Regulatory B Cells Expressing Granzyme B from Tolerant Renal Transplant Patients: Highly Differentiated B Cells with a Unique Pathway with a Specific Regulatory Profile and Strong Interactions with Immune System Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Ethics Statements
2.3. PBMC Isolation
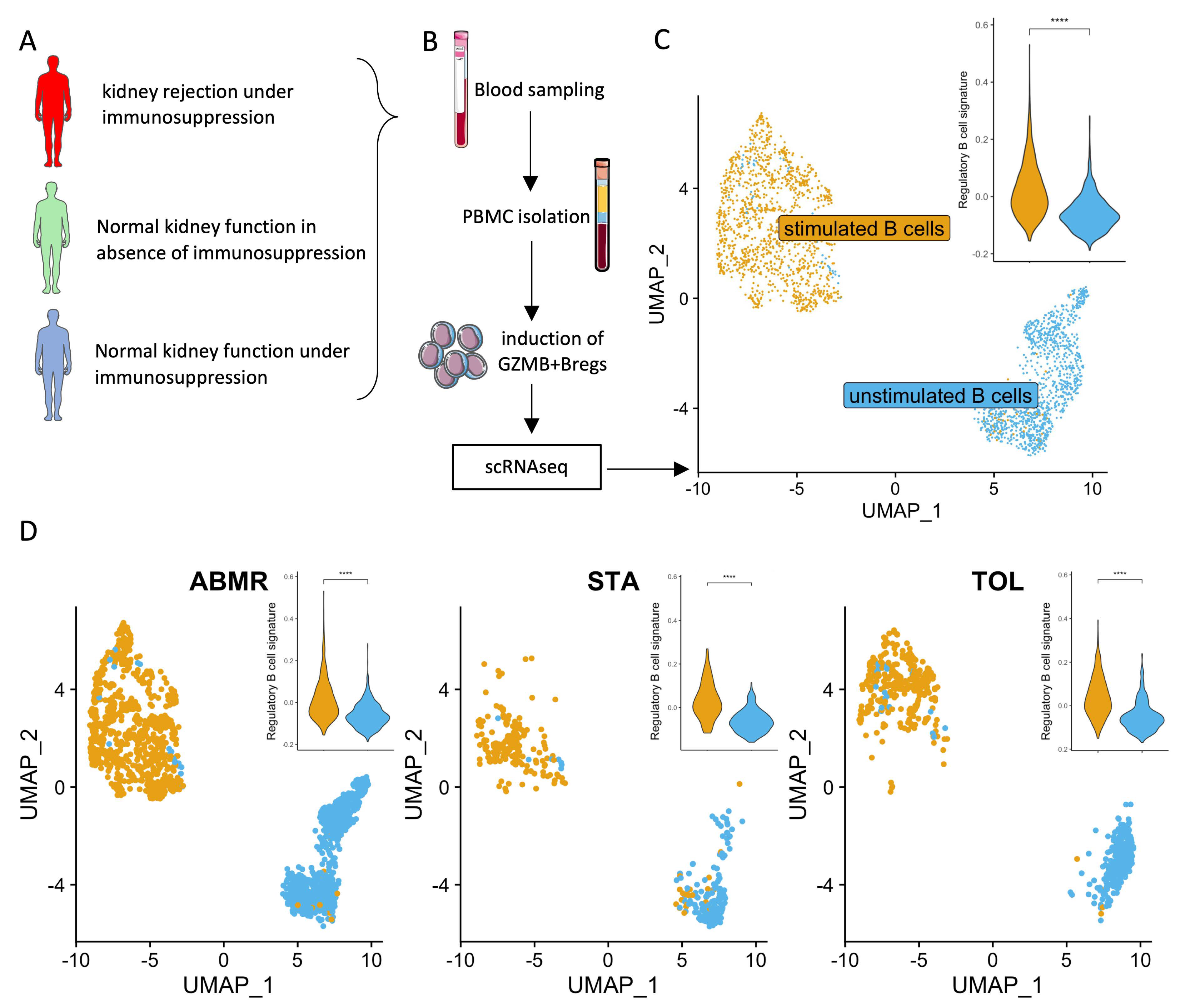
2.4. Induction of GZMB+ B Cells Ex Vivo
2.5. Cell Multiplexing and Single-Cell RNA Sequencing
2.6. scRNAseq Analysis
2.7. Biopsy Immunofluorescence Staining
3. Results
3.1. Clinical Characteristics of the Patients
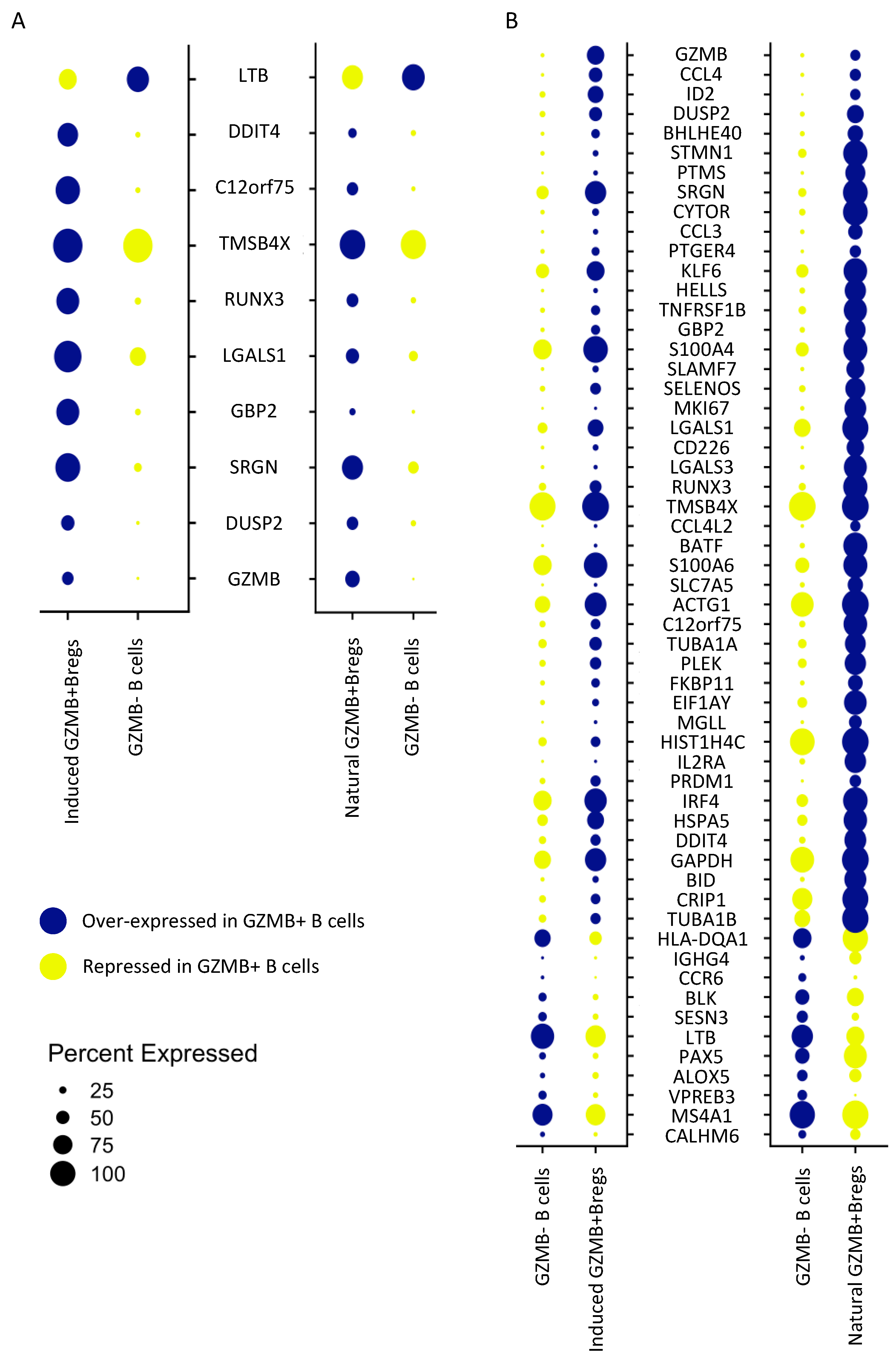
3.2. Natural GZMB+ B Cells Display a Differentiated B Cell Profile Enriched in Specific Genes Associated with NK Cells and Regulatory Functions
3.3. Natural GZMB+B Cells Are Highly Differentiated B Cells That Follow a Specific Trajectory Distinct from That of Conventional Memory B Cells, with the KLF13 Gene Overexpressed Throughout This Trajectory
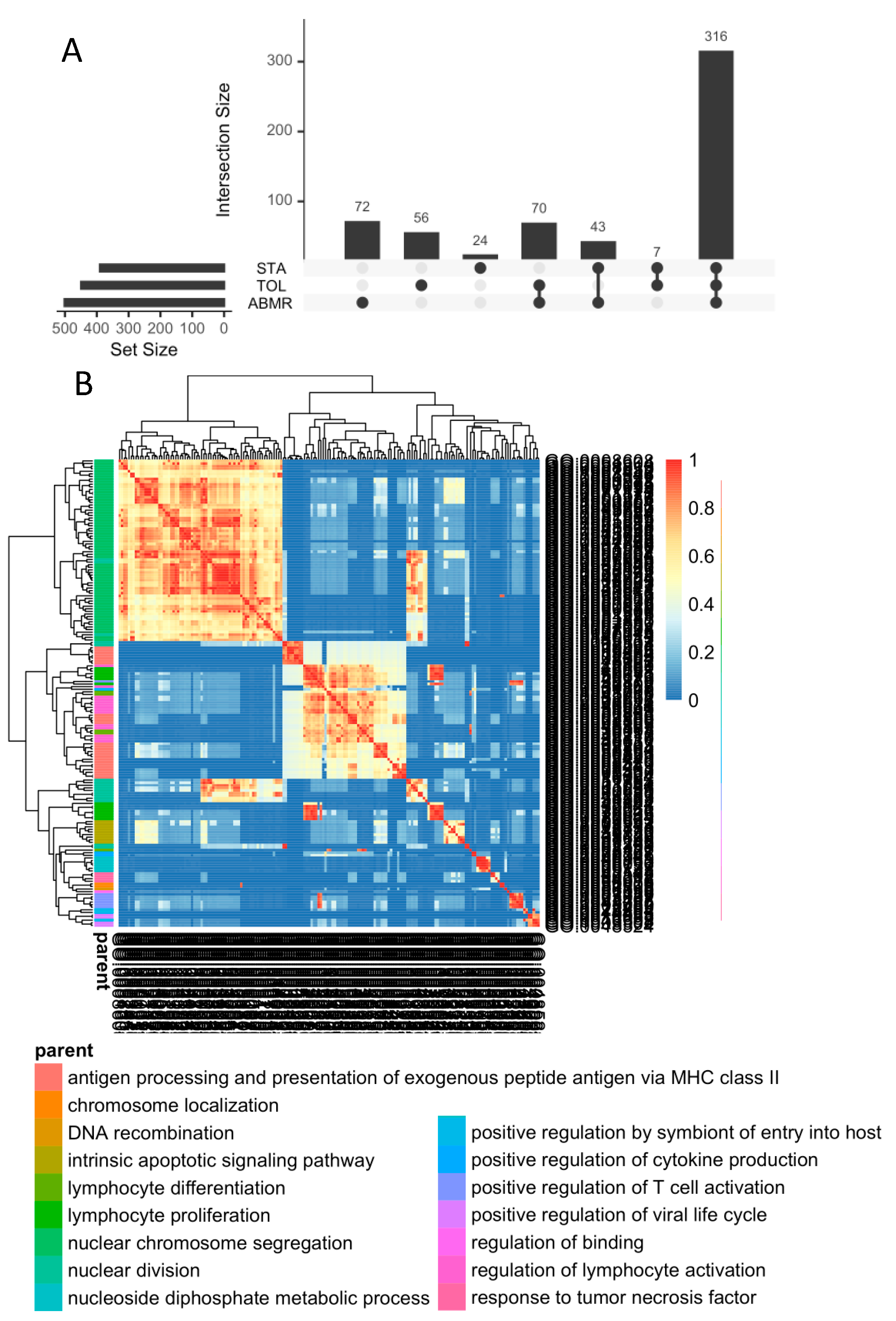
3.4. In General, GZMB+ B Cells from Tolerant Patients Exhibit a Specific Transcriptomic Profile Associated with a Decrease in the Expression of HLA Molecules, Apoptosis, and the Inflammatory Response
3.5. Ex-Vivo-Induced GZMB+ B Cells Share a “Regulatory” Signature with Natural GZMB+ B Cells in Tolerant Patients
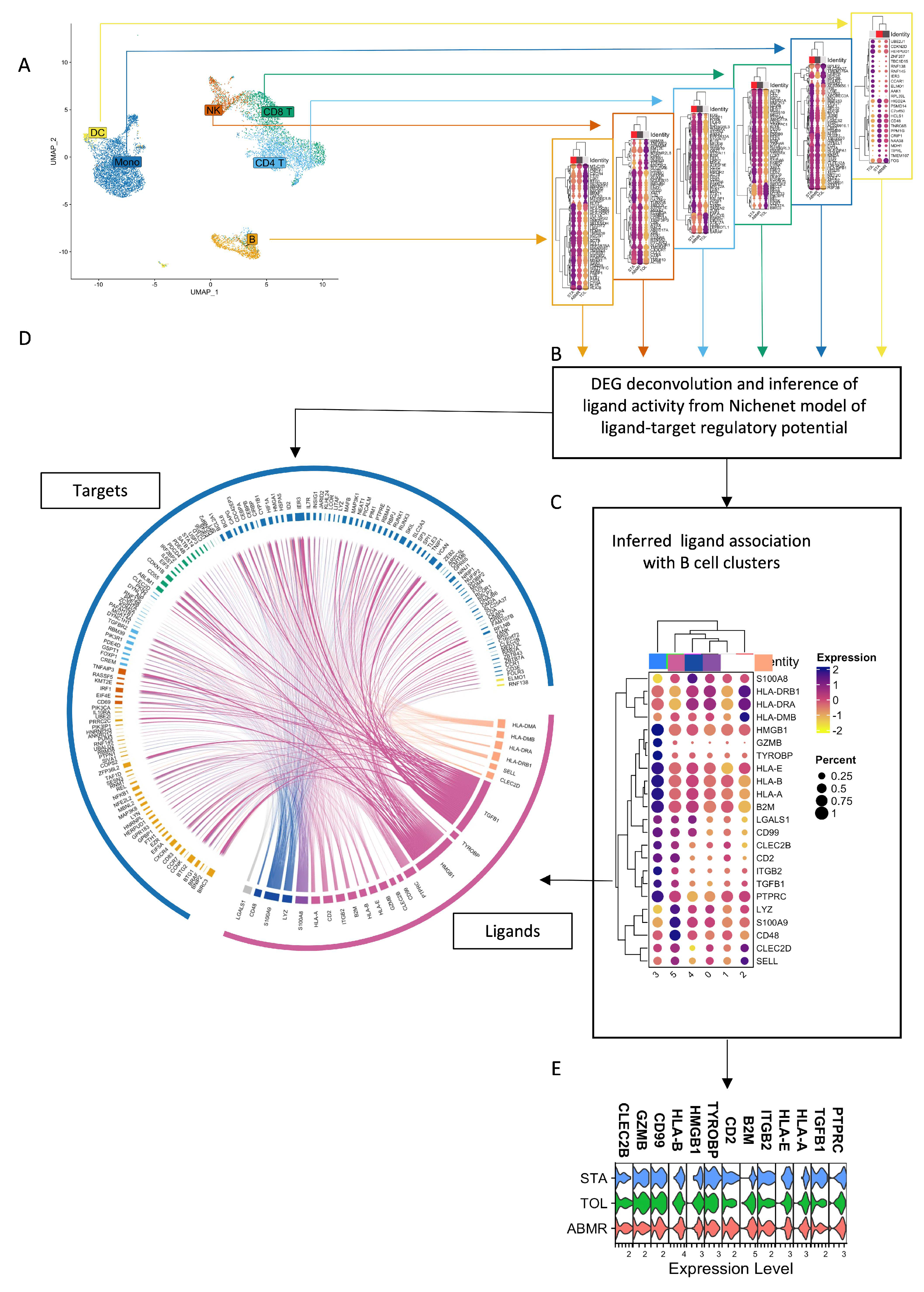
3.6. Natural GZMB+ B Cells from Tolerant Patients Interact Strongly with Peripheral Immune Cells
3.7. Under Inflammatory Conditions, GZMB+ B Cells Can Infiltrate Kidney Allografts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GZMB+ B cells: | Granzyme B positive B cells with regulatory properties. |
| Induced GZMB+ B cells: | GZMB+ B cells generated according to the in vitro protocol described in Chesneau et al. [19]. |
| Natural GZMB+ B cells: | B cells from donor’s blood or biopsies expressing GZMB without any restimulation in vitro. |
| GZMB− B cells: | B cells used to determine the characteristics of GZMB+ B cells. For the dataset generated after in vitro induction, GZMB− B cells correspond to the conditions that did not receive any stimulation (i.e., total resting B cells after 3 days), as GZMB expression is transient. |
| TOL: | Patients with no immunosuppression for at least one year and with stable creatinine < 150 mmol/L and proteinuria < 1 g/24. |
| STA: | Patients with stable graft function (creatinine < 150 mmol/L and proteinuria < 0.2 g/g creatinine) for at least 3 years under standard immunosuppression (calcineurin inhibitors [CNIs], antimetabolite ± corticosteroids). |
| ABMR: | Patients with histology-proven antibody-mediated rejection. |
| DEG: | Differentially expressed gene. |
Appendix A
References
- Adams, D.H.; Sanchez-Fueyo, A.; Samuel, D. From Immunosuppression to Tolerance. J. Hepatol. 2015, 62, S170–S185. [Google Scholar] [CrossRef] [PubMed]
- Ashton-Chess, J.; Giral, M.; Brouard, S.; Soulillou, J.-P. Spontaneous Operational Tolerance After Immunosuppressive Drug Withdrawal in Clinical Renal Allotransplantation. Transplantation 2007, 84, 1215–1219. [Google Scholar] [CrossRef] [PubMed]
- Benítez, C.; Londoño, M.-C.; Miquel, R.; Manzia, T.-M.; Abraldes, J.G.; Lozano, J.-J.; Martínez-Llordella, M.; López, M.; Angelico, R.; Bohne, F.; et al. Prospective Multicenter Clinical Trial of Immunosuppressive Drug Withdrawal in Stable Adult Liver Transplant Recipients: Hepatology. Hepatology 2013, 58, 1824–1835. [Google Scholar] [CrossRef] [PubMed]
- Taubert, R.; Danger, R.; Londoño, M.-C.; Christakoudi, S.; Martinez-Picola, M.; Rimola, A.; Manns, M.P.; Sánchez-Fueyo, A.; Jaeckel, E. Hepatic Infiltrates in Operational Tolerant Patients After Liver Transplantation Show Enrichment of Regulatory T Cells Before Proinflammatory Genes Are Downregulated. Am. J. Transplant. 2016, 16, 1285–1293. [Google Scholar] [CrossRef] [PubMed]
- Guinn, M.T.; Szuter, E.S.; Yokose, T.; Ge, J.; Rosales, I.A.; Chetal, K.; Sadreyev, R.I.; Cuenca, A.G.; Kreisel, D.; Sage, P.T.; et al. Intragraft B Cell Differentiation during the Development of Tolerance to Kidney Allografts Is Associated with a Regulatory B Cell Signature Revealed by Single Cell Transcriptomics. Am. J. Transplant. 2023, 23, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Katz, S.I.; Parker, D.; Turk, J.L. B-Cell Suppression of Delayed Hypersensitivity Reactions. Nature 1974, 251, 550–551. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Moore, D.J.; Huang, X.; Lian, M.-M.; Mohiuddin, M.; Velededeoglu, E.; Lee, M.K.; Sonawane, S.; Kim, J.; Wang, J.; et al. Cutting Edge: Transplant Tolerance Induced by Anti-CD45RB Requires B Lymphocytes. J. Immunol. 2007, 178, 6028–6032. [Google Scholar] [CrossRef]
- Clatworthy, M.R.; Watson, C.J.E.; Plotnek, G.; Bardsley, V.; Chaudhry, A.N.; Bradley, J.A.; Smith, K.G.C. B-Cell–Depleting Induction Therapy and Acute Cellular Rejection. N. Engl. J. Med. 2009, 360, 2683–2685. [Google Scholar] [CrossRef] [PubMed]
- Rosser, E.C.; Mauri, C. Regulatory B Cells: Origin, Phenotype, and Function. Immunity 2015, 42, 607–612. [Google Scholar] [CrossRef]
- Veh, J.; Ludwig, C.; Schrezenmeier, H.; Jahrsdörfer, B. Regulatory B Cells—Immunopathological and Prognostic Potential in Humans. Cells 2024, 13, 357. [Google Scholar] [CrossRef]
- Glass, M.C.; Glass, D.R.; Oliveria, J.-P.; Mbiribindi, B.; Esquivel, C.O.; Krams, S.M.; Bendall, S.C.; Martinez, O.M. Human IL-10-Producing B Cells Have Diverse States That Are Induced from Multiple B Cell Subsets. Cell Rep. 2022, 39, 110728. [Google Scholar] [CrossRef] [PubMed]
- Fillatreau, S.; Sweenie, C.H.; McGeachy, M.J.; Gray, D.; Anderton, S.M. B Cells Regulate Autoimmunity by Provision of IL-10. Nat. Immunol. 2002, 3, 944–950. [Google Scholar] [CrossRef] [PubMed]
- Iwata, Y.; Matsushita, T.; Horikawa, M.; DiLillo, D.J.; Yanaba, K.; Venturi, G.M.; Szabolcs, P.M.; Bernstein, S.H.; Magro, C.M.; Williams, A.D.; et al. Characterization of a Rare IL-10–Competent B-Cell Subset in Humans That Parallels Mouse Regulatory B10 Cells. Blood 2011, 117, 530–541. [Google Scholar] [CrossRef]
- Lin, F.; You, H.; Cao, X.; Li, T.; Hong, X.; Yang, J.; Huo, P.; Li, J.; Liu, W.; Jiang, Y. Characterization of IL-10-Producing Regulatory B Cells in Thymoma. Autoimmunity 2022, 55, 351–359. [Google Scholar] [CrossRef]
- Liu, S.; Yang, L.; Jia, S.; Zhao, R.; Jin, Z. Interleukin-35 Suppresses the Activity of Natural Killer-like B Cells in Patients with Hepatocellular Carcinoma. Int. Immunopharmacol. 2021, 100, 108161. [Google Scholar] [CrossRef]
- Lee, K.M.; Stott, R.T.; Zhao, G.; SooHoo, J.; Xiong, W.; Lian, M.M.; Fitzgerald, L.; Shi, S.; Akrawi, E.; Lei, J.; et al. TGF-β-Producing Regulatory B Cells Induce Regulatory T Cells and Promote Transplantation Tolerance: Immunomodulation. Eur. J. Immunol. 2014, 44, 1728–1736. [Google Scholar] [CrossRef]
- Zacca, E.R.; Onofrio, L.I.; Acosta, C.D.V.; Ferrero, P.V.; Alonso, S.M.; Ramello, M.C.; Mussano, E.; Onetti, L.; Cadile, I.I.; Stancich, M.I.; et al. PD-L1+ Regulatory B Cells Are Significantly Decreased in Rheumatoid Arthritis Patients and Increase after Successful Treatment. Front. Immunol. 2018, 9, 2241. [Google Scholar] [CrossRef] [PubMed]
- Durand, J.; Huchet, V.; Merieau, E.; Usal, C.; Chesneau, M.; Remy, S.; Heslan, M.; Anegon, I.; Cuturi, M.-C.; Brouard, S.; et al. Regulatory B Cells with a Partial Defect in CD40 Signaling and Overexpressing Granzyme B Transfer Allograft Tolerance in Rodents. J. Immunol. 2015, 195, 5035–5044. [Google Scholar] [CrossRef]
- Chesneau, M.; Mai, H.L.; Danger, R.; Le Bot, S.; Nguyen, T.-V.-H.; Bernard, J.; Poullaouec, C.; Guerrif, P.; Conchon, S.; Giral, M.; et al. Efficient Expansion of Human Granzyme B–Expressing B Cells with Potent Regulatory Properties. J. Immunol. 2020, 205, 2391–2401. [Google Scholar] [CrossRef]
- Jahrsdörfer, B.; Blackwell, S.E.; Wooldridge, J.E.; Huang, J.; Andreski, M.W.; Jacobus, L.S.; Taylor, C.M.; Weiner, G.J. B-Chronic Lymphocytic Leukemia Cells and Other B Cells Can Produce Granzyme B and Gain Cytotoxic Potential after Interleukin-21-Based Activation. Blood 2006, 108, 2712–2719. [Google Scholar] [CrossRef]
- Hagn, M.; Sontheimer, K.; Dahlke, K.; Brueggemann, S.; Kaltenmeier, C.; Beyer, T.; Hofmann, S.; Lunov, O.; Barth, T.F.; Fabricius, D.; et al. Human B Cells Differentiate into Granzyme B-Secreting Cytotoxic B Lymphocytes upon Incomplete T-Cell Help. Immunol. Cell Biol. 2012, 90, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Chesneau, M.; Michel, L.; Dugast, E.; Chenouard, A.; Baron, D.; Pallier, A.; Durand, J.; Braza, F.; Guerif, P.; Laplaud, D.-A.; et al. Tolerant Kidney Transplant Patients Produce B Cells with Regulatory Properties. J. Am. Soc. Nephrol. 2015, 26, 2588–2598. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.M.; Thompson-Snipes, L.; Klintmalm, G.; Demetris, A.J.; O’Leary, J.; Oh, S.; Joo, H. CD24 hi CD38 hi and CD24 hi CD27+ Human Regulatory B Cells Display Common and Distinct Functional Characteristics. J. Immunol. 2019, 203, 2110–2120. [Google Scholar] [CrossRef] [PubMed]
- Blair, P.A.; Noreña, L.Y.; Flores-Borja, F.; Rawlings, D.J.; Isenberg, D.A.; Ehrenstein, M.R.; Mauri, C. CD19+CD24hiCD38hi B Cells Exhibit Regulatory Capacity in Healthy Individuals but Are Functionally Impaired in Systemic Lupus Erythematosus Patients. Immunity 2010, 32, 129–140. [Google Scholar] [CrossRef]
- Flores-Borja, F.; Bosma, A.; Ng, D.; Reddy, V.; Ehrenstein, M.R.; Isenberg, D.A.; Mauri, C. CD19 + CD24 hi CD38 hi B Cells Maintain Regulatory T Cells While Limiting T H 1 and T H 17 Differentiation. Sci. Transl. Med. 2013, 5, 173ra23. [Google Scholar] [CrossRef] [PubMed]
- Piloni, D.; Morosini, M.; Magni, S.; Balderacchi, A.; Inghilleri, S.; Cova, E.; Oggionni, T.; Frangipane, V.; Pandolfi, L.; Scudeller, L.; et al. Peripheral CD19+CD24highCD38high B-Regulatory Cells in Lung Transplant Recipients. Transplant. Immunol. 2019, 57, 101245. [Google Scholar] [CrossRef]
- Cupi, M.L.; Sarra, M.; Marafini, I.; Monteleone, I.; Franzè, E.; Ortenzi, A.; Colantoni, A.; Sica, G.; Sileri, P.; Rosado, M.M.; et al. Plasma Cells in the Mucosa of Patients with Inflammatory Bowel Disease Produce Granzyme B and Possess Cytotoxic Activities. J. Immunol. 2014, 192, 6083–6091. [Google Scholar] [CrossRef] [PubMed]
- Hagn, M.; Ebel, V.; Sontheimer, K.; Schwesinger, E.; Lunov, O.; Beyer, T.; Fabricius, D.; Barth, T.F.E.; Viardot, A.; Stilgenbauer, S.; et al. CD5+ B Cells from Individuals with Systemic Lupus Erythematosus Express Granzyme B. Eur. J. Immunol. 2010, 40, 2060–2069. [Google Scholar] [CrossRef] [PubMed]
- Brosseau, C.; Durand, M.; Colas, L.; Durand, E.; Foureau, A.; Cheminant, M.A.; Bouchaud, G.; Castan, L.; Klein, M.; Magnan, A.; et al. CD9+ Regulatory B Cells Induce T Cell Apoptosis via IL-10 and Are Reduced in Severe Asthmatic Patients. Front. Immunol. 2018, 9, 3034. [Google Scholar] [CrossRef] [PubMed]
- Bulati, M.; Buffa, S.; Martorana, A.; Candore, G.; Lio, D.; Caruso, C.; Colonna-Romano, G. Trafficking Phenotype and Production of Granzyme B by Double Negative B Cells (IgG+IgD−CD27−) in the Elderly. Exp. Gerontol. 2014, 54, 123–129. [Google Scholar] [CrossRef]
- Chesneau, M.; Pallier, A.; Braza, F.; Lacombe, G.; Gallou, S.L.; Baron, D.; Giral, M.; Danger, R.; Guerif, P.; Aubert-Wastiaux, H.; et al. Unique B Cell Differentiation Profile in Tolerant Kidney Transplant Patients. Am. J. Transplant. 2014, 14, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Lindner, S.; Dahlke, K.; Sontheimer, K.; Hagn, M.; Kaltenmeier, C.; Barth, T.F.E.; Beyer, T.; Reister, F.; Fabricius, D.; Lotfi, R.; et al. Interleukin 21–Induced Granzyme B–Expressing B Cells Infiltrate Tumors and Regulate T Cells. Cancer Res. 2013, 73, 2468–2479. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.B.; Bi, E.; Chen, H.; Yu, J.J.; Ye, B.H. IL-21 and CD40L Synergistically Promote Plasma Cell Differentiation through Upregulation of Blimp-1 in Human B Cells. J. Immunol. 2013, 190, 1827–1836. [Google Scholar] [CrossRef] [PubMed]
- Rabani, M.; Wilde, B.; Hübbers, K.; Xu, S.; Kribben, A.; Witzke, O.; Dolff, S. IL-21 Dependent Granzyme B Production of B-Cells Is Decreased in Patients with Lupus Nephritis. Clin. Immunol. 2018, 188, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Sailliet, N.; Mai, H.-L.; Dupuy, A.; Tilly, G.; Fourgeux, C.; Braud, M.; Giral, M.; Robert, J.-M.; Degauque, N.; Danger, R.; et al. Human Granzyme B Regulatory B Cells Prevent Effector CD4+CD25- T Cell Proliferation through a Mechanism Dependent from Lymphotoxin Alpha. Front. Immunol. 2023, 14, 1183714. [Google Scholar] [CrossRef] [PubMed]
- Kaltenmeier, C.; Gawanbacht, A.; Beyer, T.; Lindner, S.; Trzaska, T.; van der Merwe, J.A.; Härter, G.; Grüner, B.; Fabricius, D.; Lotfi, R.; et al. CD4 + T Cell–Derived IL-21 and Deprivation of CD40 Signaling Favor the In Vivo Development of Granzyme B–Expressing Regulatory B Cells in HIV Patients. J. Immunol. 2015, 194, 3768–3777. [Google Scholar] [CrossRef] [PubMed]
- Dubois, F.; Limou, S.; Chesneau, M.; Degauque, N.; Brouard, S.; Danger, R. Transcriptional Meta-Analysis of Regulatory B Cells. Eur. J. Immunol. 2020, 50, 1757–1769. [Google Scholar] [CrossRef] [PubMed]
- Brouard, S.; Mansfield, E.; Braud, C.; Li, L.; Giral, M.; Hsieh, S.; Baeten, D.; Zhang, M.; Ashton-Chess, J.; Braudeau, C.; et al. Identification of a Peripheral Blood Transcriptional Biomarker Panel Associated with Operational Renal Allograft Tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 15448–15453. [Google Scholar] [CrossRef] [PubMed]
- Chesneau, M.; Mai, H.L.; Brouard, S. New Method for the Expansion of Highly Purified Human Regulatory Granzyme B-Expressing B Cells. Methods Mol. Biol. 2021, 2270, 203–216. [Google Scholar] [CrossRef]
- Stoeckius, M.; Zheng, S.; Houck-Loomis, B.; Hao, S.; Yeung, B.Z.; Mauck, W.M.; Smibert, P.; Satija, R. Cell Hashing with Barcoded Antibodies Enables Multiplexing and Doublet Detection for Single Cell Genomics. Genome Biol. 2018, 19, 224. [Google Scholar] [CrossRef]
- Abidi, A.; Laurent, T.; Bériou, G.; Bouchet-Delbos, L.; Fourgeux, C.; Louvet, C.; Triki-Marrakchi, R.; Poschmann, J.; Josien, R.; Martin, J. Characterization of Rat ILCs Reveals ILC2 as the Dominant Intestinal Subset. Front. Immunol. 2020, 11, 255. [Google Scholar] [CrossRef]
- Laurent, T.; Sinha, D.; Fourgeux, C.; Letellier, T.; Ville, S.; Bouchet-Delbos, L.; Brancherau, J.; Kerleau, C.; Brouard, S.; Blancho, G.; et al. A Gene-Expression Module in Circulating Immune Cells Is Associated with Cell Migration during Immune Diseases. bioRxiv 2023. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated Analysis of Multimodal Single-Cell Data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef] [PubMed]
- Finak, G.; McDavid, A.; Yajima, M.; Deng, J.; Gersuk, V.; Shalek, A.K.; Slichter, C.K.; Miller, H.W.; McElrath, M.J.; Prlic, M.; et al. MAST: A Flexible Statistical Framework for Assessing Transcriptional Changes and Characterizing Heterogeneity in Single-Cell RNA Sequencing Data. Genome Biol. 2015, 16, 278. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The Dynamics and Regulators of Cell Fate Decisions Are Revealed by Pseudotemporal Ordering of Single Cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef]
- Badia-i-Mompel, P.; Vélez Santiago, J.; Braunger, J.; Geiss, C.; Dimitrov, D.; Müller-Dott, S.; Taus, P.; Dugourd, A.; Holland, C.H.; Ramirez Flores, R.O.; et al. decoupleR: Ensemble of Computational Methods to Infer Biological Activities from Omics Data. Bioinform. Adv. 2022, 2, vbac016. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Browaeys, R.; Saelens, W.; Saeys, Y. NicheNet: Modeling Intercellular Communication by Linking Ligands to Target Genes. Nat. Methods 2020, 17, 159–162. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Yang, S.-Y.; Long, J.; Huang, M.-X.; Luo, P.-Y.; Bian, Z.-H.; Xu, Y.-F.; Wang, C.-B.; Yang, S.-H.; Li, L.; Selmi, C.; et al. Characterization of Organ-Specific Regulatory B Cells Using Single-Cell RNA Sequencing. Front. Immunol. 2021, 12, 711980. [Google Scholar] [CrossRef]
- Daamen, A.R.; Alajoleen, R.M.; Grammer, A.C.; Luo, X.M.; Lipsky, P.E. Single-Cell RNA Sequencing Analysis Reveals the Heterogeneity of IL-10 Producing Regulatory B Cells in Lupus-Prone Mice. Front. Immunol. 2023, 14, 1282770. [Google Scholar] [CrossRef]
- Inaba, A.; Tuong, Z.K.; Zhao, T.X.; Stewart, A.P.; Mathews, R.; Truman, L.; Sriranjan, R.; Kennet, J.; Saeb-Parsy, K.; Wicker, L.; et al. Low-Dose IL-2 Enhances the Generation of IL-10-Producing Immunoregulatory B Cells. Nat. Commun. 2023, 14, 2071. [Google Scholar] [CrossRef]
- Bigot, J.; Pilon, C.; Matignon, M.; Grondin, C.; Leibler, C.; Aissat, A.; Pirenne, F.; Cohen, J.L.; Grimbert, P. Transcriptomic Signature of the CD24 hi CD38 hi Transitional B Cells Associated with an Immunoregulatory Phenotype in Renal Transplant Recipients. Am. J. Transplant. 2016, 16, 3430–3442. [Google Scholar] [CrossRef] [PubMed]
- van de Veen, W.; Stanic, B.; Yaman, G.; Wawrzyniak, M.; Söllner, S.; Akdis, D.G.; Rückert, B.; Akdis, C.A.; Akdis, M. IgG4 Production Is Confined to Human IL-10–Producing Regulatory B Cells That Suppress Antigen-Specific Immune Responses. J. Allergy Clin. Immunol. 2013, 131, 1204–1212. [Google Scholar] [CrossRef]
- Lin, W.; Cerny, D.; Chua, E.; Duan, K.; Yi, J.T.J.; Shadan, N.B.; Lum, J.; Maho-Vaillant, M.; Zolezzi, F.; Wong, S.C.; et al. Human Regulatory B Cells Combine Phenotypic and Genetic Hallmarks with a Distinct Differentiation Fate. J. Immunol. 2014, 193, 2258–2266. [Google Scholar] [CrossRef]
- Kessel, A.; Haj, T.; Peri, R.; Snir, A.; Melamed, D.; Sabo, E.; Toubi, E. Human CD19+CD25high B Regulatory Cells Suppress Proliferation of CD4+ T Cells and Enhance Foxp3 and CTLA-4 Expression in T-Regulatory Cells. Autoimmun. Rev. 2012, 11, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Manickam, C.; Nwanze, C.; Ram, D.R.; Shah, S.V.; Smith, S.; Jones, R.; Hueber, B.; Kroll, K.; Varner, V.; Goepfert, P.; et al. Progressive Lentivirus Infection Induces Natural Killer Cell Receptor-Expressing B Cells in the Gastrointestinal Tract. AIDS 2018, 32, 1571–1578. [Google Scholar] [CrossRef] [PubMed]
- Cogswell, A.; Jo, S.; Ferguson, N.; Gupta, K.; Barker, E. Presence of Natural Killer B Cells in Simian Immunodeficiency Virus-Infected Colon That Have Properties and Functions Similar to Those of Natural Killer Cells and B Cells but Are a Distinct Cell Population. J. Virol. 2022, 96, e00235-22. [Google Scholar] [CrossRef]
- Hagn, M.; Schwesinger, E.; Ebel, V.; Sontheimer, K.; Maier, J.; Beyer, T.; Syrovets, T.; Laumonnier, Y.; Fabricius, D.; Simmet, T.; et al. Human B Cells Secrete Granzyme B When Recognizing Viral Antigens in the Context of the Acute Phase Cytokine IL-21. J. Immunol. 2009, 183, 1838–1845. [Google Scholar] [CrossRef]
- Bae, H.; Lee, H.; Ko, E.J.; Kim, C.; Lee, S.; Yang, C.W.; Oh, E.; Chung, B.H. Discovery of Cellular and Genetic Signatures of Immune Tolerance in Kidney Transplant Recipients through Single Cell RNA Sequencing Analysis. HLA 2023, 102, 316–330. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, P.K.; Buchkovich, N.J. Human Cytomegalovirus Decreases Major Histocompatibility Complex Class II by Regulating Class II Transactivator Transcript Levels in a Myeloid Cell Line. J. Virol. 2020, 94, e01901–e01919. [Google Scholar] [CrossRef] [PubMed]
- Le, T.A.; Chu, V.T.; Lino, A.C.; Schrezenmeier, E.; Kressler, C.; Hamo, D.; Rajewsky, K.; Dörner, T.; Dang, V.D. Efficient CRISPR-Cas9-Mediated Mutagenesis in Primary Human B Cells for Identifying Plasma Cell Regulators. Mol. Ther.-Nucleic Acids 2022, 30, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Ruer-Laventie, J.; Simoni, L.; Schickel, J.; Soley, A.; Duval, M.; Knapp, A.; Marcellin, L.; Lamon, D.; Korganow, A.; Martin, T.; et al. Overexpression of Fkbp11, a Feature of Lupus B Cells, Leads to B Cell Tolerance Breakdown and Initiates Plasma Cell Differentiation. Immun. Inflam. Dis. 2015, 3, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Arabpour, M.; Rasolmali, R.; Talei, A.-R.; Mehdipour, F.; Ghaderi, A. Granzyme B Production by Activated B Cells Derived from Breast Cancer-Draining Lymph Nodes. Mol. Immunol. 2019, 114, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Zheremyan, E.A.; Ustiugova, A.S.; Uvarova, A.N.; Karamushka, N.M.; Stasevich, E.M.; Gogoleva, V.S.; Bogolyubova, A.V.; Mitkin, N.A.; Kuprash, D.V.; Korneev, K.V. Differentially Activated B Cells Develop Regulatory Phenotype and Show Varying Immunosuppressive Features: A Comparative Study. Front. Immunol. 2023, 14, 1178445. [Google Scholar] [CrossRef] [PubMed]
- Shankar, S.; Stolp, J.; Juvet, S.C.; Beckett, J.; Macklin, P.S.; Issa, F.; Hester, J.; Wood, K.J. Ex Vivo-Expanded Human CD19+TIM-1+ Regulatory B Cells Suppress Immune Responses in Vivo and Are Dependent upon the TIM-1/STAT3 Axis. Nat. Commun. 2022, 13, 3121. [Google Scholar] [CrossRef] [PubMed]
- Zeng, N.; Jian, Z.; Zhu, W.; Xu, J.; Fan, Y.; Xiao, F. KLF13 Overexpression Protects Sepsis-induced Myocardial Injury and LPS -induced Inflammation and Apoptosis. Int. J. Exp. Pathol. 2023, 104, 23–32. [Google Scholar] [CrossRef]
- Outram, S.V.; Gordon, A.R.; Hager-Theodorides, A.L.; Metcalfe, J.; Crompton, T.; Kemp, P. KLF13 Influences Multiple Stages of Both B and T Cell Development. Cell Cycle 2008, 7, 2047–2055. [Google Scholar] [CrossRef]
- Huang, B.; Ahn, Y.-T.; McPherson, L.; Clayberger, C.; Krensky, A.M. Interaction of PRP4 with Krüppel-Like Factor 13 Regulates CCL5 Transcription. J. Immunol. 2007, 178, 7081–7087. [Google Scholar] [CrossRef]
- Hamano, K.; Rawsthorne, M.-A.; Bushell, A.R.; Morris, P.J.; Wood, K.J. Evidence that the continued presence of the organ graft and not peripheral donor microchimerism is essential for maintenance of tolerance to alloantigen in vivo in anti-CD4 treated recipients1,2. Transplantation 1996, 62, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Logtenberg, M.E.W.; Scheeren, F.A.; Schumacher, T.N. The CD47-SIRPα Immune Checkpoint. Immunity 2020, 52, 742–752. [Google Scholar] [CrossRef] [PubMed]
- Amati, A.-L.; Zakrzewicz, A.; Siebers, R.; Wilker, S.; Heldmann, S.; Zakrzewicz, D.; Hecker, A.; McIntosh, J.M.; Padberg, W.; Grau, V. Chemokines (CCL3, CCL4, and CCL5) Inhibit ATP-Induced Release of IL-1 β by Monocytic Cells. Mediat. Inflamm. 2017, 2017, 1434872. [Google Scholar] [CrossRef]
- Bystry, R.S.; Aluvihare, V.; Welch, K.A.; Kallikourdis, M.; Betz, A.G. B Cells and Professional APCs Recruit Regulatory T Cells via CCL4. Nat. Immunol. 2001, 2, 1126–1132. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.J.; Willsmore, Z.; Laddach, R.; Crescioli, S.; Chauhan, J.; Cheung, A.; Black, A.; Geh, J.L.C.; MacKenzie Ross, A.D.; Healy, C.; et al. Enriched Circulating and Tumor-Resident TGF-β + Regulatory B Cells in Patients with Melanoma Promote FOXP3 + Tregs. OncoImmunology 2022, 11, 2104426. [Google Scholar] [CrossRef] [PubMed]
- Boivin, W.A.; Shackleford, M.; Vanden Hoek, A.; Zhao, H.; Hackett, T.L.; Knight, D.A.; Granville, D.J. Granzyme B Cleaves Decorin, Biglycan and Soluble Betaglycan, Releasing Active Transforming Growth Factor-Β1. PLoS ONE 2012, 7, e33163. [Google Scholar] [CrossRef]
- Lamarthée, B.; Callemeyn, J.; Van Herck, Y.; Antoranz, A.; Anglicheau, D.; Boada, P.; Becker, J.U.; Debyser, T.; De Smet, F.; De Vusser, K.; et al. Transcriptional and Spatial Profiling of the Kidney Allograft Unravels a Central Role for FcyRIII+ Innate Immune Cells in Rejection. Nat. Commun. 2023, 14, 4359. [Google Scholar] [CrossRef]
- Magil, A.B. Monocytes/Macrophages in Renal Allograft Rejection. Transplant. Rev. 2009, 23, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Kwan, T.; Wu, H.; Chadban, S.J. Macrophages in Renal Transplantation: Roles and Therapeutic Implications. Cell. Immunol. 2014, 291, 58–64. [Google Scholar] [CrossRef]
- Zhang, H.; Cavazzoni, C.B.; Hanson, B.L.; Bechu, E.D.; Podestà, M.A.; Azzi, J.; Blazar, B.R.; Chong, A.S.; Kreisel, D.; Alessandrini, A.; et al. Transcriptionally Distinct B Cells Infiltrate Allografts After Kidney Transplantation. Transplantation 2023, 107, e47–e57. [Google Scholar] [CrossRef]
- Guzel, H.G.; Yilmaz, V.T.; Koksoy, S.; Kocak, H.; Kisaoglu, A.; Soylu, M.; Akkaya, B.; Demiryilmaz, I.; Aydinli, B.; Suleymanlar, G. Regulatory B Cells Profile in Kidney Transplant Recipients with Chronic-Active Antibody-Mediated Rejection. Transplant. Proc. 2023, 55, 1140–1146. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.; Chadban, S.J.; Loh, Y.W.; Kwan, T.K.-T.; Wang, C.; Singer, J.; Niewold, P.; Ling, Z.; Spiteri, A.; Getts, D.; et al. Targeting Inflammatory Monocytes by Immune-Modifying Nanoparticles Prevents Acute Kidney Allograft Rejection. Kidney Int. 2022, 102, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Malone, A.F. Monocytes and Macrophages in Kidney Transplantation and Insights from Single Cell RNA-Seq Studies. Kidney360 2021, 2, 1654–1659. [Google Scholar] [CrossRef] [PubMed]
- Filippone, E.J.; Farber, J.L. The Implications of B-Lineage Cells in Kidney Allografts. Transplantation 2020, 104, 2011–2023. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Ming, Y.; Yang, C. Regulatory B Cells: The Cutting Edge of Immune Tolerance in Kidney Transplantation. Cell Death Dis. 2018, 9, 109. [Google Scholar] [CrossRef] [PubMed]
- Coelho, V.; Saitovitch, D.; Kalil, J.; Silva, H.M. Rethinking the Multiple Roles of B Cells in Organ Transplantation. Curr. Opin. Organ. Transplant. 2013, 18, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Barnett, N.; Dorling, A.; Mamode, N. B Cells in Renal Transplantation: Pathological Aspects and Therapeutic Interventions. Nephrol. Dial. Transplant. 2011, 26, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Von Vietinghoff, S.; Kurts, C. Regulation and Function of CX3CR1 and Its Ligand CX3CL1 in Kidney Disease. Cell Tissue Res. 2021, 385, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, J.; Huang, M.; Kosonen, R.; Lee, J.E. CCR4 and CCR5 Involvement in Monocyte-Derived Macrophage Migration in Neuroinflammation. Front. Immunol. 2022, 13, 876033. [Google Scholar] [CrossRef]
- Roussey-Kesler, G.; Giral, M.; Moreau, A.; Subra, J.-F.; Legendre, C.; Noël, C.; Pillebout, E.; Brouard, S.; Soulillou, J.-P. Clinical Operational Tolerance after Kidney Transplantation. Am. J. Transplant. 2006, 6, 736–746. [Google Scholar] [CrossRef]
- Demetris, A.J.; Lunz Iii, J.G.; Randhawa, P.; Wu, T.; Nalesnik, M.; Thomson, A.W. Monitoring of Human Liver and Kidney Allograft Tolerance: A Tissue/Histopathology Perspective. Transplant. Int. 2009, 22, 120–141. [Google Scholar] [CrossRef] [PubMed]
- Becker, L.E.; De Oliveira Biazotto, F.; Conrad, H.; Schaier, M.; Kihm, L.P.; Gross-Weissmann, M.-L.; Waldherr, R.; Bierhaus, A.; Nawroth, P.P.; Zeier, M.; et al. Cellular Infiltrates and NFκB Subunit C-Rel Signaling in Kidney Allografts of Patients With Clinical Operational Tolerance. Transplantation 2012, 94, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Gallon, L.; Mathew, J.M.; Bontha, S.V.; Dumur, C.I.; Dalal, P.; Nadimpalli, L.; Maluf, D.G.; Shetty, A.A.; Ildstad, S.T.; Leventhal, J.R.; et al. Intragraft Molecular Pathways Associated with Tolerance Induction in Renal Transplantation. J. Am. Soc. Nephrol. 2018, 29, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Vanikar, A.V.; Trivedi, H.L.; Thakkar, U.G. Six Years’ Experience of Tolerance Induction in Renal Transplantation Using Stem Cell Therapy. Clin. Immunol. 2018, 187, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-Q.; Zhu, Y.; Qi, M.; Zeng, Y.; Liu, Z.-J.; Ding, C.; Zhang, T.; Li, X.-L.; Han, D.-D.; He, Q. Granzyme B+ B Cells Detected by Single-Cell Sequencing Are Associated with Prognosis in Patients with Intrahepatic Cholangiocarcinoma Following Liver Transplantation. Cancer Immunol. Immunother. 2024, 73, 58. [Google Scholar] [CrossRef]
- Mueller, S.N.; Mackay, L.K. Tissue-Resident Memory T Cells: Local Specialists in Immune Defence. Nat. Rev. Immunol. 2016, 16, 79–89. [Google Scholar] [CrossRef]
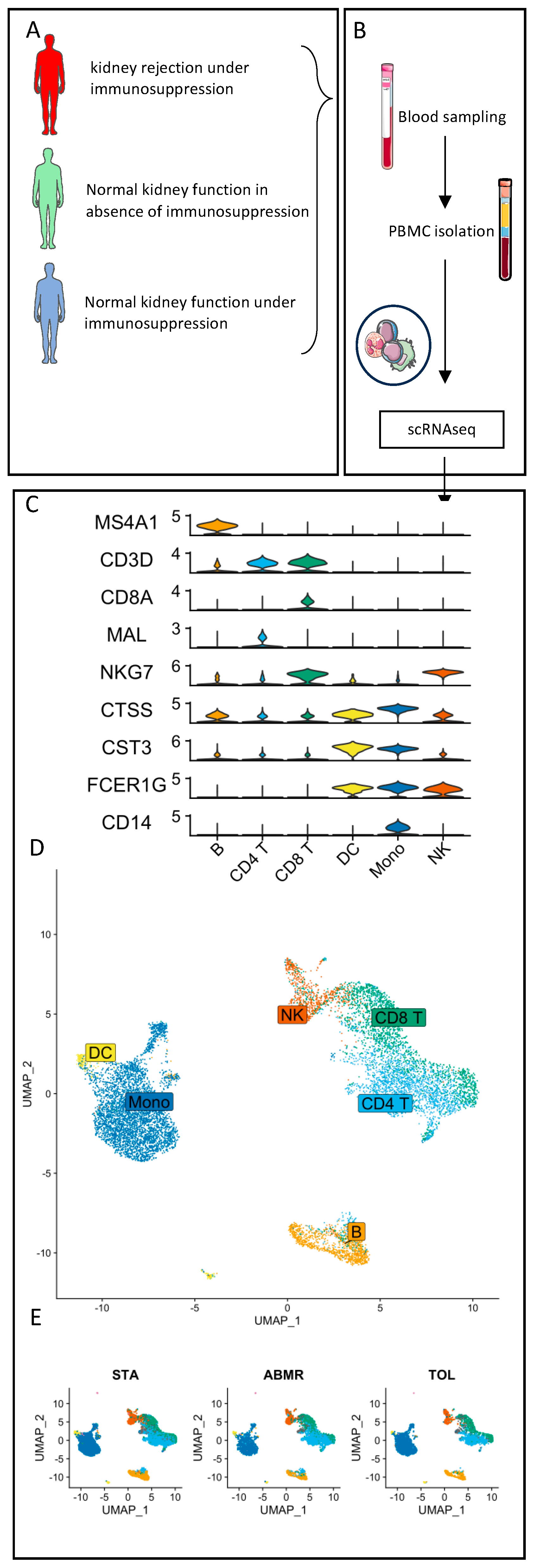
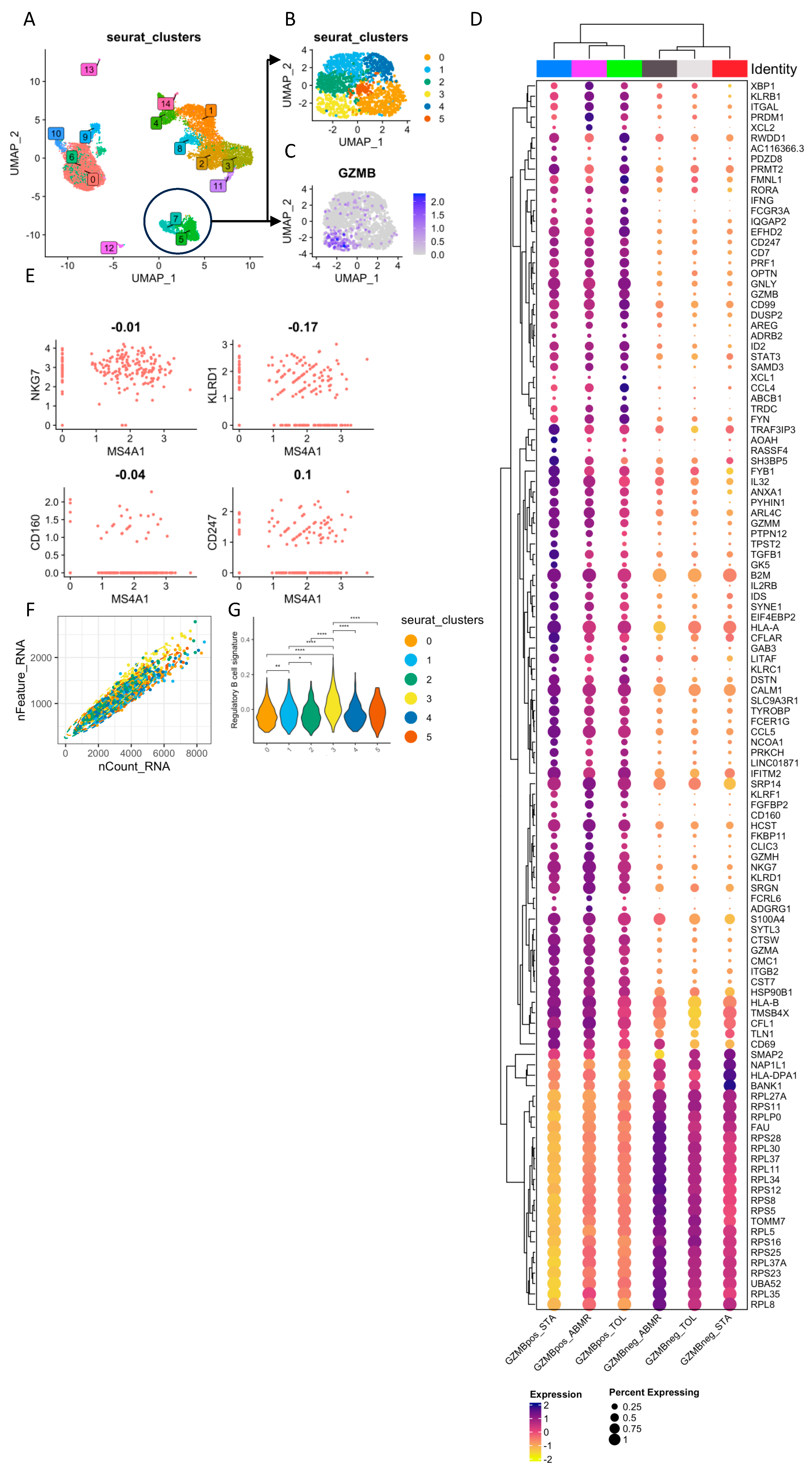
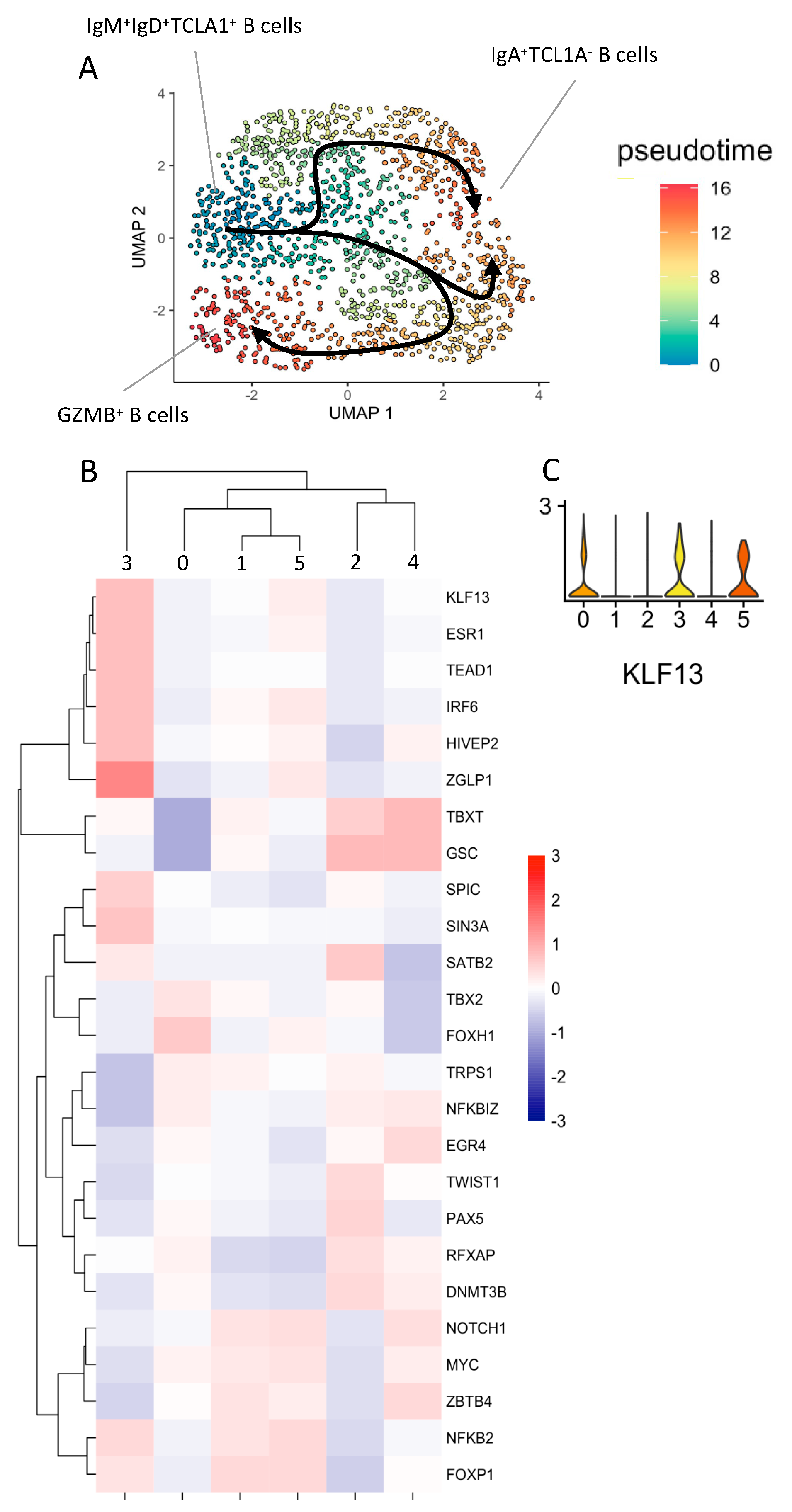
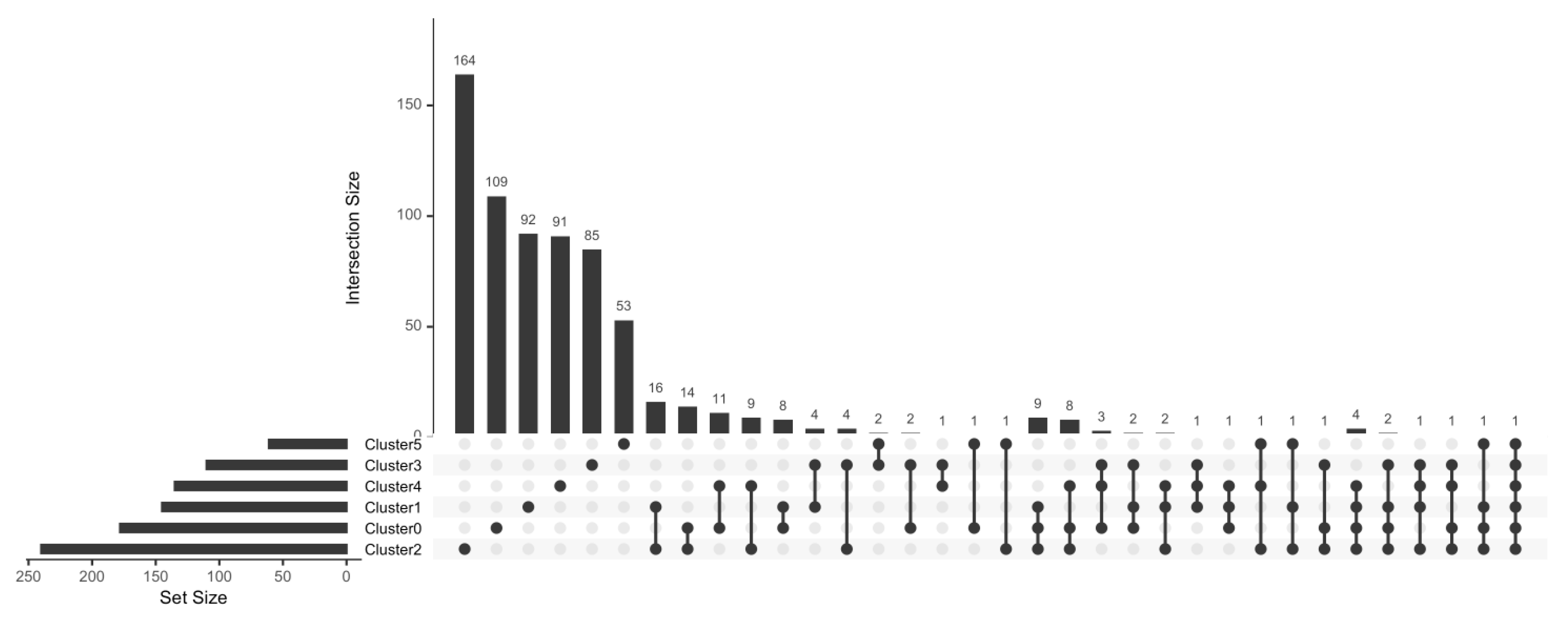

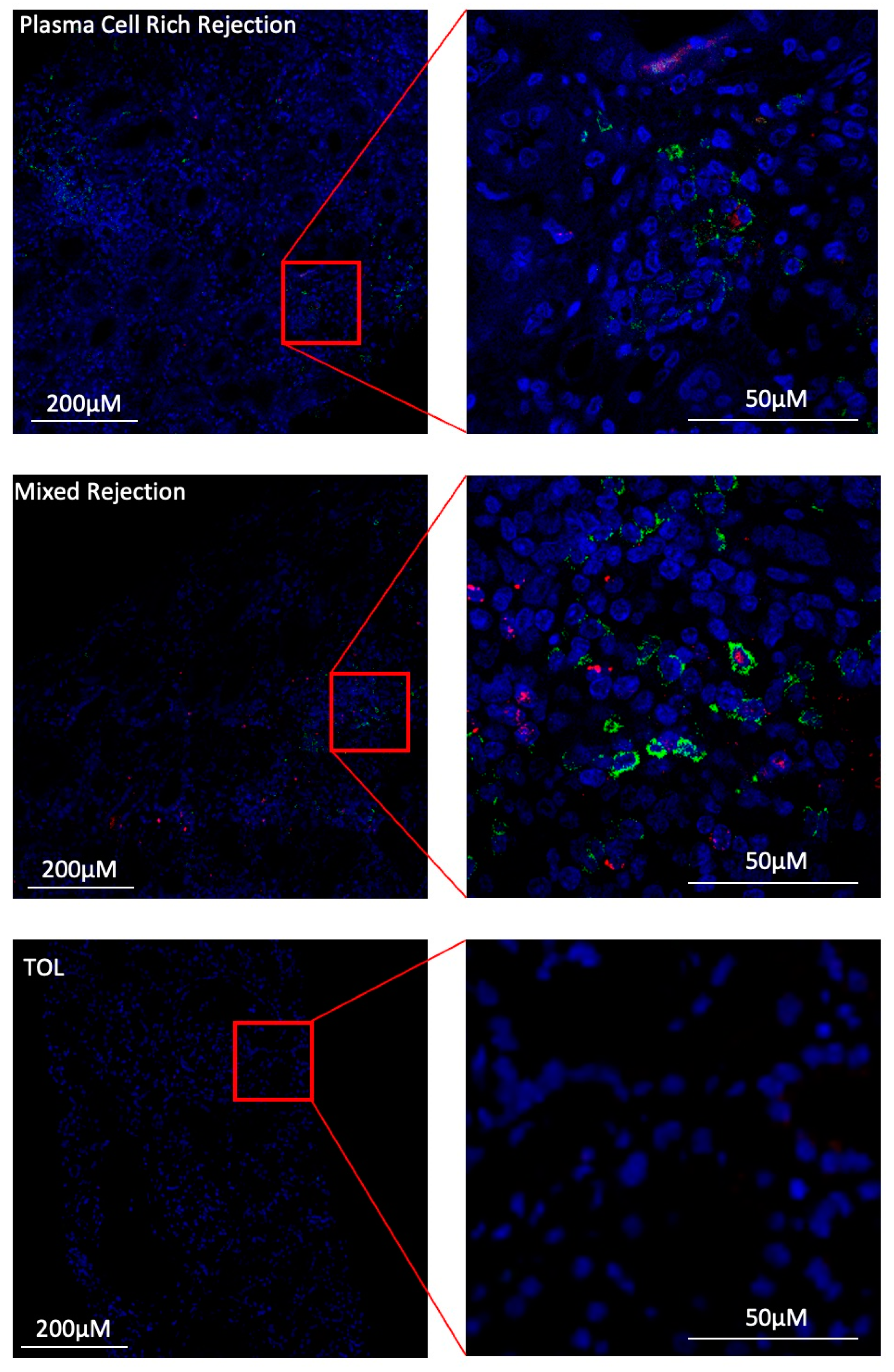
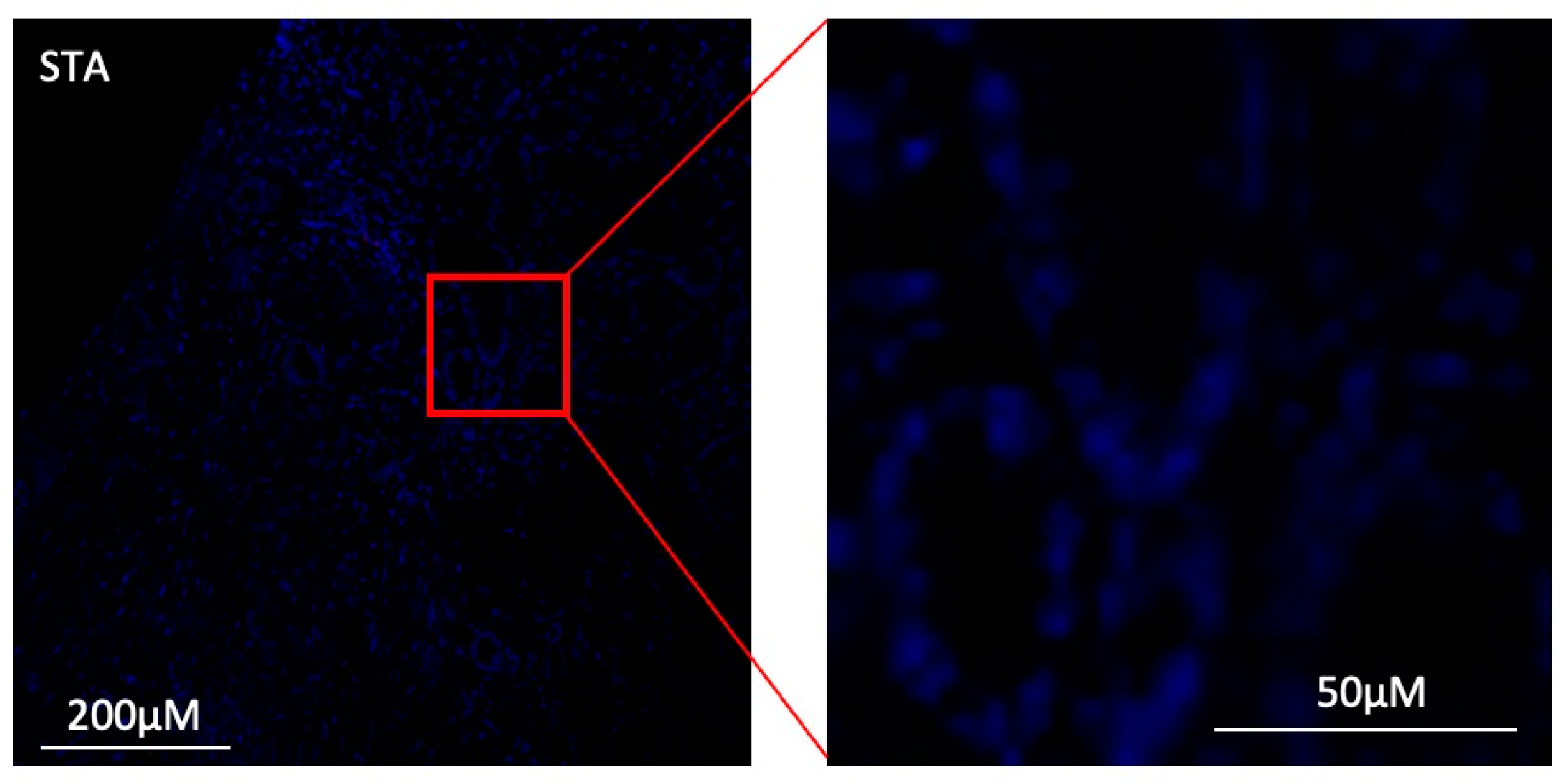
| STA (n = 4) | ABMR (n = 5) | TOL (n = 4) | |
|---|---|---|---|
| Sex (M/F) | 3/1 | 2/3 | 3/1 |
| Age at the time of analysis (years) | 46.25 ± 17 | 55.4 ± 4 | 45.75 ± 19.1 |
| Donor (LD/NLD) | 0/4 | 1/4 | 2/2 |
| Time post-transplantation (days) | 1056 ± 1381 | 2178 ± 2783 | 6891 ± 3709 |
| Creatinemia (μMol) | 85.25 ± 23.01 | 159.6 ± 42.20 | 126.25 ± 52.81 |
| Proteinuria (g/g) | 0 ± 0 | 0.405 ± 0.276 | 0.05 ± 0.01 |
| Mismatched HLA * | 4/5/5/5 | 2/4/2/1/3 | 4/3/0/0 |
| Immunosuppression at sampling ** | 4 | 5 | 0 |
| CNIs | 4 | 5 | 0 |
| Steroids | 2 | 5 | 0 |
| Antiproliferative | 4 | 4 | 0 |
| Sender B Cell Cluster | Ligand-Receptor Pairs | Target Cells |
|---|---|---|
| B cell Cluster 0 (GZMB−) | / | / |
| B cell Cluster 1 (GZMB−) | RPS19-C5AR1 | Mono |
| B cell Cluster 2 (GZMB−) | HLA-DMA-CD4/HLA-DMA-CD74/HLA-DPB1-CD4/HLA-DRB1-CD37/HLA-DRB1-CD53/HLA-DRB1-CD81/HLA-DRB1-CD82 HLA-DQA1-CD4/SELL-SELPLG/CLEC2D-KLRB1/HLA-DQB1-CD4/HLA-DRA-CD4/HLA-DRA-CD37/HLA-DRA-CD53/HLA-DRA-CD63/HLA-DRA-CD81/HLA-DRA-CD82/HLA-DMB-CD4/HLA-DMB-CD74 | CD4 T/CD8 T/NK/Mono/B/DC |
| B cell Cluster 3 (GZMB+) | CLEC2B-KLRF1/CALR-TAP1/CALR-TAP2/CALR-LRP1/TYROBP-TREM1/TYROBP-KLRD1/HLA-F-CD8A/HLA-F-LILRB1/HLA-F-LILRB2/CD99-CD99/HSP90B1-LRP1/ITGB2-ICAM1/ITGB2-ICAM3/ITGB2-CD82/ITGB2-CD226/B2M-TAP1/B2M-TAP2/B2M-LRP1/B2M-LILRB1/CD7-ADAM15/GZMB-IGF2R/GZMB-MCL1/HLA-B-CD8A/HLA-B-LILRB1/HLA-B-LILRB2/HLA-B-KLRD1/HMGB1-TLR2/HMGB1-TLR4/HMGB1-CXCR4/HMGB1-CD163/HLA-E-CD8A/HLA-E-KLRD1/CD2-CD53/ANXA1-FPR1/TGFB1-TGFBR1/TGFB1-TGFBR2/TGFB1-TGFBR3/TGFB1-APP/TGFB1-ITGB1/HLA-A-CD8A/HLA-A-LILRB1/HLA-A-LILRB2/HLA-A-KLRD1/HLA-A-APLP2/HLA-C-LILRB1/HLA-C-LILRB2/PTPRC-CD4/PTPRC-CD247/CD47-SIRPA/CCL4-CCR1/CCL5-CCR1 | CD4 T/CD8 T/NK/Mono/B/DC |
| B cell Cluster 4 (GZMB−) | S100A8-CD36/S100A8-CD68/S100A8-CD69/S100A8-ITGB2 | CD4 T/CD8 T/NK/Mono/B/DC |
| B cell Cluster 5 (GZMB−) | LYZ-ITGAL/S100A9-CD36/S100A9-CD68/S100A9-ITGB2/S100A9-TLR4/CD48-CD2 | CD4 T/CD8 T/NK/Mono/B/DC |
| General expression | LGALS1-CD69/LGALS1-ITGB1/LGALS1-PTPRC | CD4 T/CD8 T/NK/Mono/B/DC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sailliet, N.; Dupuy, A.; Brinas, F.; Renaudin, K.; Colas, L.; Kerleau, C.; Nguyen, T.-V.-H.; Fourgeux, C.; Poschmann, J.; Gosset, C.; et al. Regulatory B Cells Expressing Granzyme B from Tolerant Renal Transplant Patients: Highly Differentiated B Cells with a Unique Pathway with a Specific Regulatory Profile and Strong Interactions with Immune System Cells. Cells 2024, 13, 1287. https://doi.org/10.3390/cells13151287
Sailliet N, Dupuy A, Brinas F, Renaudin K, Colas L, Kerleau C, Nguyen T-V-H, Fourgeux C, Poschmann J, Gosset C, et al. Regulatory B Cells Expressing Granzyme B from Tolerant Renal Transplant Patients: Highly Differentiated B Cells with a Unique Pathway with a Specific Regulatory Profile and Strong Interactions with Immune System Cells. Cells. 2024; 13(15):1287. https://doi.org/10.3390/cells13151287
Chicago/Turabian StyleSailliet, Nicolas, Amandine Dupuy, François Brinas, Karine Renaudin, Luc Colas, Clarisse Kerleau, Thi-Van-Ha Nguyen, Cynthia Fourgeux, Jérémie Poschmann, Clément Gosset, and et al. 2024. "Regulatory B Cells Expressing Granzyme B from Tolerant Renal Transplant Patients: Highly Differentiated B Cells with a Unique Pathway with a Specific Regulatory Profile and Strong Interactions with Immune System Cells" Cells 13, no. 15: 1287. https://doi.org/10.3390/cells13151287





