, A.D.; Badura, A. The cerebellum, sensitive periods, and autism. Neuron 2014, 83, 518–532. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, C.J.; Limperopoulos, C. Structure-function relationships in the developing cerebellum: Evidence from early-life cerebellar injury and neurodevelopmental disorders. Semin. Fetal Neonatal Med. 2016, 21, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Vorstman, J.A.S.; Scherer, S.W. Contemplating syndromic autism. Genet. Med. 2023, 25, 100919. [Google Scholar] [CrossRef]
- Ziats, C.A.; Patterson, W.G.; Friez, M. Syndromic Autism Revisited: Review of the Literature and Lessons Learned. Pediatr. Neurol. 2021, 114, 21–25. [Google Scholar] [CrossRef]
- Sztainberg, Y.; Zoghbi, H.Y. Lessons learned from studying syndromic autism spectrum disorders. Nat. Neurosci. 2016, 19, 1408–1417. [Google Scholar] [CrossRef]
- Vyas, Y.; Cheyne, J.E.; Lee, K.; Jung, Y.; Cheung, P.Y.; Montgomery, J.M. Shankopathies in the Developing Brain in Autism Spectrum Disorders. Front. Neurosci. 2021, 15, 775431. [Google Scholar] [CrossRef]
- Phelan, K.; McDermid, H.E. The 22q13.3 Deletion Syndrome (Phelan-McDermid Syndrome). Mol. Syndromol. 2012, 2, 186–201. [Google Scholar] [CrossRef] [PubMed]
- Kloth, A.D.; Badura, A.; Li, A.; Cherskov, A.; Connolly, S.G.; Giovannucci, A.; Bangash, M.A.; Grasselli, G.; Penagarikano, O.; Piochon, C.; et al. Cerebellar associative sensory learning defects in five mouse autism models. eLife 2015, 4, e06085. [Google Scholar] [CrossRef]
- Ta, D.; Downs, J.; Baynam, G.; Wilson, A.; Richmond, P.; Leonard, H. A brief history of MECP2 duplication syndrome: 20-years of clinical understanding. Orphanet J. Rare Dis. 2022, 17, 131. [Google Scholar] [CrossRef]
- Good, K.V.; Vincent, J.B.; Ausió, J. MeCP2: The Genetic Driver of Rett Syndrome Epigenetics. Front. Genet. 2021, 12, 620859. [Google Scholar] [CrossRef] [PubMed]
- Balmer, D.; Goldstine, J.; Rao, Y.M.; LaSalle, J.M. Elevated methyl-CpG-binding protein 2 expression is acquired during postnatal human brain development and is correlated with alternative polyadenylation. J. Mol. Med. 2003, 81, 61–68. [Google Scholar] [CrossRef]
- Tillotson, R.; Bird, A. The Molecular Basis of MeCP2 Function in the Brain. J. Mol. Biol. 2020, 432, 1602–1623. [Google Scholar] [CrossRef]
- Chen, R.Z.; Akbarian, S.; Tudor, M.; Jaenisch, R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 2001, 27, 327–331. [Google Scholar] [CrossRef]
- Kanner, L. Autistic disturbances of affective contact. Nerv. Child 1943, 2, 217–250. [Google Scholar]
- De Rubeis, S.; He, X.; Goldberg, A.P.; Poultney, C.S.; Samocha, K.; Ercument Cicek, A.; Kou, Y.; Liu, L.; Fromer, M.; Walker, S.; et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014, 515, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Krumm, N.; Turner, T.N.; Baker, C.; Vives, L.; Mohajeri, K.; Witherspoon, K.; Raja, A.; Coe, B.P.; Stessman, H.A.; He, Z.X.; et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015, 47, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Gaugler, T.; Klei, L.; Sanders, S.J.; Bodea, C.A.; Goldberg, A.P.; Lee, A.B.; Mahajan, M.; Manaa, D.; Pawitan, Y.; Reichert, J.; et al. Most genetic risk for autism resides with common variation. Nat. Genet. 2014, 46, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Hogart, A.; Wu, D.; LaSalle, J.M.; Schanen, N.C. The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol. Dis. 2010, 38, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Maranga, C.; Fernandes, T.G.; Bekman, E.; da Rocha, S.T. Angelman syndrome: A journey through the brain. FEBS J. 2020, 287, 2154–2175. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Liu, Y.; Moreno, S.; Baudry, M.; Bi, X. Imbalanced Mechanistic Target of Rapamycin C1 and C2 Activity in the Cerebellum of Angelman Syndrome Mice Impairs Motor Function. J. Neurosci. 2015, 35, 4706–4718. [Google Scholar] [CrossRef] [PubMed]
- Egawa, K.; Kitagawa, K.; Inoue, K.; Takayama, M.; Takayama, C.; Saitoh, S.; Kishino, T.; Kitagawa, M.; Fukuda, A. Decreased tonic inhibition in cerebellar granule cells causes motor dysfunction in a mouse model of Angelman syndrome. Sci. Transl. Med. 2012, 4, 163ra157. [Google Scholar] [CrossRef] [PubMed]
- Vicari, S.; Napoli, E.; Cordeddu, V.; Menghini, D.; Alesi, V.; Loddo, S.; Novelli, A.; Tartaglia, M. Copy number variants in autism spectrum disorders. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2019, 92, 421–427. [Google Scholar] [CrossRef]
- Grove, J.; Ripke, S.; Als, T.D.; Mattheisen, M.; Walters, R.K.; Won, H.; Pallesen, J.; Agerbo, E.; Andreassen, O.A.; Anney, R.; et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019, 51, 431–444. [Google Scholar] [CrossRef]
- Matoba, N.; Liang, D.; Sun, H.; Aygün, N.; McAfee, J.C.; Davis, J.E.; Raffield, L.M.; Qian, H.; Piven, J.; Li, Y.; et al. Common genetic risk variants identified in the SPARK cohort support DDHD2 as a candidate risk gene for autism. Transl. Psychiatry 2020, 10, 265. [Google Scholar] [CrossRef]
- Visel, A.; Rubin, E.M.; Pennacchio, L.A. Genomic views of distant-acting enhancers. Nature 2009, 461, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.; Badayeva, Y.; Yeung, J.; Wu, J.; Abdalla-Wyse, A.; Yang, E.; Consortium, F.; Trost, B.; Scherer, S.W.; Goldowitz, D. Temporal analysis of enhancers during mouse cerebellar development reveals dynamic and novel regulatory functions. eLife 2022, 11, e74207. [Google Scholar] [CrossRef] [PubMed]
- Yeung, J.; Ha, T.J.; Swanson, D.J.; Goldowitz, D. A Novel and Multivalent Role of Pax6 in Cerebellar Development. J. Neurosci. Off. J. Soc. Neurosci. 2016, 36, 9057–9069. [Google Scholar] [CrossRef] [PubMed]
- Iossifov, I.; O’Roak, B.J.; Sanders, S.J.; Ronemus, M.; Krumm, N.; Levy, D.; Stessman, H.A.; Witherspoon, K.T.; Vives, L.; Patterson, K.E.; et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014, 515, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Ruzzo, E.K.; Perez-Cano, L.; Jung, J.Y.; Wang, L.K.; Kashef-Haghighi, D.; Hartl, C.; Singh, C.; Xu, J.; Hoekstra, J.N.; Leventhal, O.; et al. Inherited and De Novo Genetic Risk for Autism Impacts Shared Networks. Cell 2019, 178, 850–866.e26. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Sanders, S.J.; Liu, L.; De Rubeis, S.; Lim, E.T.; Sutcliffe, J.S.; Schellenberg, G.D.; Gibbs, R.A.; Daly, M.J.; Buxbaum, J.D.; et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013, 9, e1003671. [Google Scholar] [CrossRef]
- Sanders, S.J.; He, X.; Willsey, A.J.; Ercan-Sencicek, A.G.; Samocha, K.E.; Cicek, A.E.; Murtha, M.T.; Bal, V.H.; Bishop, S.L.; Dong, S.; et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 2015, 87, 1215–1233. [Google Scholar] [CrossRef] [PubMed]
- Buxbaum, J.D.; Daly, M.J.; Devlin, B.; Lehner, T.; Roeder, K.; State, M.W. The autism sequencing consortium: Large-scale, high-throughput sequencing in autism spectrum disorders. Neuron 2012, 76, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Satterstrom, F.K.; Kosmicki, J.A.; Wang, J.; Breen, M.S.; De Rubeis, S.; An, J.Y.; Peng, M.; Collins, R.; Grove, J.; Klei, L.; et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2020, 180, 568–584.e23. [Google Scholar] [CrossRef]
- Zhou, X.; Feliciano, P.; Shu, C.; Wang, T.; Astrovskaya, I.; Hall, J.B.; Obiajulu, J.U.; Wright, J.R.; Murali, S.C.; Xu, S.X.; et al. Integrating de novo and inherited variants in 42,607 autism cases identifies mutations in new moderate-risk genes. Nat. Genet. 2022, 54, 1305–1319. [Google Scholar] [CrossRef]
- Fu, J.M.; Satterstrom, F.K.; Peng, M.; Brand, H.; Collins, R.L.; Dong, S.; Wamsley, B.; Klei, L.; Wang, L.; Hao, S.P.; et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 2022, 54, 1320–1331. [Google Scholar] [CrossRef]
- Trost, B.; Thiruvahindrapuram, B.; Chan, A.J.S.; Engchuan, W.; Higginbotham, E.J.; Howe, J.L.; Loureiro, L.O.; Reuter, M.S.; Roshandel, D.; Whitney, J.; et al. Genomic architecture of autism from comprehensive whole-genome sequence annotation. Cell 2022, 185, 4409–4427.e18. [Google Scholar] [CrossRef]
- Parikshak, N.N.; Swarup, V.; Belgard, T.G.; Irimia, M.; Ramaswami, G.; Gandal, M.J.; Hartl, C.; Leppa, V.; Ubieta, L.T.; Huang, J.; et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 2016, 540, 423–427. [Google Scholar] [CrossRef]
- Hagerman, R.J.; Berry-Kravis, E.; Hazlett, H.C.; Bailey, D.B., Jr.; Moine, H.; Kooy, R.F.; Tassone, F.; Gantois, I.; Sonenberg, N.; Mandel, J.L.; et al. Fragile X syndrome. Nat. Rev. Dis. Primers 2017, 3, 17065. [Google Scholar] [CrossRef]
- Milla, L.A.; Corral, L.; Rivera, J.; Zuñiga, N.; Pino, G.; Nunez-Parra, A.; Cea-Del Rio, C.A. Neurodevelopment and early pharmacological interventions in Fragile X Syndrome. Front. Neurosci. 2023, 17, 1213410. [Google Scholar] [CrossRef]
- Richter, J.D.; Zhao, X. The molecular biology of FMRP: New insights into fragile X syndrome. Nat. Rev. Neurosci. 2021, 22, 209–222. [Google Scholar] [CrossRef]
- Chen, Y.S.; Guo, L.; Han, M.; Zhang, S.M.; Chen, Y.Q.; Zou, J.; Bai, S.Y.; Cheng, G.R.; Zeng, Y. Cerebellum neuropathology and motor skill deficits in fragile X syndrome. Int. J. Dev. Neurosci. Off. J. Int. Soc. Dev. Neurosci. 2022, 82, 557–568. [Google Scholar] [CrossRef]
- Koekkoek, S.K.; Yamaguchi, K.; Milojkovic, B.A.; Dortland, B.R.; Ruigrok, T.J.; Maex, R.; De Graaf, W.; Smit, A.E.; VanderWerf, F.; Bakker, C.E.; et al. Deletion of FMR1 in Purkinje cells enhances parallel fiber LTD, enlarges spines, and attenuates cerebellar eyelid conditioning in Fragile X syndrome. Neuron 2005, 47, 339–352. [Google Scholar] [CrossRef]
- Gibson, J.M.; Vazquez, A.H.; Yamashiro, K.; Jakkamsetti, V.; Ren, C.; Lei, K.; Dentel, B.; Pascual, J.M.; Tsai, P.T. Cerebellar contribution to autism-relevant behaviors in fragile X syndrome models. Cell Rep. 2023, 42, 113533. [Google Scholar] [CrossRef] [PubMed]
- Rolland, T.; Cliquet, F.; Anney, R.J.L.; Moreau, C.; Traut, N.; Mathieu, A.; Huguet, G.; Duan, J.; Warrier, V.; Portalier, S.; et al. Phenotypic effects of genetic variants associated with autism. Nat. Med. 2023, 29, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Ellis, S.E.; Ashar, F.N.; Moes, A.; Bader, J.S.; Zhan, J.; West, A.B.; Arking, D.E. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat. Commun. 2014, 5, 5748. [Google Scholar] [CrossRef] [PubMed]
- Voineagu, I.; Wang, X.; Johnston, P.; Lowe, J.K.; Tian, Y.; Horvath, S.; Mill, J.; Cantor, R.M.; Blencowe, B.J.; Geschwind, D.H. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 2011, 474, 380–384. [Google Scholar] [CrossRef]
- Zerbo, O.; Leong, A.; Barcellos, L.; Bernal, P.; Fireman, B.; Croen, L.A. Immune mediated conditions in autism spectrum disorders. Brain Behav. Immun. 2015, 46, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Arenella, M.; Fanelli, G.; Kiemeney, L.A.; McAlonan, G.; Murphy, D.G.; Bralten, J. Genetic relationship between the immune system and autism. Brain Behav. Immun. Health 2023, 34, 100698. [Google Scholar] [CrossRef]
- Murtaza, N.; Uy, J.; Singh, K.K. Emerging proteomic approaches to identify the underlying pathophysiology of neurodevelopmental and neurodegenerative disorders. Mol. Autism 2020, 11, 27. [Google Scholar] [CrossRef]
- Broek, J.A.C.; Guest, P.C.; Rahmoune, H.; Bahn, S. Proteomic analysis of post mortem brain tissue from autism patients: Evidence for opposite changes in prefrontal cortex and cerebellum in synaptic connectivity-related proteins. Mol. Autism 2014, 5, 41. [Google Scholar] [CrossRef]
- Abraham, J.R.; Szoko, N.; Barnard, J.; Rubin, R.A.; Schlatzer, D.; Lundberg, K.; Li, X.; Natowicz, M.R. Proteomic Investigations of Autism Brain Identify Known and Novel Pathogenetic Processes. Sci. Rep. 2019, 9, 13118. [Google Scholar] [CrossRef] [PubMed]
- Pintacuda, G.; Hsu, Y.-H.H.; Tsafou, K.; Li, K.W.; Martín, J.M.; Riseman, J.; Biagini, J.C.; Ching, J.K.T.; Mena, D.; Gonzalez-Lozano, M.A.; et al. Protein interaction studies in human induced neurons indicate convergent biology underlying autism spectrum disorders. Cell Genom. 2023, 3, 100250. [Google Scholar] [CrossRef]
- Ramnani, N. The primate cortico-cerebellar system: Anatomy and function. Nat. Rev. Neurosci. 2006, 7, 511–522. [Google Scholar] [CrossRef]
- Van Overwalle, F.; Manto, M.; Cattaneo, Z.; Clausi, S.; Ferrari, C.; Gabrieli, J.D.E.; Guell, X.; Heleven, E.; Lupo, M.; Ma, Q.; et al. Consensus Paper: Cerebellum and Social Cognition. Cerebellum 2020, 19, 833–868. [Google Scholar] [CrossRef]
- Stoodley, C.J.; Schmahmann, J.D. Evidence for topographic organization in the cerebellum of motor control versus cognitive and affective processing. Cortex 2010, 46, 831–844. [Google Scholar] [CrossRef]
- Stoodley, C.J.; Tsai, P.T. Adaptive Prediction for Social Contexts: The Cerebellar Contribution to Typical and Atypical Social Behaviors. Annu. Rev. Neurosci. 2021, 44, 475–493. [Google Scholar] [CrossRef]
- Mapelli, L.; Soda, T.; D’Angelo, E.; Prestori, F. The Cerebellar Involvement in Autism Spectrum Disorders: From the Social Brain to Mouse Models. Int. J. Mol. Sci. 2022, 23, 3894. [Google Scholar] [CrossRef]
- Fetit, R.; Hillary, R.F.; Price, D.J.; Lawrie, S.M. The neuropathology of autism: A systematic review of post-mortem studies of autism and related disorders. Neurosci. Biobehav. Rev. 2021, 129, 35–62. [Google Scholar] [CrossRef]
- De Zeeuw, C.I.; Lisberger, S.G.; Raymond, J.L. Diversity and dynamism in the cerebellum. Nat. Neurosci. 2021, 24, 160–167. [Google Scholar] [CrossRef]
- Hull, C.; Regehr, W.G. The Cerebellar Cortex. Annu. Rev. Neurosci. 2022, 45, 151–175. [Google Scholar] [CrossRef]
- Haldipur, P.; Bharti, U.; Alberti, C.; Sarkar, C.; Gulati, G.; Iyengar, S.; Gressens, P.; Mani, S. Preterm delivery disrupts the developmental program of the cerebellum. PLoS ONE 2011, 6, e23449. [Google Scholar] [CrossRef]
- Aldinger, K.A.; Thomson, Z.; Phelps, I.G.; Haldipur, P.; Deng, M.; Timms, A.E.; Hirano, M.; Santpere, G.; Roco, C.; Rosenberg, A.B.; et al. Spatial and cell type transcriptional landscape of human cerebellar development. Nat. Neurosci. 2021, 24, 1163–1175. [Google Scholar] [CrossRef]
- Zhong, S.; Wang, M.; Huang, L.; Chen, Y.; Ge, Y.; Zhang, J.; Shi, Y.; Dong, H.; Zhou, X.; Wang, B.; et al. Single-cell epigenomics and spatiotemporal transcriptomics reveal human cerebellar development. Nat. Commun. 2023, 14, 7613. [Google Scholar] [CrossRef] [PubMed]
- Haldipur, P.; Millen, K.J.; Aldinger, K.A. Human Cerebellar Development and Transcriptomics: Implications for Neurodevelopmental Disorders. Annu. Rev. Neurosci. 2022, 45, 515–531. [Google Scholar] [CrossRef]
- Haldipur, P.; Aldinger, K.A.; Bernardo, S.; Deng, M.; Timms, A.E.; Overman, L.M.; Winter, C.; Lisgo, S.N.; Razavi, F.; Silvestri, E.; et al. Spatiotemporal expansion of primary progenitor zones in the developing human cerebellum. Science 2019, 366, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Courchesne, E.; Hesselink, J.R.; Jernigan, T.L.; Yeung-Courchesne, R. Abnormal neuroanatomy in a nonretarded person with autism. Unusual findings with magnetic resonance imaging. Arch. Neurol. 1987, 44, 335–341. [Google Scholar] [CrossRef]
- Scott, J.A.; Schumann, C.M.; Goodlin-Jones, B.L.; Amaral, D.G. A comprehensive volumetric analysis of the cerebellum in children and adolescents with autism spectrum disorder. Autism Res. Off. J. Int. Soc. Autism Res. 2009, 2, 246–257. [Google Scholar] [CrossRef]
- Webb, S.J.; Sparks, B.F.; Friedman, S.D.; Shaw, D.W.; Giedd, J.; Dawson, G.; Dager, S.R. Cerebellar vermal volumes and behavioral correlates in children with autism spectrum disorder. Psychiatry Res. 2009, 172, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Hiremath, C.; Khokhar, S.K.; Bansal, E.; Sagar, K.J.V.; Padmanabha, H.; Girimaji, A.S.; Narayan, S.; Kishore, M.T.; Yamini, B.K.; et al. Altered cerebellar lobular volumes correlate with clinical deficits in siblings and children with ASD: Evidence from toddlers. J. Transl. Med. 2023, 21, 246. [Google Scholar] [CrossRef] [PubMed]
- D’Mello, A.M.; Crocetti, D.; Mostofsky, S.H.; Stoodley, C.J. Cerebellar gray matter and lobular volumes correlate with core autism symptoms. NeuroImage Clin. 2015, 7, 631–639. [Google Scholar] [CrossRef]
- Laidi, C.; Floris, D.L.; Tillmann, J.; Elandaloussi, Y.; Zabihi, M.; Charman, T.; Wolfers, T.; Durston, S.; Moessnang, C.; Dell’Acqua, F.; et al. Cerebellar Atypicalities in Autism? Biol. Psychiatry 2022, 92, 674–682. [Google Scholar] [CrossRef]
- Limperopoulos, C.; Bassan, H.; Gauvreau, K.; Robertson, R.L., Jr.; Sullivan, N.R.; Benson, C.B.; Avery, L.; Stewart, J.; Soul, J.S.; Ringer, S.A.; et al. Does cerebellar injury in premature infants contribute to the high prevalence of long-term cognitive, learning, and behavioral disability in survivors? Pediatrics 2007, 120, 584–593. [Google Scholar] [CrossRef]
- Limperopoulos, C.; Chilingaryan, G.; Guizard, N.; Robertson, R.L.; Du Plessis, A.J. Cerebellar injury in the premature infant is associated with impaired growth of specific cerebral regions. Pediatr. Res. 2010, 68, 145–150. [Google Scholar] [CrossRef]
- Fatemi, S.H.; Halt, A.R.; Realmuto, G.; Earle, J.; Kist, D.A.; Thuras, P.; Merz, A. Purkinje cell size is reduced in cerebellum of patients with autism. Cell. Mol. Neurobiol. 2002, 22, 171–175. [Google Scholar] [CrossRef]
- Skefos, J.; Cummings, C.; Enzer, K.; Holiday, J.; Weed, K.; Levy, E.; Yuce, T.; Kemper, T.; Bauman, M. Regional alterations in purkinje cell density in patients with autism. PLoS ONE 2014, 9, e81255. [Google Scholar] [CrossRef]
- Fatemi, S.H.; Folsom, T.D.; Reutiman, T.J.; Thuras, P.D. Expression of GABA(B) receptors is altered in brains of subjects with autism. Cerebellum 2009, 8, 64–69. [Google Scholar] [CrossRef]
- Nicolini, C.; Fahnestock, M. The valproic acid-induced rodent model of autism. Exp. Neurol. 2018, 299, 217–227. [Google Scholar] [CrossRef]
- Guerra, M.; Medici, V.; Weatheritt, R.; Corvino, V.; Palacios, D.; Geloso, M.C.; Farini, D.; Sette, C. Fetal exposure to valproic acid dysregulates the expression of autism-linked genes in the developing cerebellum. Transl. Psychiatry 2023, 13, 114. [Google Scholar] [CrossRef]
- Roux, S.; Bailly, Y.; Bossu, J.L. Regional and sex-dependent alterations in Purkinje cell density in the valproate mouse model of autism. Neuroreport 2019, 30, 82–88. [Google Scholar] [CrossRef]
- Meyza, K.Z.; Blanchard, D.C. The BTBR mouse model of idiopathic autism—Current view on mechanisms. Neurosci. Biobehav. Rev. 2017, 76, 99–110. [Google Scholar] [CrossRef]
- Kiffmeyer, E.A.; Cosgrove, J.A.; Siganos, J.K.; Bien, H.E.; Vipond, J.E.; Vogt, K.R.; Kloth, A.D. Deficits in cerebellum-dependent learning and cerebellar morphology in male and female BTBR autism model mice. NeuroSci 2022, 3, 624–644. [Google Scholar] [CrossRef]
- Xiao, R.; Zhong, H.; Li, X.; Ma, Y.; Zhang, R.; Wang, L.; Zang, Z.; Fan, X. Abnormal Cerebellar Development Is Involved in Dystonia-Like Behaviors and Motor Dysfunction of Autistic BTBR Mice. Front. Cell Dev. Biol. 2020, 8, 231. [Google Scholar] [CrossRef]
- Greco, C.M.; Navarro, C.S.; Hunsaker, M.R.; Maezawa, I.; Shuler, J.F.; Tassone, F.; Delany, M.; Au, J.W.; Berman, R.F.; Jin, L.W.; et al. Neuropathologic features in the hippocampus and cerebellum of three older men with fragile X syndrome. Mol. Autism 2011, 2, 2. [Google Scholar] [CrossRef] [PubMed]
- Huber, K.M. The fragile X-cerebellum connection. Trends Neurosci. 2006, 29, 183–185. [Google Scholar] [CrossRef] [PubMed]
- Ertan, G.; Arulrajah, S.; Tekes, A.; Jordan, L.; Huisman, T.A. Cerebellar abnormality in children and young adults with tuberous sclerosis complex: MR and diffusion weighted imaging findings. J. Neuroradiol. 2010, 37, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Tsai, P.T.; Hull, C.; Chu, Y.; Greene-Colozzi, E.; Sadowski, A.R.; Leech, J.M.; Steinberg, J.; Crawley, J.N.; Regehr, W.G.; Sahin, M. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature 2012, 488, 647–651. [Google Scholar] [CrossRef] [PubMed]
- D’Mello, A.M.; Stoodley, C.J. Cerebro-cerebellar circuits in autism spectrum disorder. Front. Neurosci. 2015, 9, 408. [Google Scholar] [CrossRef] [PubMed]
- Mostofsky, S.H.; Powell, S.K.; Simmonds, D.J.; Goldberg, M.C.; Caffo, B.; Pekar, J.J. Decreased connectivity and cerebellar activity in autism during motor task performance. Brain J. Neurol. 2009, 132, 2413–2425. [Google Scholar] [CrossRef] [PubMed]
- Unruh, K.E.; Bartolotti, J.V.; McKinney, W.S.; Schmitt, L.M.; Sweeney, J.A.; Mosconi, M.W. Functional connectivity of cortical-cerebellar networks in relation to sensorimotor behavior and clinical features in autism spectrum disorder. Cereb. Cortex 2023, 33, 8990–9002. [Google Scholar] [CrossRef] [PubMed]
- Jack, A.; Pelphrey, K.A. Annual Research Review: Understudied populations within the autism spectrum—Current trends and future directions in neuroimaging research. J. Child Psychol. Psychiatry Allied Discip. 2017, 58, 411–435. [Google Scholar] [CrossRef] [PubMed]
- Kana, R.K.; Maximo, J.O.; Williams, D.L.; Keller, T.A.; Schipul, S.E.; Cherkassky, V.L.; Minshew, N.J.; Just, M.A. Aberrant functioning of the theory-of-mind network in children and adolescents with autism. Mol. Autism 2015, 6, 59. [Google Scholar] [CrossRef]
- Oldehinkel, M.; Mennes, M.; Marquand, A.; Charman, T.; Tillmann, J.; Ecker, C.; Dell’Acqua, F.; Brandeis, D.; Banaschewski, T.; Baumeister, S.; et al. Altered Connectivity Between Cerebellum, Visual, and Sensory-Motor Networks in Autism Spectrum Disorder: Results from the EU-AIMS Longitudinal European Autism Project. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2019, 4, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Cerliani, L.; Mennes, M.; Thomas, R.M.; Di Martino, A.; Thioux, M.; Keysers, C. Increased Functional Connectivity Between Subcortical and Cortical Resting-State Networks in Autism Spectrum Disorder. JAMA Psychiatry 2015, 72, 767–777. [Google Scholar] [CrossRef]
- Khan, A.J.; Nair, A.; Keown, C.L.; Datko, M.C.; Lincoln, A.J.; Müller, R.-A. Cerebro-cerebellar Resting-State Functional Connectivity in Children and Adolescents with Autism Spectrum Disorder. Biol. Psychiatry 2015, 78, 625–634. [Google Scholar] [CrossRef]
- Verly, M.; Verhoeven, J.; Zink, I.; Mantini, D.; Peeters, R.; Deprez, S.; Emsell, L.; Boets, B.; Noens, I.; Steyaert, J.; et al. Altered functional connectivity of the language network in ASD: Role of classical language areas and cerebellum. NeuroImage Clin. 2014, 4, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Okada, N.J.; Liu, J.; Tsang, T.; Nosco, E.; McDonald, N.M.; Cummings, K.K.; Jung, J.; Patterson, G.; Bookheimer, S.Y.; Green, S.A.; et al. Atypical cerebellar functional connectivity at 9 months of age predicts delayed socio-communicative profiles in infants at high and low risk for autism. J. Child Psychol. Psychiatry Allied Discip. 2022, 63, 1002–1016. [Google Scholar] [CrossRef] [PubMed]
- Marek, S.; Siegel, J.S.; Gordon, E.M.; Raut, R.V.; Gratton, C.; Newbold, D.J.; Ortega, M.; Laumann, T.O.; Adeyemo, B.; Miller, D.B.; et al. Spatial and Temporal Organization of the Individual Human Cerebellum. Neuron 2018, 100, 977–993.e7. [Google Scholar] [CrossRef] [PubMed]
- Clifford, H.; Dulneva, A.; Ponting, C.P.; Haerty, W.; Becker, E.B.E. A gene expression signature in developing Purkinje cells predicts autism and intellectual disability co-morbidity status. Sci. Rep. 2019, 9, 485. [Google Scholar] [CrossRef] [PubMed]
- Sepp, M.; Leiss, K.; Murat, F.; Okonechnikov, K.; Joshi, P.; Leushkin, E.; Spanig, L.; Mbengue, N.; Schneider, C.; Schmidt, J.; et al. Cellular development and evolution of the mammalian cerebellum. Nature 2024, 625, 788–796. [Google Scholar] [CrossRef] [PubMed]
- Sydnor, L.M.; Aldinger, K.A. Structure, Function, and Genetics of the Cerebellum in Autism. J. Psychiatry Brain Sci. 2022, 7, e220008. [Google Scholar] [CrossRef]
- Brandenburg, C.; Griswold, A.J.; Van Booven, D.J.; Kilander, M.B.C.; Frei, J.A.; Nestor, M.W.; Dykxhoorn, D.M.; Pericak-Vance, M.A.; Blatt, G.J. Transcriptomic analysis of isolated and pooled human postmortem cerebellar Purkinje cells in autism spectrum disorders. Front. Genet. 2022, 13, 944837. [Google Scholar] [CrossRef] [PubMed]
- Ament, S.A.; Cortes-Gutierrez, M.; Herb, B.R.; Mocci, E.; Colantuoni, C.; McCarthy, M.M. A single-cell genomic atlas for maturation of the human cerebellum during early childhood. Sci. Transl. Med. 2023, 15, eade1283. [Google Scholar] [CrossRef] [PubMed]
- Prata, J.; Machado, A.S.; von Doellinger, O.; Almeida, M.I.; Barbosa, M.A.; Coelho, R.; Santos, S.G. The Contribution of Inflammation to Autism Spectrum Disorders: Recent Clinical Evidence. Methods Mol. Biol. 2019, 2011, 493–510. [Google Scholar] [CrossRef]
- Ha, S.; Lee, D.; Cho, Y.S.; Chung, C.; Yoo, Y.E.; Kim, J.; Lee, J.; Kim, W.; Kim, H.; Bae, Y.C.; et al. Cerebellar Shank2 Regulates Excitatory Synapse Density, Motor Coordination, and Specific Repetitive and Anxiety-Like Behaviors. J. Neurosci. Off. J. Soc. Neurosci. 2016, 36, 12129–12143. [Google Scholar] [CrossRef]
- Peter, S.; ten Brinke, M.M.; Stedehouder, J.; Reinelt, C.M.; Wu, B.; Zhou, H.; Zhou, K.; Boele, H.-J.; Kushner, S.A.; Lee, M.G.; et al. Dysfunctional cerebellar Purkinje cells contribute to autism-like behaviour in Shank2-deficient mice. Nat. Commun. 2016, 7, 12627. [Google Scholar] [CrossRef] [PubMed]
- Watson, L.M.; Wong, M.M.K.; Vowles, J.; Cowley, S.A.; Becker, E.B.E. A Simplified Method for Generating Purkinje Cells from Human-Induced Pluripotent Stem Cells. Cerebellum 2018, 17, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Behesti, H.; Kocabas, A.; Buchholz, D.E.; Carroll, T.S.; Hatten, M.E. Altered temporal sequence of transcriptional regulators in the generation of human cerebellar granule cells. eLife 2021, 10, e67074. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, M.; Tochitsky, I.; Buchholz, D.E.; Winden, K.; Kujala, V.; Kapur, K.; Cataltepe, D.; Turner, D.; Han, M.-J.; Woolf, C.J.; et al. Purkinje cells derived from TSC patients display hypoexcitability and synaptic deficits associated with reduced FMRP levels and reversed by rapamycin. Mol. Psychiatry 2018, 23, 2167–2183. [Google Scholar] [CrossRef] [PubMed]
- Muguruma, K.; Nishiyama, A.; Kawakami, H.; Hashimoto, K.; Sasai, Y. Self-Organization of Polarized Cerebellar Tissue in 3D Culture of Human Pluripotent Stem Cells. Cell Rep. 2015, 10, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Nayler, S.; Agarwal, D.; Curion, F.; Bowden, R.; Becker, E.B.E. High-resolution transcriptional landscape of xeno-free human induced pluripotent stem cell-derived cerebellar organoids. Sci. Rep. 2021, 11, 12959. [Google Scholar] [CrossRef] [PubMed]
- Atamian, A.; Birtele, M.; Hosseini, N.; Nguyen, T.; Seth, A.; Del Dosso, A.; Paul, S.; Tedeschi, N.; Taylor, R.; Coba, M.P.; et al. Human cerebellar organoids with functional Purkinje cells. Cell Stem Cell 2024, 31, 39–51.e6. [Google Scholar] [CrossRef] [PubMed]
- Levy, R.J.; Paşca, S.P. What Have Organoids and Assembloids Taught Us About the Pathophysiology of Neuropsychiatric Disorders? Biol. Psychiatry 2023, 93, 632–641. [Google Scholar] [CrossRef]
- Schaaf, C.P.; Betancur, C.; Yuen, R.K.C.; Parr, J.R.; Skuse, D.H.; Gallagher, L.; Bernier, R.A.; Buchanan, J.A.; Buxbaum, J.D.; Chen, C.A.; et al. A framework for an evidence-based gene list relevant to autism spectrum disorder. Nat. Rev. Genet. 2020, 21, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Jacob, S.; Wolff, J.J.; Steinbach, M.S.; Doyle, C.B.; Kumar, V.; Elison, J.T. Neurodevelopmental heterogeneity and computational approaches for understanding autism. Transl. Psychiatry 2019, 9, 63–74. [Google Scholar] [CrossRef]
- Uddin, M.; Wang, Y.; Woodbury-Smith, M. Artificial intelligence for precision medicine in neurodevelopmental disorders. NPJ Digit. Med. 2019, 2, 112–121. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).