Enhancing Photosynthesis and Plant Productivity through Genetic Modification
Abstract
:1. Introduction
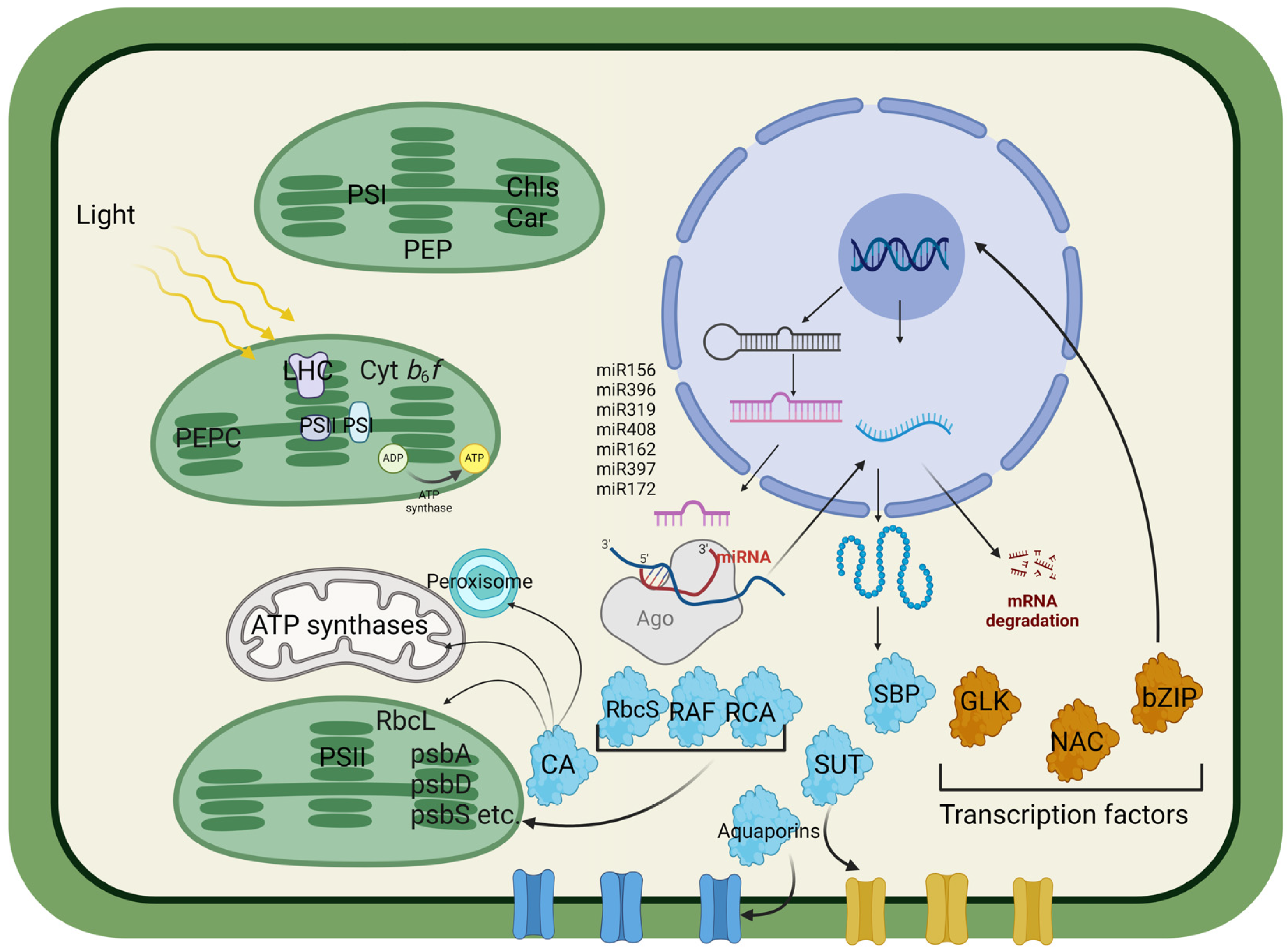
2. Chloroplast Component Modifications
2.1. Optimization of Light Harvesting and Pigment Content
2.2. Photosystem II
2.3. C. cytochrome b6f Complex
2.4. Photosystem I
2.5. Electron Transport Chain
2.6. Chloroplast ATP Synthase
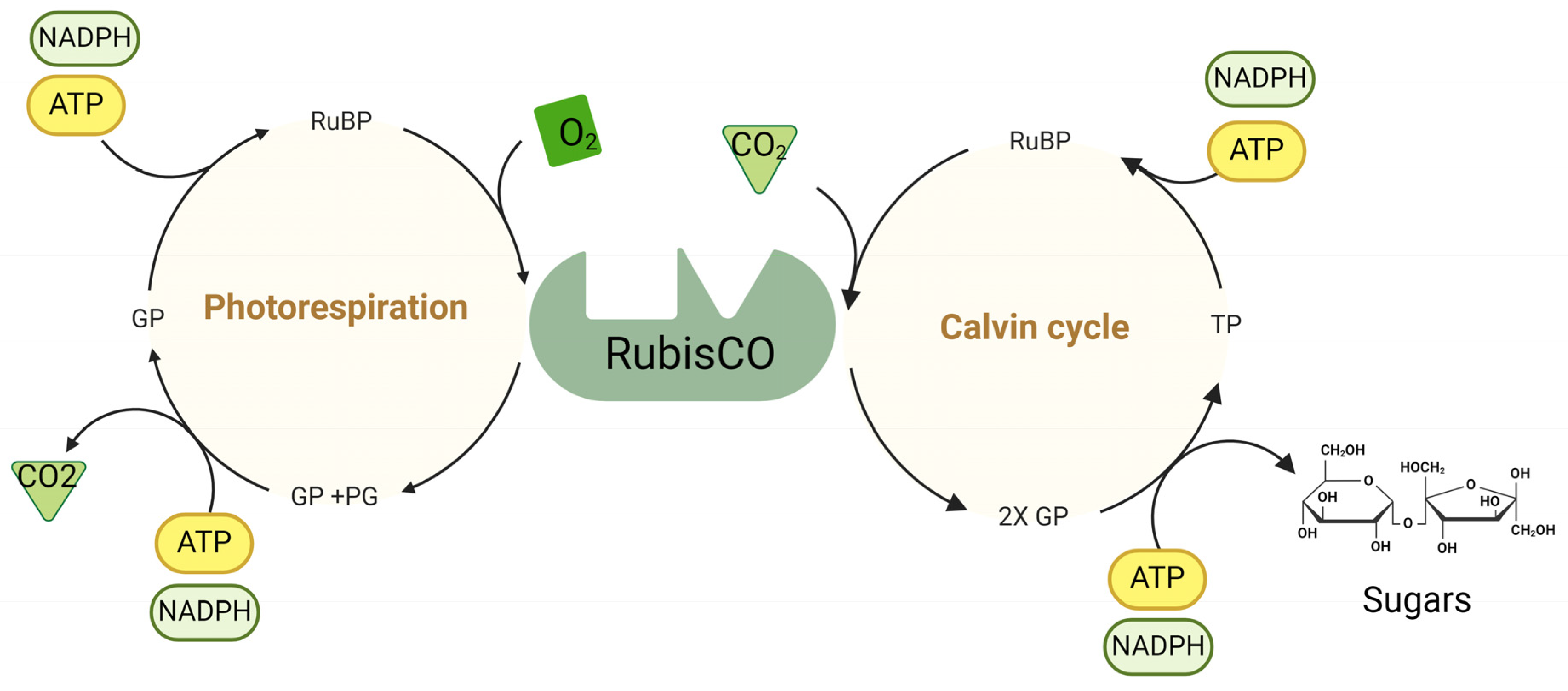
3. Carbon Assimilation Efficiency
3.1. RuBisCO as a Target to Improve Carbon Assimilation Efficiency
3.2. Rubisco Assembly Factors
3.3. Sedoheptulose-1,7-Bisphosphatase
3.4. Genes Involved in C4-Type Photosynthesis
3.5. Photorespiratory
4. Cellular Transport and Regulation
4.1. Sucrose Transporters
4.2. Aquaporins
4.3. Carbonic Anhydrase
5. Gene Regulation
5.1. High Pigment Epistasis 1
5.2. Transcription Factors
5.3. MicroRNAs
6. Environmental Stressors Limiting Photosynthesis
6.1. Temperature Stress
6.2. Light Stress
6.3. Water and Salt Stresses
6.4. Biotic Stress
6.5. Cross-Adaptation
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leister, D. Genetic Engineering, Synthetic Biology and the Light Reactions of Photosynthesis. Plant Physiol. 2018, 179, 778–793. [Google Scholar] [CrossRef]
- Renger, G. Primary Processes of Photosynthesis: Principles and Apparatus; Royal Society of Chemistry: London, UK, 2007. [Google Scholar]
- Foyer, C.H.; Neukermans, J.; Queval, G.; Noctor, G.; Harbinson, J. Photosynthetic Control of Electron Transport and the Regulation of Gene Expression. J. Exp. Bot. 2012, 63, 1637–1661. [Google Scholar] [CrossRef]
- Rochaix, J.-D. Regulation of photosynthetic electron transport. Biochim. Et Biophys. Acta (BBA)-Bioenerg. 2011, 1807, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zeng, H.; Xu, F.; Yan, F.; Xu, W. H+-ATPases in Plant Growth and Stress Responses. Annu. Rev. Plant Biol. 2022, 73, 495–521. [Google Scholar] [CrossRef]
- Simkin, A.J.; McAusland, L.; Lawson, T.; Raines, C.A. Overexpression of the RieskeFeS Protein Increases Electron Transport Rates and Biomass Yield. Plant Physiol. 2017, 175, 134–145. [Google Scholar] [CrossRef]
- Hajirezaei, M.; Peisker, M.; Tschiersch, H.; Palatnik, J.F.; Valle, E.M.; Carrillo, N.; Sonnewald, U. Small Changes in the Activity of Chloroplastic NADP+-dependent Ferredoxin Oxidoreductase Lead to Impaired Plant Growth and Restrict Photosynthetic Activity of Transgenic Tobacco Plants. Plant J. 2002, 29, 281–293. [Google Scholar] [CrossRef]
- Sakoda, K.; Yamori, W.; Shimada, T.; Sugano, S.S.; Hara-Nishimura, I.; Tanaka, Y. Higher Stomatal Density Improves Photosynthetic Induction and Biomass Production in Arabidopsis under Fluctuating Light. Front. Plant Sci. 2020, 11, 589603. [Google Scholar] [CrossRef] [PubMed]
- Yamori, W.; Kusumi, K.; Iba, K.; Terashima, I. Increased Stomatal Conductance Induces Rapid Changes to Photosynthetic Rate in Response to Naturally Fluctuating Light Conditions in Rice. Plant Cell Environ. 2020, 43, 1230–1240. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Hashimoto-Sugimoto, M.; Iba, K.; Terashima, I.; Yamori, W. Improved Stomatal Opening Enhances Photosynthetic Rate and Biomass Production in Fluctuating Light. J. Exp. Bot. 2020, 71, 2339–2350. [Google Scholar] [CrossRef]
- Tsuchihira, A.; Hanba, Y.T.; Kato, N.; Doi, T.; Kawazu, T.; Maeshima, M. Effect of Overexpression of Radish Plasma Membrane Aquaporins on Water-Use Efficiency, Photosynthesis and Growth of Eucalyptus Trees. Tree Physiol. 2010, 30, 417–430. [Google Scholar] [CrossRef]
- Hanba, Y.T.; Shibasaka, M.; Hayashi, Y.; Hayakawa, T.; Kasamo, K.; Terashima, I.; Katsuhara, M. Overexpression of the Barley Aquaporin HvPIP2; 1 Increases Internal CO2 Conductance and CO2 Assimilation in the Leaves of Transgenic Rice Plants. Plant Cell Physiol. 2004, 45, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wang, K.; Yuan, W.; Xu, W.; Liu, S.; Kronzucker, H.J.; Chen, G.; Miao, R.; Zhang, M.; Ding, M.; et al. Overexpression of Rice Aquaporin OsPIP1;2 Improves Yield by Enhancing Mesophyll CO2 Conductance and Phloem Sucrose Transport. J. Exp. Bot. 2019, 70, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, N.N.; Ignatova, L.K.; Fedorchuk, T.P.; Ivanov, B.N. Carbonic Anhydrases in Photosynthetic Cells of Higher Plants. Biochem. Mosc. 2015, 80, 674–687. [Google Scholar] [CrossRef]
- Momayyezi, M.; McKown, A.D.; Bell, S.C.S.; Guy, R.D. Emerging Roles for Carbonic Anhydrase in Mesophyll Conductance and Photosynthesis. Plant J. 2020, 101, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Wang, P.; Korkaric, M.; Capper, R.G.; Saunders, N.J.; Langdale, J.A. GLK Transcription Factors Coordinate Expression of the Photosynthetic Apparatus in Arabidopsis. Plant Cell 2009, 21, 1109–1128. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Mitsuda, N.; Ohtani, M.; Ohme-Takagi, M.; Kato, K.; Demura, T. VASCULAR-RELATED NAC-DOMAIN 7 Directly Regulates the Expression of a Broad Range of Genes for Xylem Vessel Formation. Plant J. 2011, 66, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA 2/Ethylene Responsive Factor (AP 2/ERF) Transcription Factors: Mediators of Stress Responses and Developmental Programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Stracke, R.; Favory, J.-J.; Gruber, H.; Bartelniewoehner, L.; Bartels, S.; Binkert, M.; Funk, M.; Weisshaar, B.; Ulm, R. The Arabidopsis bZIP Transcription Factor HY5 Regulates Expression of the PFG1/MYB12 Gene in Response to Light and Ultraviolet-B Radiation. Plant Cell Environ. 2010, 33, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.-Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental Functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis Thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef]
- Liebsch, D.; Palatnik, J.F. MicroRNA miR396, GRF Transcription Factors and GIF Co-Regulators: A Conserved Plant Growth Regulatory Module with Potential for Breeding and Biotechnology. Curr. Opin. Plant Biol. 2020, 53, 31–42. [Google Scholar] [CrossRef]
- Simkin, A.J.; Kapoor, L.; Doss, C.G.P.; Hofmann, T.A.; Lawson, T.; Ramamoorthy, S. The Role of Photosynthesis Related Pigments in Light Harvesting, Photoprotection and Enhancement of Photosynthetic Yield in Planta. Photosynth. Res. 2022, 152, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Ashikhmin, A.; Pashkovskiy, P.; Kosobryukhov, A.; Khudyakova, A.; Abramova, A.; Vereshchagin, M.; Bolshakov, M.; Kreslavski, V. The Role of Pigments and Cryptochrome 1 in the Adaptation of Solanum Lycopersicum Photosynthetic Apparatus to High-Intensity Blue Light. Antioxidants 2024, 13, 605. [Google Scholar] [CrossRef] [PubMed]
- Rodermel, S. Pathways of Plastid-to-Nucleus Signaling. Trends Plant Sci. 2001, 6, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Saavedra, C.; Stange, C. Biosynthesis of Carotenoids in Plants: Enzymes and Color. In Carotenoids in Nature; Stange, C., Ed.; Subcellular Biochemistry; Springer International Publishing: Cham, Switzerland, 2016; Volume 79, pp. 35–69. ISBN 978-3-319-39124-3. [Google Scholar]
- Moreno, J.C.; Mi, J.; Agrawal, S.; Kössler, S.; Turečková, V.; Tarkowská, D.; Thiele, W.; Al-Babili, S.; Bock, R.; Schöttler, M.A. Expression of a Carotenogenic Gene Allows Faster Biomass Production by Redesigning Plant Architecture and Improving Photosynthetic Efficiency in Tobacco. Plant J. 2020, 103, 1967–1984. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.C.; Cerda, A.; Simpson, K.; Lopez-Diaz, I.; Carrera, E.; Handford, M.; Stange, C. Increased Nicotiana Tabacum Fitness through Positive Regulation of Carotenoid, Gibberellin and Chlorophyll Pathways Promoted by Daucus Carota Lycopene β-Cyclase (Dclcyb1) Expression. J. Exp. Bot. 2016, 67, 2325–2338. [Google Scholar] [CrossRef] [PubMed]
- Melis, A. Solar Energy Conversion Efficiencies in Photosynthesis: Minimizing the Chlorophyll Antennae to Maximize Efficiency. Plant Sci. 2009, 177, 272–280. [Google Scholar] [CrossRef]
- Ort, D.R.; Zhu, X.; Melis, A. Optimizing antenna size to maximize photosynthetic efficiency. Plant Physiol. 2011, 155, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Kirst, H.; Gabilly, S.T.; Niyogi, K.K.; Lemaux, P.G.; Melis, A. Photosynthetic Antenna Engineering to Improve Crop Yields. Planta 2017, 245, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Zhou, Z.; Li, Z.; Chen, Y.; Wang, Z.; Zhang, H. Rice (Oryza sativa L.) with Reduced Chlorophyll Content Exhibit Higher Photosynthetic Rate and Efficiency, Improved Canopy Light Distribution, and Greater Yields than Normally Pigmented Plants. Field Crops Res. 2017, 200, 58–70. [Google Scholar] [CrossRef]
- Cardona, T.; Shao, S.; Nixon, P.J. Enhancing Photosynthesis in Plants: The Light Reactions. Essays Biochem. 2018, 62, 85–94. [Google Scholar] [CrossRef]
- Ho, M.-Y.; Shen, G.; Canniffe, D.P.; Zhao, C.; Bryant, D.A. Light-Dependent Chlorophyll f Synthase Is a Highly Divergent Paralogueg of PsbA of Photosystem II. Science 2016, 353, 9178. [Google Scholar] [CrossRef]
- Nürnberg, D.J.; Morton, J.; Santabarbara, S.; Telfer, A.; Joliot, P.; Antonaru, L.A.; Ruban, A.V.; Cardona, T.; Krausz, E.; Boussac, A. Photochemistry beyond the Red Limit in Chlorophyll F-Containing Photosystems. Science 2018, 360, 1210–1213. [Google Scholar] [CrossRef] [PubMed]
- Drewry, D.T.; Kumar, P.; Long, S.P. Simultaneous Improvement in Productivity, Water Use, and Albedo through Crop Structural Modification. Glob. Chang. Biol. 2014, 20, 1955–1967. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Zhang, G.; Zhu, X.-G. Optimal Crop Canopy Architecture to Maximizse Canopy Photosynthetic CO2 Uptake under Elevated CO2—A Theoretical Study Using a Mechanistic Model of Canopy Photosynthesis. Funct. Plant Biol. 2013, 40, 108–124. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Morinaka, Y.; Ohnishi, T.; Sunohara, H.; Fujioka, S.; Ueguchi-Tanaka, M.; Mizutani, M.; Sakata, K.; Takatsuto, S.; Yoshida, S.; et al. Erect Leaves Caused by Brassinosteroid Deficiency Increase Biomass Production and Grain Yield in Rice. Nat. Biotechnol. 2006, 24, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Hirotsu, N.; Ujiie, K.; Perera, I.; Iri, A.; Kashiwagi, T.; Ishimaru, K. Partial Loss-of-Function of NAL1 Alters Canopy Photosynthesis by Changing the Contribution of Upper and Lower Canopy Leaves in Rice. Sci. Rep. 2017, 7, 15958. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, X.; Zhong, X.; Chang, S.; Qian, Q.; Zhang, Y.; Zhu, X. Partially Functional NARROW LEAF1 Balances Leaf Photosynthesis and Plant Architecture for Greater Rice Yield. Plant Physiol. 2022, 189, 772–789. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, W.; Ren, B.; Zhao, B.; Zhang, J.; Liu, P.; Zhang, Z. High Temperature Reduces Photosynthesis in Maize Leaves by Damaging Chloroplast Ultrastructure and Photosystem II. J. Agron. Crop Sci. 2020, 206, 548–564. [Google Scholar] [CrossRef]
- Ruban, A.V. Nonphotochemical Chlorophyll Fluorescence Quenching: Mechanism and Effectiveness in Protecting Plants from Photodamage. Plant Physiol. 2016, 170, 1903–1916. [Google Scholar] [CrossRef]
- Kato, Y.; Miura, E.; Ido, K.; Ifuku, K.; Sakamoto, W. The Variegated Mutants Lacking Chloroplastic FtsHs Are Defective in D1 Degradation and Accumulate Reactive Oxygen Species. Plant Physiol. 2009, 151, 1790–1801. [Google Scholar] [CrossRef]
- Niyogi, K.K.; Truong, T.B. Evolution of Flexible Nonnon-Photochemical Quenching Mechanisms That Regulate Light Harvesting in Oxygenic Photosynthesis. Curr. Opin. Plant Biol. 2013, 16, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Havaux, M.; Dall’Osto, L.; Bassi, R. Zeaxanthin Has Enhanced Antioxidant Capacity with Respect to All Other Xanthophylls in Arabidopsis Leaves and Functions Independent of Binding to PSII Antennae. Plant Physiol. 2007, 145, 1506–1520. [Google Scholar] [CrossRef] [PubMed]
- Dall’Osto, L.; Lico, C.; Alric, J.; Giuliano, G.; Havaux, M.; Bassi, R. Lutein Is Needed for Efficient Chlorophyll Triplet Quenching in the Major LHCII Antenna Complex of Higher Plants and Effective Photoprotection in Vivounder Strong Light. BMC Plant Biol. 2006, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Welc, R.; Luchowski, R.; Kluczyk, D.; Zubik-Duda, M.; Grudzinski, W.; Maksim, M.; Reszczynska, E.; Sowinski, K.; Mazur, R.; Nosalewicz, A.; et al. Mechanisms Shaping the Synergism of Zeaxanthin and PsbS in Photoprotective Energy Dissipation in the Photosynthetic Apparatus of Plants. Plant J. 2021, 107, 418–433. [Google Scholar] [CrossRef]
- Hubbart, S.; Smillie, I.R.A.; Heatley, M.; Swarup, R.; Foo, C.C.; Zhao, L.; Murchie, E.H. Enhanced Thylakoid Photoprotection Can Increase Yield and Canopy Radiation Use Efficiency in Rice. Commun. Biol. 2018, 1, 22. [Google Scholar] [CrossRef]
- Nelson, A. Investigation of the Role of Overexpression of PsbS Under Stress Inducible and Constitutive Promoters to Improve Water Use Efficiency; University of Nebraska: Lincoln, NE, USA, 2022. [Google Scholar]
- Lehretz, G.G.; Schneider, A.; Leister, D.; Sonnewald, U. High Non-photochemical Quenching of VPZ Transgenic Potato Plants Limits CO2 Assimilation under High Light Conditions and Reduces Tuber Yield under Fluctuating Light. J. Integr. Plant Biol. 2022, 64, 1821–1832. [Google Scholar] [CrossRef]
- Tikhonov, A.N. The Cytochrome B6f Complex at the Crossroad of Photosynthetic Electron Transport Pathways. Plant Physiol. Biochem. 2014, 81, 163–183. [Google Scholar] [CrossRef]
- Schöttler, M.A.; Tóth, S.Z.; Boulouis, A.; Kahlau, S. Photosynthetic Complex Stoichiometry Dynamics in Higher Plants: Biogenesis, Function, and Turnover of ATP Synthase and the Cytochrome B6f Complex. J. Exp. Bot. 2014, 66, 2373–2400. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, M.; Lopez-Calcagno, P.E.; Raines, C.A.; Furbank, R.T.; von Caemmerer, S. Overexpression of the Rieske FeS Protein of the Cytochrome B6f Complex Increases C4 Photosynthesis in Setaria Viridis. Commun. Biol. 2019, 2, 314. [Google Scholar] [CrossRef]
- Yamori, W.; Kondo, E.; Sugiura, D.; Terashima, I.; Suzuki, Y.; Makino, A. Enhanced Leaf Photosynthesis as a Target to Increase Grain Yield: Insights from Transgenic Rice Lines with Variable Rieske FeS Protein Content in the Cytochrome b6/f Complex. Plant Cell Environ. 2016, 39, 80–87. [Google Scholar] [CrossRef]
- Yamori, W.; Takahashi, S.; Makino, A.; Price, G.D.; Badger, M.R.; von Caemmerer, S. The Roles of ATP Synthase and the Cytochrome b 6/f Complexes in Limiting Chloroplast Electron Transport and Determining Photosynthetic Capacity. Plant Physiol. 2010, 155, 956–962. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, M.; Heyno, E.; Woodford, R.; Massey, B.; Birke, H.C.; von Caemmerer, S. Enhanced Abundance and Activity of the Chloroplast ATP Synthase in Rice through the Overexpression of the AtpD Subunit. J. Exp. Bot. 2022, 73, 6891–6901. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, K. Structure and Function of Plant-Type Ferredoxins. Photosynth. Res. 2004, 81, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Hanke, G.; Mulo, P. Plant Type Ferredoxins and Ferredoxin-dependent Metabolism. Plant Cell Environ. 2013, 36, 1071–1084. [Google Scholar] [CrossRef] [PubMed]
- Voss, I.; Koelmann, M.; Wojtera, J.; Holtgrefe, S.; Kitzmann, C.; Backhausen, J.E.; Scheibe, R. Knockout of Major Leaf Ferredoxin Reveals New Redox-regulatory Adaptations in Arabidopsis Thaliana. Physiol. Plant. 2008, 133, 584–598. [Google Scholar] [CrossRef] [PubMed]
- Okutani, S.; Hanke, G.T.; Satomi, Y.; Takao, T.; Kurisu, G.; Suzuki, A.; Hase, T. Three Maize Leaf Ferredoxin: NADPH Oxidoreductases Vary in Subchloroplast Location, Expression, and Interaction with Ferredoxin. Plant Physiol. 2005, 139, 1451–1459. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhong, D.; Yang, X.; Zhao, Y.; Dai, L.; Zeng, D.; Wang, Q.; Gao, L.; Li, S. ZmFdC2 Encoding a Ferredoxin Protein With C-Terminus Extension Is Indispensable for Maize Growth. Front. Plant Sci. 2021, 12, 646359. [Google Scholar] [CrossRef] [PubMed]
- Tognetti, V.B.; Palatnik, J.F.; Fillat, M.F.; Melzer, M.; Hajirezaei, M.-R.; Valle, E.M.; Carrillo, N. Functional Replacement of Ferredoxin by a Cyanobacterial Flavodoxin in Tobacco Confers Broad-Range Stress Tolerance. Plant Cell 2006, 18, 2035–2050. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhang, Y.; Wang, N.; He, H.; Wen, B.; Zhang, R.; Fu, X.; Xiao, W.; Li, D.; Li, L.; et al. Genome-Wide Identification and Characterization of the Prunus persicaPrunus Persica Ferredoxin Gene Family and Its Role in Improving Heat Tolerance. Plant Physiol. Biochem. 2022, 179, 108–119. [Google Scholar] [CrossRef]
- Pierella Karlusich, J.J.; Lodeyro, A.F.; Carrillo, N. The Long Goodbye: The Rise and Fall of Flavodoxin during Plant Evolution. J. Exp. Bot. 2014, 65, 5161–5178. [Google Scholar] [CrossRef]
- Hagemann, M.; Jeanjean, R.; Fulda, S.; Havaux, M.; Joset, F.; Erdmann, N. Flavodoxin Accumulation Contributes to Enhanced Cyclic Electron Flow around Photosystem I in Salt-Stressed Cells of Synechocystis Sp. Strain PCC 6803. Physiol. Plant. 1999, 105, 670–678. [Google Scholar] [CrossRef]
- Niazian, M.; Sadat-Noori, S.A.; Tohidfar, M.; Mortazavian, S.M.M.; Sabbatini, P. Betaine Aldehyde Dehydrogenase (BADH) vs. Flavodoxin (Fld): Two Important Genes for Enhancing Plants Stress Tolerance and Productivity. Front. Plant Sci. 2021, 12, 650215. [Google Scholar] [CrossRef] [PubMed]
- Erdner, D.L.; Price, N.M.; Doucette, G.J.; Peleato, M.L.; Anderson, D.M. Characterization of Ferredoxin and Flavodoxin as Markers of Iron Limitation in Marine Phytoplankton. Mar. Ecol. Prog. Ser. 1999, 184, 43–53. [Google Scholar] [CrossRef]
- Mayta, M.L.; Arce, R.C.; Zurbriggen, M.D.; Valle, E.M.; Hajirezaei, M.-R.; Zanor, M.I.; Carrillo, N. Expression of a Chloroplast-Targeted Cyanobacterial Flavodoxin in Tomato Plants Increases Harvest Index by Altering Plant Size and Productivity (Original Research. Front. Plant Sci. 2019, 10, 1432. [Google Scholar] [CrossRef] [PubMed]
- Shahbazi, M.; Tohidfar, M.; Aliniaeifard, S.; Yazdanpanah, F.; Bosacchi, M. Transgenic Tobacco Coco-Expressing Flavodoxin and Betaine Aldehyde Dehydrogenase Confers Cadmium Tolerance through Boosting Antioxidant Capacity. Protoplasma 2022, 259, 965–979. [Google Scholar] [CrossRef] [PubMed]
- Nazari, M.; Tohidfar, M.; Ramshini, H.; Vahdati, K. Molecular and Morphological Evaluation of Transgenic Persian Walnut Plants Harboring Fld Gene under Osmotic Stress Condition. Mol. Biol. Rep. 2022, 49, 433–441. [Google Scholar] [CrossRef]
- Eberhard, S.; Finazzi, G.; Wollman, F.A. The Dynamics of Photosynthesis. Annu. Rev. Genet. 2008, 42, 463–515. [Google Scholar] [CrossRef]
- Barkan, A.-C.; Goldschmidtclermont, M. Participation of Nuclear Genes in Chloroplast Gene Expression. Biochimie 2000, 82, 559–572. [Google Scholar] [CrossRef]
- Yamori, W.; Shikanai, T. Physiological Functions of Cyclic Electron Transport Around Photosystem I in Sustaining Photosynthesis and Plant Growth. Annu. Rev. Plant Biol. 2016, 67, 81–106. [Google Scholar] [CrossRef]
- Pashkovskiy, P.; Vereshchagin, M.; Kreslavski, V.; Ivanov, Y.; Kumachova, T.; Ryabchenko, A.; Voronkov, A.; Kosobryukhov, A.; Kuznetsov, V.; Allakhverdiev, S.I. Effect of Phytochrome Deficiency on Photosynthesis, Light-Related Genes Expression and Flavonoid Accumulation in Solanum Lycopersicum under Red and Blue Light. Cells 2022, 11, 3437. [Google Scholar] [CrossRef]
- Basso, L.; Sakoda, K.; Kobayashi, R.; Yamori, W.; Shikanai, T. Flavodiiron Proteins Enhance the Rate of CO2 Assimilation in Arabidopsis under Fluctuating Light Intensity. Plant Physiol. 2022, 189, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Wada, S.; Yamamoto, H.; Suzuki, Y.; Yamori, W.; Shikanai, T.; Makino, A. Flavodiiron Protein Substitutes for Cyclic Electron Flow without Competing CO2 Assimilation in Rice. Plant Physiol. 2018, 176, 1509–1518. [Google Scholar] [CrossRef] [PubMed]
- Kohzuma, K.; Froehlich, J.E.; Davis, G.A.; Temple, J.A.; Minhas, D.; Dhingra, A.; Cruz, J.A.; Kramer, D.M. The Role of Light–Dark Regulation of the Chloroplast ATP Synthase. Front. Plant Sci. 2017, 8, 274864. [Google Scholar] [CrossRef] [PubMed]
- Rott, M.; Martins, N.F.; Thiele, W.; Lein, W.; Bock, R.; Kramer, D.M.; Schöttler, M.A. ATP Synthase Repression in Tobacco Restricts Photosynthetic Electron Transport, CO2 Assimilation, and Plant Growth by Overacidification of the Thylakoid Lumen. Plant Cell 2011, 23, 304–321. [Google Scholar] [CrossRef] [PubMed]
- Boekema, E.J.; Lücken, U. The Structure of the CF1 Part of the ATP-Synthase Complex from Chloroplasts. In Oxygenic Photosynthesis: The Light Reactions; Ort, D.R., Yocum, C.F., Heichel, I.F., Eds.; Advances in Photosynthesis and Respiration; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2004; Volume 4, pp. 487–492. ISBN 978-0-7923-3683-9. [Google Scholar]
- Fristedt, R.; Martins, N.F.; Strenkert, D.; Clarke, C.A.; Suchoszek, M.; Thiele, W.; Schöttler, M.A.; Merchant, S.S. The Thylakoid Membrane Protein CGL160 Supports CF1CF0 ATP Synthase Accumulation in Arabidopsis Thaliana. PLoS ONE 2015, 10, e0121658. [Google Scholar] [CrossRef] [PubMed]
- Parry, M.A.J.; Andralojc, P.J.; Scales, J.C.; Salvucci, M.E.; Carmo-Silva, A.E.; Alonso, H.; Whitney, S.M. Rubisco Activity and Regulation as Targets for Crop Improvement. J. Exp. Bot. 2012, 64, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-H.; Tang, M.; Jin, X.-Q.; Li, H.; Chen, L.-S.; Wang, Q.-L.; Sun, A.-Z.; Yi, Y.; Guo, F.-Q. Regulation of Calvin-Benson Cycle Enzymes under High Temperature Stress. aBIOTECH 2022, 3, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Bracher, A.; Whitney, S.M.; Hartl, F.-H.; Hayer-Hartl, M. Biogenesis and Metabolic Maintenance of Rubisco. Annu. Rev. Plant Biol. 2017, 68, 29–60. [Google Scholar] [CrossRef] [PubMed]
- Whitney, S.M.; Sharwood, R.E.; Orr, D.; White, S.J.; Alonso, H.; Galmés, J. Isoleucine 309 Acts as a C4 Catalytic Switch That Increases Ribulose-1,5-B1, 5-Bisphosphate Carboxylase/Oxygenase (Rubisco) Carboxylation Rate in Flaveria. Proc. Natl. Acad. Sci. USA 2011, 108, 14688–14693. [Google Scholar] [CrossRef] [PubMed]
- Yamori, W. Strategies for Engineering Photosynthesis for Enhanced Plant Biomass Production; Physiological, Molecular Breeding and Genetic Perspectives: Rice Improvement; Springer Nature: Berlin/Heidelberg, Germany, 2021. [Google Scholar]
- Ishikawa, C.; Hatanaka, T.; Misoo, S.; Miyake, C.; Fukayama, H. Functional Incorporation of Sorghum Small Subunit Increases the Catalytic Turnover Rate of Rubisco in Transgenic Rice. Plant Physiol. 2011, 156, 1603–1611. [Google Scholar] [CrossRef]
- Matsumura, H.; Shiomi, K.; Yamamoto, A.; Taketani, Y.; Kobayashi, N.; Yoshizawa, T.; Tanaka, S.-I.; Yoshikawa, H.; Endo, M.; Fukayama, H. Hybrid Rubisco with Complete Replacement of Rice Rubisco Small Subunits by Sorghum Counterparts Confers C4 Plant-like High Catalytic Activity. Mol. Plant 2020, 13, 1570–1581. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.T.; Occhialini, A.; Andralojc, P.J.; Parry, M.A.; Hanson, M.R. A Faster Rubisco with Potential to Increase Photosynthesis in Crops. Nature 2014, 513, 547–550. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.T.; Hanson, M.R. Red Algal Rubisco Fails to Accumulate in Transplastomic Tobacco Expressing Griffithsia Monilis RbcL and RbcS Genes. Plant Direct 2018, 2, e00045. [Google Scholar] [CrossRef]
- Vitlin Gruber, A.; Feiz, L. Rubisco Assembly in the Chloroplast. Front. Mol. Biosci. 2018, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Salesse-Smith, C.E.; Sharwood, R.E.; Busch, F.A.; Kromdijk, J.; Bardal, V.; Stern, D.B. Overexpression of Rubisco Subunits with RAF1 Increases Rubisco Content in Maize. Nat. Plants 2018, 4, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Fan, Q.; Tang, Y.; Sun, Y.; Wang, L.; Wei, M.; Chang, Y. Overexpression of DfRaf from Fragrant Woodfern (Dryopteris fragrans) Enhances High-Temperature Tolerance in Tobacco (Nicotiana tabacum). Genes 2022, 13, 1212. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, M.E.; Crafts-Brandner, S.J. Relationship between the Heat Tolerance of Photosynthesis and the Thermal Stability of Rubisco Activase in Plants from Contrasting Thermal Environments. Plant Physiol. 2004, 134, 1460–1470. [Google Scholar] [CrossRef] [PubMed]
- Wijewardene, I.; Shen, G.; Zhang, H. Enhancing Crop Yield by Using Rubisco Activase to Improve Photosynthesis under Elevated Temperatures. Stress Biol. 2021, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Degen, G.E.; Worrall, D.-S.; Carmo-Silva, E. An Isoleucine Residue Acts as a Thermal and Regulatory Switch in Wheat Rubisco Activase. Plant J. 2020, 103, 742–751. [Google Scholar] [CrossRef] [PubMed]
- Kurek, I.; Chang, T.K.; Bertain, S.M.; Madrigal, A.; Liu, L.; Lassner, M.W.; Zhu, G. Enhanced Thermostability of Arabidopsis Rubisco Activase Improves Photosynthesis and Growth Rates under Moderate Heat Stress. Plant Cell 2007, 19, 3230–3241. [Google Scholar] [CrossRef]
- Yamori, W.; Masumoto, C.; Fukayama, H.; Makino, A. Rubisco Activase Is a Key Regulator of Non-steady-state Photosynthesis at Any Leaf Temperature and, to a Lesser Extent, of Steady-state Photosynthesis at High Temperature. Plant J. 2012, 71, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.; Liu, P.; Jiang, Z.; Ai, X. Overexpression of the Rubisco Activase Gene Improves Growth and Low Temperature and Weak Light Tolerance in Cucumis Sativus. Physiol. Plant. 2017, 161, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Scafaro, A.P.; Atwell, B.J.; Muylaert, S.; Reusel, B.V.; Ruiz, G.A.; van Rie, J.; Gallé, A. A Thermotolerant Variant of Rubisco Activase from a Wild Relative Improves Growth and Seed Yield in Rice under Heat Stress. Front. Plant Sci. 2018, 9, 1663. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Liu, J.; Zhang, Z.; Xi, Y.; Li, S.; Xiong, L.; Xing, Y. A Long Transcript Mutant of the Rubisco Activase Gene RCA Upregulated by the Transcription Factor Ghd2 Enhances Drought Tolerance in Rice. Plant J. 2022, 110, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Sakoda, K.; Fukayama, H.; Kondo, E.; Suzuki, Y.; Makino, A.; Terashima, I.; Yamori, W. Overexpression of Both Rubisco and Rubisco Activase Rescues Rice Photosynthesis and Biomass under Heat Stress. Plant Cell Environ. 2021, 44, 2308–2320. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Mueller-Cajar, O.; Yamori, W. Improving Plant Heat Tolerance through Modification of Rubisco Activase in C3 Plants to Secure Crop Yield and Food Security in a Future Warming World. J. Exp. Bot. 2023, 74, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, S.; Lawson, T.; Fryer, M.; Zakhleniuk, O.V.; Lloyd, J.C.; Raines, C.A. Increased Sedoheptulose-1,7-Bisphosphatase Activity in Transgenic Tobacco Plants Stimulates Photosynthesis and Growth from an Early Stage in Development. Plant Physiol. 2005, 138, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, D.M.; Locke, A.M.; Khozaei, M.; Raines, C.A.; Long, S.P.; Ort, D.R. Over-Expressing the C3 Photosynthesis Cycle Enzyme Sedoheptulose-1-7 Bisphosphatase Improves Photosynthetic Carbon Gain and Yield under Fully Open Air CO2 fumigation (FACE). BMC Plant Biol. 2011, 11, 123. [Google Scholar] [CrossRef] [PubMed]
- Driever, S.M.; Simkin, A.J.; Alotaibi, S.; Fisk, S.J.; Madgwick, P.J.; Sparks, C.A.; Jones, H.D.; Lawson, T.; Parry, M.A.J.; Raines, C.A. Increased SBPase Activity Improves Photosynthesis and Grain Yield in Wheat Grown in Greenhouse Conditions. Phil. Trans. R. Soc. B 2017, 372, 20160384. [Google Scholar] [CrossRef]
- Tamoi, M.; Nagaoka, M.; Miyagawa, Y.; Shigeoka, S. Contribution of Fructose-1, 6-Bisphosphatase and Sedoheptulose-1,7-Bisphosphatase to the Photosynthetic Rate and Carbon Flow in the Calvin Cycle in Transgenic Plants. Plant Cell Physiol. 2006, 47, 380–390. [Google Scholar] [CrossRef]
- Ku, M.S.; Agarie, S.; Nomura, M.; Fukayama, H.; Tsuchida, H.; Ono, K.; Hirose, S.; Toki, S.; Miyao, M.; Matsuoka, M. High-Level Expression of Maize Phosphoenolpyruvate Carboxylase in Transgenic Rice Plants. Nat. Biotechnol. 1999, 17, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; VanBuren, R.; Wai, C.M.; Tang, H.; Schatz, M.C.; Bowers, J.E.; Lyons, E.; Wang, M.-L.; Chen, J.; Biggers, E. The Pineapple Genome and the Evolution of CAM Photosynthesis. Nat. Genet. 2015, 47, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Hu, R.; Yin, H.; Jenkins, J.; Shu, S.; Tang, H.; Liu, D.; Weighill, D.A.; Cheol Yim, W.; Ha, J. The Kalanchoë Genome Provides Insights into Convergent Evolution and Building Blocks of Crassulacean Acid Metabolism. Nat. Commun. 2017, 8, 1899. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Guo, H.-B.; Weston, D.J.; Borland, A.M.; Ranjan, P.; Abraham, P.E.; Jawdy, S.S.; Wachira, J.; Tuskan, G.A.; Tschaplinski, T.J.; et al. Diel Rewiring and Positive Selection of Ancient Plant Proteins Enabled Evolution of CAM Photosynthesis in Agave. BMC Genom. 2018, 19, 588. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Hu, R.; Zhang, J.; Guo, H.-B.; Cheng, H.; Li, L.; Borland, A.M.; Qin, H.; Chen, J.-G.; Muchero, W. Overexpression of an Agave Phospho Enol Pyruvate Carboxylase Improves Plant Growth and Stress Tolerance. Cells 2021, 10, 582. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Tang, D.; Xie, M.; Zhang, J.; Zhai, L.; Mao, J.; Luo, C.; Lipzen, A.; Zhang, Y.; Savage, E. Agave REVEILLE1 Regulates the Onset and Release of Seasonal Dormancy in Populus. Plant Physiol. 2023, 191, 1492–1504. [Google Scholar] [CrossRef]
- Yang, X.; Cushman, J.C.; Borland, A.M.; Edwards, E.J.; Wullschleger, S.D.; Tuskan, G.A.; Owen, N.A.; Griffiths, H.; Smith, J.A.C.; De Paoli, H.C.; et al. A Roadmap for Research on Crassulacean Acid Metabolism (CAM) to Enhance Sustainable Food and Bioenergy Production in a Hotter, Drier World. New Phytol. 2015, 207, 491–504. [Google Scholar] [CrossRef]
- Fukayama, H.; Hatch, M.D.; Tamai, T.; Tsuchida, H.; Sudoh, S.; Furbank, R.T.; Miyao, M. Activity Regulation and Physiological Impacts of Maize C 4-Specific Phospho Enol Pyruvate Carboxylase Overproduced in Transgenic Rice Plants. Photosynth. Res. 2003, 77, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, T.D. Estimating the Rate of Photorespiration in Leaves. Physiol. Plant. 1988, 73, 147–152. [Google Scholar] [CrossRef]
- Peterhansel, C.; Horst, I.; Niessen, M.; Blume, C.; Kebeish, R.; Kürkcüoglu, S.; Kreuzaler, F. Photorespiration. Arab. Book/Am. Soc. Plant Biol. 2010, 8, e0130. [Google Scholar] [CrossRef]
- Ogren, W.L. Photorespiration: Pathways, Regulation, and Modification. Annu. Rev. Plant Physiol. 1984, 35, 415–442. [Google Scholar] [CrossRef]
- Ehlers, I.; Augusti, A.; Betson, T.R.; Nilsson, M.B.; Marshall, J.D.; Schleucher, J. Detecting Long-Term Metabolic Shifts Using Isotopomers: CO2-Driven Suppression of Photorespiration in C3 Plants over the 20th Century. Proc. Natl. Acad. Sci. USA 2015, 112, 15585–15590. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.-R.; Wang, L.-M.; Lin, X.-L.; Yao, Z.; Xu, H.-W.; Zhu, C.-H.; Teng, H.-Y.; Cui, L.-L.; Liu, E.E.; Zhang, J.-J.; et al. Engineering a New Chloroplastic Photorespiratory Bypass to Increase Photosynthetic Efficiency and Productivity in Rice. Mol. Plant 2019, 12, 199–214. [Google Scholar] [CrossRef]
- Walker, B.J.; VanLoocke, A.; Bernacchi, C.J.; Ort, D.R. The Costs of Photorespiration to Food Production Now and in the Future. Annu. Rev. Plant Biol. 2016, 67, 107–129. [Google Scholar] [CrossRef]
- Peterhansel, C.; Krause, K.; Braun, H.P.; Espie, G.; Fernie, A.; Hanson, D.; Keech, O.; Maurino, V.; Mielewczik, M.; Sage, R.F. Engineering Photorespiration: Current State and Future Possibilities. Plant Biol. 2013, 15, 754–758. [Google Scholar] [CrossRef]
- Kebeish, R.; Niessen, M.; Thiruveedhi, K.; Bari, R.; Hirsch, H.-J.; Rosenkranz, R.; Stäbler, N.; Schönfeld, B.; Kreuzaler, F.; Peterhänsel, C. Chloroplastic Photorespiratory Bypass Increases Photosynthesis and Biomass Production in Arabidopsis thalianaArabidopsis Thaliana. Nat. Biotechnol. 2007, 25, 593–599. [Google Scholar] [CrossRef]
- Nayak, L.; Panda, D.; Dash, G.K.; Lal, M.K.; Swain, P.; Baig, M.; Kumar, A. A Chloroplast Glycolate Catabolic Pathway Bypassing the Endogenous Photorespiratory Cycle Enhances Photosynthesis, Biomass and Yield in Rice (Oryza sativa L.). Plant Sci. 2022, 314, 111103. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Madgwick, P.J.; Powers, S.J.; Keys, A.J.; Lea, P.J.; Parry, M.A. An Engineered Pathway for Glyoxylate Metabolism in Tobacco Plants Aimed to Avoid the Release of Ammonia in Photorespiration. BMC Biotechnol. 2011, 11, 111. [Google Scholar] [CrossRef]
- Maier, A.; Fahnenstich, H.; Caemmerer, S.; Engqvist, M.K.; Weber, A.P.; Flügge, U.-I.; Maurino, V.G. Transgenic Introduction of a Glycolate Oxidative Cycle into A. Thaliana Chloroplasts Leads to Growth Improvement. Front. Plant Sci. 2012, 3, 38. [Google Scholar] [CrossRef]
- Shi, X.; Bloom, A. Photorespiration: The Futile Cycle? Plants 2021, 10, 908. [Google Scholar] [CrossRef]
- South, P.F.; Cavanagh, A.P.; Liu, H.W.; Ort, D.R. Synthetic Glycolate Metabolism Pathways Stimulate Crop Growth and Productivity in the Field. Science 2019, 363, 9077. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-M.; Shen, B.-R.; Li, B.-D.; Zhang, C.-L.; Lin, M.; Tong, P.-P.; Cui, L.-L.; Zhang, Z.-S.; Peng, X.-X. A Synthetic Photorespiratory Shortcut Enhances Photosynthesis to Boost Biomass and Grain Yield in Rice. Mol. Plant 2020, 13, 1802–1815. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhong, X.; Lin, D.; Wu, K.; Wu, Z.; Zhang, Z.; Peng, X. Grain Quality Affected by Introducing Photorespiratory Bypasses into Rice. Agronomy 2022, 12, 566. [Google Scholar] [CrossRef]
- Kuhn, C.; Hajirezaei, M.-R.; Fernie, A.R.; Roessner-Tunali, U.; Czechowski, T.; Hirner, B.; Frommer, W.B. The Sucrose Transporter StSUT1 Localizes to Sieve Elements in Potato Tuber Phloem and Influences Tuber Physiology and Development. Plant Physiol. 2003, 131, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Aluko, O.O.; Li, C.; Wang, Q.; Liu, H. Sucrose Utilization for Improved Crop Yields: A Review Article. Int. J. Mol. Sci. 2021, 22, 4704. [Google Scholar] [CrossRef] [PubMed]
- Leggewie, G.; Kolbe, A.; Lemoine, R.; Roessner, U.; Lytovchenko, A.; Zuther, E.; Kehr, J.; Frommer, W.B.; Riesmeier, J.W.; Willmitzer, L. Overexpression of the Sucrose Transporter So SUT1 in Potato Results in Alterations in Leaf Carbon Partitioning and in Tuber Metabolism but Has Little Impact on Tuber Morphology. Planta 2003, 217, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Sakurai-Ishikawa, J.; Murai-Hatano, M.; Hayashi, H.; Ahamed, A.; Fukushi, K.; Matsumoto, T.; Kitagawa, Y. Transpiration from Shoots Triggers Diurnal Changes in Root Aquaporin Expression. Plant Cell Environ. 2011, 34, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Aharon, R.; Shahak, Y.; Wininger, S.; Bendov, R.; Kapulnik, Y.; Galili, G. Overexpression of a Plasma Membrane Aquaporin in Transgenic Tobacco Improves Plant Vigor under Favorable Growth Conditions but Not under Drought or Salt Stress. Plant Cell 2003, 15, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Siefritz, F.; Tyree, M.T.; Lovisolo, C.; Schubert, A.; Kaldenhoff, R. PIP1 Plasma Membrane Aquaporins in Tobacco: From Cellular Effects to Function in Plants. Plant Cell 2002, 14, 869–876. [Google Scholar] [CrossRef]
- Chen, J.; Yue, K.; Shen, L.; Zheng, C.; Zhu, Y.; Han, K.; Kai, L. Aquaporins and CO2 Diffusion across Biological Membrane. Front. Physiol. 2023, 14, 1205290. [Google Scholar] [CrossRef]
- Groszmann, M.; Osborn, H.L.; Evans, J.R. Carbon Dioxide and Water Transport through Plant Aquaporins. Plant Cell Environ. 2017, 40, 938–961. [Google Scholar] [CrossRef] [PubMed]
- Kaldenhoff, R.; Kai, L.; Uehlein, N. Aquaporins and Membrane Diffusion of CO2 in Living Organisms. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 1592–1595. [Google Scholar] [CrossRef] [PubMed]
- Uehlein, N.; Lovisolo, C.; Siefritz, F.; Kaldenhoff, R. The Tobacco Aquaporin NtAQP1 Is a Membrane CO2 Pore with Physiological Functions. Nature 2003, 425, 734–737. [Google Scholar] [CrossRef]
- Heckwolf, M.; Pater, D.; Hanson, D.T.; Kaldenhoff, R. The Arabidopsis Thaliana Aquaporin AtPIP1;2 Is a Physiologically Relevant CO2 Transport Facilitator. Plant J. 2011, 67, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Z.; Liu, F.; Sun, L.; Hao, F. Versatile Roles of Aquaporins in Plant Growth and Development. Int. J. Mol. Sci. 2020, 21, 9485. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, M.; Osborn, H.; Groszmann, M.; Bala, S.; Bowerman, A.; McGaughey, S.; Byrt, C.; Alonso-cantabrana, H.; Tyerman, S.; Furbank, R.T.; et al. Expression of a CO2-Permeable Aquaporin Enhances Mesophyll Conductance in the C4 Species Setaria Viridis. eLife 2021, 10, e70095. [Google Scholar] [CrossRef]
- Postaire, O.; Tournaire-Roux, C.; Grondin, A.; Boursiac, Y.; Morillon, R.; Schäffner, A.R.; Maurel, C. A PIP1 Aquaporin Contributes to Hydrostatic Pressure-Induced Water Transport in Both the Root and Rosette of Arabidopsis. Plant Physiol. 2010, 152, 1418–1430. [Google Scholar] [CrossRef]
- Studer, A.J.; Gandin, A.; Kolbe, A.R.; Wang, L.; Cousins, A.B.; Brutnell, T.P. A Limited Role for Carbonic Anhydrase in C4 Photosynthesis as Revealed by a Ca1ca2 Double Mutant in Maize. Plant Physiol. 2014, 165, 608–617. [Google Scholar] [CrossRef]
- Moroney, J.V.; Bartlett, S.G.; Samuelsson, G. Carbonic Anhydrases in Plants and Algae. Plant Cell Environ. 2001, 24, 141–153. [Google Scholar] [CrossRef]
- Fabre, N.; Reiter, I.M.; Becuwe-Linka, N.; Genty, B.; Rumeau, D. Characterization and Expression Analysis of Genes Encoding α and β Carbonic Anhydrases in Arabidopsis. Plant Cell Environ. 2007, 30, 617–629. [Google Scholar] [CrossRef]
- Rudenko, N.N.; Borisova-Mubarakshina, M.M.; Ignatova, L.K.; Fedorchuk, T.P.; Nadeeva-Zhurikova, E.M.; Ivanov, B.N. Role of Plant Carbonic Anhydrases under Stress Conditions. Plant Stress Physiol. 2021, 4, 7–38. [Google Scholar]
- Jin, H.; Li, M.; Duan, S.; Fu, M.; Dong, X.; Liu, B.; Feng, D.; Wang, J.; Wang, H.-B. Optimization of light-harvesting pigment improves photosynthetic efficiency. Plant Physiol. 2016, 172, 1720–1731. [Google Scholar] [CrossRef]
- Chen, M.; Ji, M.; Wen, B.; Liu, L.; Li, S.; Chen, X.; Gao, D.; Li, L. GOLDEN 2-LIKE Transcription Factors of Plants. Front. Plant Sci. 2016, 7, 1509. [Google Scholar] [CrossRef] [PubMed]
- Ailizati, A.; Nagahage, I.S.P.; Miyagi, A.; Ishikawa, T.; Kawai-Yamada, M.; Demura, T.; Yamaguchi, M. An Arabidopsis NAC Domain Transcriptional Activator VND7 Negatively Regulates VNI2 Expression. Plant Biotechnol. 2021, 38, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Omranian, N.; Neumetzler, L.; Wang, T.; Herter, T.; Usadel, B.; Persson, S. A Transcriptional and Metabolic Framework for Secondary Wall Formation in Arabidopsis. Plant Physiol. 2016, 172, 1334–1351. [Google Scholar] [PubMed]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual Function of an Arabidopsis Transcription Factor DREB2A in Water-Stress-Responsive and Heat-Stress-Responsive Gene Expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, M.; Li, L.; Ma, Y. Functions and Application of the AP2/ERF Transcription Factor Family in Crop Improvement F. JIPB 2011, 53, 570–585. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-C.; Chi, C.; Jin, L.-J.; Zhu, J.; Yu, J.-Q.; Zhou, Y.-H. The bZip Transcription Factor HY5 Mediates CRY1a-Induced Anthocyanin Biosynthesis in Tomato. Plant Cell Environ. 2018, 41, 1762–1775. [Google Scholar] [CrossRef] [PubMed]
- Pashkovskiy, P.P.; Ryazansky, S.S. Biogenesis, Evolution, and Functions of Plant microRNAs. Biochemistry 2013, 78, 627–637. [Google Scholar] [CrossRef]
- Jerome Jeyakumar, J.M.; Ali, A.; Wang, W.-M.; Thiruvengadam, M. Characterizing the Role of the miR156-SPL Network in Plant Development and Stress Response. Plants 2020, 9, 1206. [Google Scholar] [CrossRef]
- Wang, L.; Tian, Y.; Shi, W.; Yu, P.; Hu, Y.; Lv, J.; Fu, C.; Fan, M.; Bai, M.-Y. The miR396-GRFs Module Mediates the Prevention of Photo-Oxidative Damage by Brassinosteroids during Seedling de-Etiolation in Arabidopsis. Plant Cell 2020, 32, 2525–2542. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Wu, H.; Zhang, T.; Ge, X.; Wang, T.; Zhou, W.; Zhang, L.; Ma, D.; Wang, A. Genome-Wide Identification of TCP Transcription Factors Family in Sweet Potato Reveals Significant Roles of miR319-Targeted TCPs in Leaf Anatomical Morphology. Front. Plant Sci. 2021, 12, 686698. [Google Scholar] [CrossRef] [PubMed]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of Jasmonate Biosynthesis and Senescence by miR319 Targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Huang, D.; Guo, Z.; Kuang, Z.; Zhang, H.; Xie, X.; Ma, Z.; Gao, S.; Lerdau, M.T.; Chu, C.; et al. Overexpression of microRNA408 Enhances Photosynthesis, Growth, and Seed Yield in Diverse Plants. JIPB 2018, 60, 323–340. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Burd, S.; Lers, A. Mi408 Is Involved in Abiotic Stress Responses in Arabidopsis. Plant J. 2015, 84, 169–187. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Bhunia, R.K.; Rajam, M.V. MicroRNAs as Potential Targets for Improving Rice Yield via Plant Architecture Modulation: Recent Studies and Future Perspectives. J. Biosci. 2020, 45, 116. [Google Scholar] [CrossRef]
- Tang, J.; Chu, C. MicroRNAs in Crop Improvement: Fine-Tuners for Complex Traits. Nat. Plants 2017, 3, 17077. [Google Scholar] [CrossRef]
- Pan, L.; Zhao, H.; Yu, Q.; Bai, L.; Dong, L. miR397/Laccase Gene Mediated Network Improves Tolerance to Fenoxaprop-P-Ethyl in Beckmannia Syzigachne and Oryza Sativa. Front. Plant Sci. 2017, 8, 879. [Google Scholar] [CrossRef]
- Zhang, Y.; Shan, X.; Zhao, Q.; Shi, F. The MicroRNA397a-LACCASE17 Module Regulates Lignin Biosynthesis in Medicago ruthenica (L.). Front. Plant Sci. 2022, 13, 978515. [Google Scholar] [CrossRef]
- Waheed, S.; Zeng, L. The Critical Role of miRNAs in Regulation of Flowering Time and Flower Development. Genes 2020, 11, 319. [Google Scholar] [CrossRef]
- Pashkovskiy, P.; Kreslavskii, V.; Khudyakova, A.; Kosobryukhov, A.; Kuznetsov, V.; Allakhverdiev, S. Influence of Phytochromes on microRNA Expression, Phenotype, and Photosynthetic Activity in A. Thaliana Phy Mutants under Light with Different Spectral Composition. Photosynthetica 2022, 61, 138–147. [Google Scholar] [CrossRef]
- Centritto, M.; Brilli, F.; Fodale, R.; Loreto, F. Different Sensitivity of Isoprene Emission, Respiration and Photosynthesis to High Growth Temperature Coupled with Drought Stress in Black Poplar (Populus nigra) Saplings. Tree Physiol. 2011, 31, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Wise, R.; Olson, A.; Schrader, S.; Sharkey, T.D. Electron Transport Is the Functional Limitation of Photosynthesis in Field-grown Pima Cotton Plants at High Temperature. Plant Cell Environ. 2004, 27, 717–724. [Google Scholar] [CrossRef]
- Ristic, Z.; Bukovnik, U.; Momčilović, I.; Fu, J.; Prasad, P.V. Heat-Induced Accumulation of Chloroplast Protein Synthesis Elongation Factor, EF-Tu, in Winter Wheat. J. Plant Physiol. 2008, 165, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.-L.; Chen, J.-H.; He, N.-Y.; Guo, F.-Q. Metabolic Reprogramming in Chloroplasts under Heat Stress in Plants. Int. J. Mol. Sci. 2018, 19, 849. [Google Scholar] [CrossRef] [PubMed]
- Lípová, L.; Krchňák, P.; Komenda, J.; Ilík, P. Heat-Induced Disassembly and Degradation of Chlorophyll-Containing Protein Complexes in Vivo. Biochim. Et Biophys. Acta (BBA) Bioenerg. 2010, 1797, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Reda, F.; Mandoura, H.M. Response of Enzymes Activities, Photosynthetic Pigments, Proline to Low or High Temperature Stressed Wheat Plant (Triticum aestivum L.) in the Presence or Absence of Exogenous Proline or Cysteine. Int. J. Acad. Res. 2011, 3, 108–115. [Google Scholar]
- Dutta, S.; Mohanty, S.; Tripathy, B.C. Role of Temperature Stress on Chloroplast Biogenesis and Protein Import in Pea. Plant Physiol. 2009, 150, 1050–1061. [Google Scholar] [CrossRef] [PubMed]
- Mathur, S.; Agrawal, D.; Jajoo, A. Photosynthesis: Response to High Temperature Stress. J. Photochem. Photobiol. B Biol. 2014, 137, 116–126. [Google Scholar] [CrossRef]
- Mazur, R.; Gieczewska, K.; Kowalewska, Ł.; Kuta, A.; Proboszcz, M.; Gruszecki, W.I.; Mostowska, A.; Garstka, M. Specific Composition of Lipid Phases Allows Retaining an Optimal Thylakoid Membrane Fluidity in Plant Response to Low-Temperature Treatment. Front. Plant Sci. 2020, 11, 723. [Google Scholar] [CrossRef]
- Muhammad, I.; Shalmani, A.; Ali, M.; Yang, Q.-H.; Ahmad, H.; Li, F.B. Mechanisms Regulating the Dynamics of Photosynthesis under Abiotic Stresses. Front. Plant Sci. 2021, 11, 615942. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Bai, X.; Ran, F.; Zhang, C.; Yan, Y.; Li, P.; Chen, H. Effects of Combined Extreme Cold and Drought Stress on Growth, Photosynthesis, and Physiological Characteristics of Cool-Season Grasses. Sci. Rep. 2024, 14, 116. [Google Scholar] [CrossRef] [PubMed]
- Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A.; Min, T.; Hao, Z.; Lu, Y.; Chen, J.; Yang, L. ICE-CBF-COR Signaling Cascade and Its Regulation in Plants Responding to Cold Stress. Int. J. Mol. Sci. 2022, 23, 1549. [Google Scholar] [CrossRef] [PubMed]
- Taghvaei, M.M.; Lahiji, H.S.; Golfazani, M.M. Evaluation of Expression Changes, Proteins Interaction Network, and microRNAs Targeting Catalase and Superoxide Dismutase Genes under Cold Stress in Rapeseed (Brassica napus L.). OCL 2022, 29, 3. [Google Scholar] [CrossRef]
- Zhang, H.; Dong, J.; Zhao, X.; Zhang, Y.; Ren, J.; Xing, L.; Jiang, C.; Wang, X.; Wang, J.; Zhao, S. Research Progress in Membrane Lipid Metabolism and Molecular Mechanism in Peanut Cold Tolerance. Front. Plant Sci. 2019, 10, 838. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Ming, Y.; Wang, H.-B.; Jin, H.-L. Strategies for Adaptation to High Light in Plants. aBIOTECH 2024. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, F.; Hao, L.; Yu, J.; Guo, L.; Zhou, H.; Ma, C.; Zhang, X.; Xu, M. Elevated CO2 Concentration Induces Photosynthetic Down-Regulation with Changes in Leaf Structure, Non-Structural Carbohydrates and Nitrogen Content of Soybean. BMC Plant Biol. 2019, 19, 255. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.; Jiang, C.; Chen, L.; Paul, A.; Chatterjee, A.; Shen, G. Achieving Abiotic Stress Tolerance in Plants through Antioxidative Defense Mechanisms. Front. Plant Sci. 2023, 14, 1110622. [Google Scholar] [CrossRef]
- Abramova, A.; Vereshchagin, M.; Kulkov, L.; Kreslavski, V.D.; Kuznetsov, V.V.; Pashkovskiy, P. Potential Role of Phytochromes A and B and Cryptochrome 1 in the Adaptation of Solanum Lycopersicum to UV-B Radiation. Int. J. Mol. Sci. 2023, 24, 13142. [Google Scholar] [CrossRef] [PubMed]
- Lingwan, M.; Pradhan, A.A.; Kushwaha, A.K.; Dar, M.A.; Bhagavatula, L.; Datta, S. Photoprotective Role of Plant Secondary Metabolites: Biosynthesis, Photoregulation, and Prospects of Metabolic Engineering for Enhanced Protection under Excessive Light. Environ. Exp. Bot. 2023, 209, 105300. [Google Scholar] [CrossRef]
- Ashikhmin, A.; Bolshakov, M.; Pashkovskiy, P.; Vereshchagin, M.; Khudyakova, A.; Shirshikova, G.; Kozhevnikova, A.; Kosobryukhov, A.; Kreslavski, V.; Kuznetsov, V.; et al. The Adaptive Role of Carotenoids and Anthocyanins in Solanum Lycopersicum Pigment Mutants under High Irradiance. Cells 2023, 3, 2569. [Google Scholar] [CrossRef] [PubMed]
- Pashkovskiy, P.; Kreslavski, V.; Khudyakova, A.; Ashikhmin, A.; Bolshakov, M.; Kozhevnikova, A.; Kosobryukhov, A.; Kuznetsov, V.V.; Allakhverdiev, S.I. Effect of High-Intensity Light on the Photosynthetic Activity, Pigment Content and Expression of Light-Dependent Genes of Photomorphogenetic Solanum Lycopersicum Hp Mutants. Plant Physiol. Biochem. 2021, 167, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Urban, L.; Aarrouf, J.; Bidel, L.P. Assessing the Effects of Water Deficit on Photosynthesis Using Parameters Derived from Measurements of Leaf Gas Exchange and of Chlorophyll a Fluorescence. Front. Plant Sci. 2017, 8, 2068. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lu, M.; Wang, Y.; Wang, Y.; Liu, Z.; Chen, S. Response Mechanism of Plants to Drought Stress. Horticulturae 2021, 7, 50. [Google Scholar] [CrossRef]
- Hussain, S.; Rao, M.J.; Anjum, M.A.; Ejaz, S.; Zakir, I.; Ali, M.A.; Ahmad, N.; Ahmad, S. Oxidative Stress and Antioxidant Defense in Plants Under Drought Conditions. In Plant Abiotic Stress Tolerance; Hasanuzzaman, M., Hakeem, K.R., Nahar, K., Alharby, H.F., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 207–219. ISBN 978-3-030-06117-3. [Google Scholar]
- Kordrostami, M.; Rabiei, B. Salinity Stress Tolerance in Plants: Physiological, Molecular, and Biotechnological Approaches. In Plant Abiotic Stress Tolerance; Hasanuzzaman, M., Hakeem, K.R., Nahar, K., Alharby, H.F., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 101–127. ISBN 978-3-030-06117-3. [Google Scholar]
- Hameed, A.; Ahmed, M.Z.; Hussain, T.; Aziz, I.; Ahmad, N.; Gul, B.; Nielsen, B.L. Effects of Salinity Stress on Chloroplast Structure and Function. Cells 2021, 10, 2023. [Google Scholar] [CrossRef] [PubMed]
- Billah, M.; Aktar, S.; Sikder, R.K.; Ahammed, G.J.; Hu, W.; Li, F.; Yang, Z. Exploring Regulatory Roles of Plant Thylakoid-Bound Proteins Involved in Abiotic Stress Responses. J. Plant Growth Regul. 2024, 43, 1570–1591. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Z.; Sui, N. Sensitivity and Responses of Chloroplasts to Salt Stress in Plants. Front. Plant Sci. 2024, 15, 1374086. [Google Scholar] [CrossRef] [PubMed]
- Muthurajan, R.; Ramanathan, V.; Bansilal Shillak, A.; Madhuri Pralhad, S.; Shankarrao, C.N.; Rahman, H.; Kambale, R.; Nallathambi, J.; Tamilselvan, S.; Madasamy, P. Controlled Over-Expression of AtDREB1A Enhances Tolerance against Drought and Salinity in Rice. Agronomy 2021, 11, 159. [Google Scholar] [CrossRef]
- Pabuayon, I.C.M.; Jiang, J.; Qian, H.; Chung, J.-S.; Shi, H. Gain-of-Function Mutations of AtNHX1 Suppress Sos1 Salt Sensitivity and Improve Salt Tolerance in Arabidopsis. Stress Biol. 2021, 1, 14. [Google Scholar] [CrossRef]
- Guerzoni, J.T.S.; Belintani, N.G.; Moreira, R.M.P.; Hoshino, A.A.; Domingues, D.S.; Filho, J.C.B.; Vieira, L.G.E. Stress-Induced Δ1-Pyrroline-5-Carboxylate Synthetase (P5CS) Gene Confers Tolerance to Salt Stress in Transgenic Sugarcane. Acta Physiol. Plant. 2014, 36, 2309–2319. [Google Scholar] [CrossRef]
- Dave, A.; Agarwal, P.; Agarwal, P.K. Mechanism of High Affinity Potassium Transporter (HKT) towards Improved Crop Productivity in Saline Agricultural Lands. 3 Biotech 2022, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Afzal, M.Z.; Jia, Q.; Ibrahim, A.K.; Niyitanga, S.; Zhang, L. Mechanisms and Signaling Pathways of Salt Tolerance in Crops: Understanding from the Transgenic Plants. Trop. Plant Biol. 2020, 13, 297–320. [Google Scholar] [CrossRef]
- Iqbal, Z.; Iqbal, M.S.; Hashem, A.; Abd_Allah, E.F.; Ansari, M.I. Plant Defense Responses to Biotic Stress and Its Interplay with Fluctuating Dark/Light Conditions. Front. Plant Sci. 2021, 12, 631810. [Google Scholar] [CrossRef] [PubMed]
- Romero-Rodríguez, B.; Petek, M.; Jiao, C.; Križnik, M.; Zagorščak, M.; Fei, Z.; Bejarano, E.R.; Gruden, K.; Castillo, A.G. Transcriptional and Epigenetic Changes during Tomato Yellow Leaf Curl Virus Infection in Tomato. BMC Plant Biol. 2023, 23, 651. [Google Scholar] [CrossRef] [PubMed]
- Piau, M.; Schmitt-Keichinger, C. The Hypersensitive Response to Plant Viruses. Viruses 2023, 15, 2000. [Google Scholar] [CrossRef] [PubMed]
- El-Sappah, A.H.; Qi, S.; Soaud, S.A.; Huang, Q.; Saleh, A.M.; Abourehab, M.A.S.; Wan, L.; Cheng, G.; Liu, J.; Ihtisham, M. Natural Resistance of Tomato Plants to Tomato Yellow Leaf Curl Virus. Front. Plant Sci. 2022, 13, 1081549. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Tang, C.; Wang, X.; Sun, S.; Zhao, J.; Kang, Z.; Wang, X. An Effector Protein of the Wheat Stripe Rust Fungus Targets Chloroplasts and Suppresses Chloroplast Function. Nat. Commun. 2019, 10, 5571. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-Recognition Receptors Are Required for NLR-Mediated Plant Immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Yang, H.; Luo, P. Changes in Photosynthesis Could Provide Important Insight into the Interaction between Wheat and Fungal Pathogens. Int. J. Mol. Sci. 2021, 22, 8865. [Google Scholar] [CrossRef]
- Shao, W.; Shi, G.; Chu, H.; Du, W.; Zhou, Z.; Wuriyanghan, H. Development of an NLR-ID Toolkit and Identification of Novel Disease-Resistance Genes in Soybean. Plants 2024, 13, 668. [Google Scholar] [CrossRef]
- Visakorpi, K.; Gripenberg, S.; Malhi, Y.; Bolas, C.; Oliveras, I.; Harris, N.; Rifai, S.; Riutta, T. Small-scale Indirect Plant Responses to Insect Herbivory Could Have Major Impacts on Canopy Photosynthesis and Isoprene Emission. New Phytol. 2018, 220, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M.; Chuang, W. Plant Resistance to Aphid Feeding: Behavioral, Physiological, Genetic and Molecular Cues Regulate Aphid Host Selection and Feeding. Pest Manag. Sci. 2014, 70, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Villalobos-López, M.A.; Arroyo-Becerra, A.; Quintero-Jiménez, A.; Iturriaga, G. Biotechnological Advances to Improve Abiotic Stress Tolerance in Crops. Int. J. Mol. Sci. 2022, 23, 12053. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Mishra, M.; Dhankher, O.P.; Singla-Pareek, S.L.; Pareek, A. Molecular Chaperones: Key Players of Abiotic Stress Response in Plants. In Genetic Enhancement of Crops for Tolerance to Abiotic Stress: Mechanisms and Approaches, Vol. I; Rajpal, V.R., Sehgal, D., Kumar, A., Raina, S.N., Eds.; Sustainable Development and Biodiversity; Springer International Publishing: Cham, Switzerland, 2019; Volume 20, pp. 125–165. ISBN 978-3-319-91955-3. [Google Scholar]
- Suprasanna, P.; Nikalje, G.C.; Rai, A.N. Osmolyte Accumulation and Implications in Plant Abiotic Stress Tolerance. In Osmolytes and Plants Acclimation to Changing Environment: Emerging Omics Technologies; Iqbal, N., Nazar, R., Khan, N.A., Eds.; Springer India: New Delhi, India, 2016; pp. 1–12. ISBN 978-81-322-2615-4. [Google Scholar]
- Yamchi, A.; Jazii, F.R.; Mousavi, A.; Karkhane, A.A. Proline Accumulation in Transgenic Tobacco as a Result of Expression of Arabidopsis 1-Pyrroline-5-Carboxylate Synthetase (P5CS) During Osmotic Stress. J. Plant Biochem. Biotechnol. 2007, 16, 9–15. [Google Scholar] [CrossRef]
- Su, J.; Wu, R. Stress-Inducible Synthesis of Proline in Transgenic Rice Confers Faster Growth under Stress Conditions than That with Constitutive Synthesis. Plant Sci. 2004, 166, 941–948. [Google Scholar] [CrossRef]
- Zameer, R.; Fatima, K.; Azeem, F.; ALgwaiz, H.I.; Sadaqat, M.; Rasheed, A.; Batool, R.; Shah, A.N.; Zaynab, M.; Shah, A.A. Genome-Wide Characterization of Superoxide Dismutase (SOD) Genes in Daucus Carota: Novel Insights into Structure, Expression, and Binding Interaction with Hydrogen Peroxide (H2O2) under Abiotic Stress Condition. Front. Plant Sci. 2022, 13, 870241. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Li, Q.; Park, S.-C.; Wang, X.; Liu, Y.; Zhang, Y.; Tang, W.; Kou, M.; Ma, D. Overexpression of CuZnSOD and APX Enhance Salt Stress Tolerance in Sweet Potato. Plant Physiol. Biochem. 2016, 109, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.C.; Salvi, P.; Kamble, N.U.; Joshi, P.K.; Majee, M.; Arora, S. Ectopic Overexpression of Cytosolic Ascorbate Peroxidase Gene (Apx1) Improves Salinity Stress Tolerance in Brassica Juncea by Strengthening Antioxidative Defense Mechanism. Acta Physiol. Plant. 2020, 42, 45. [Google Scholar] [CrossRef]
- Sottosanto, J.B.; Saranga, Y.; Blumwald, E. Impact of AtNHX1, a Vacuolar Na+/H+ Antiporter, upon Gene Expression during Short- and Long-Term Salt Stress in Arabidopsis Thaliana. BMC Plant Biol. 2007, 7, 18. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, J.; Wu, X.; Dong, L. Na+/K+ Balance and Transport Regulatory Mechanisms in Weedy and Cultivated Rice (Oryza sativa L.) Under Salt Stress. BMC Plant Biol. 2018, 18, 375. [Google Scholar] [CrossRef]
- Lata, C.; Bhutty, S.; Bahadur, R.P.; Majee, M.; Prasad, M. Association of an SNP in a Novel DREB2-like Gene SiDREB2 with Stress Tolerance in Foxtail Millet [Setaria italica (L.)]. J. Exp. Bot. 2011, 62, 3387–3401. [Google Scholar] [CrossRef] [PubMed]
- Hoang, X.L.T.; Nguyen, Y.-N.H.; Thao, N.P.; Tran, L.-S.P. NAC Transcription Factors in Drought and Salinity Tolerance. In Salt and Drought Stress Tolerance in Plants; Hasanuzzaman, M., Tanveer, M., Eds.; Signaling and Communication in Plants; Springer International Publishing: Cham, Switzerland, 2020; pp. 351–366. ISBN 978-3-030-40276-1. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nazari, M.; Kordrostami, M.; Ghasemi-Soloklui, A.A.; Eaton-Rye, J.J.; Pashkovskiy, P.; Kuznetsov, V.; Allakhverdiev, S.I. Enhancing Photosynthesis and Plant Productivity through Genetic Modification. Cells 2024, 13, 1319. https://doi.org/10.3390/cells13161319
Nazari M, Kordrostami M, Ghasemi-Soloklui AA, Eaton-Rye JJ, Pashkovskiy P, Kuznetsov V, Allakhverdiev SI. Enhancing Photosynthesis and Plant Productivity through Genetic Modification. Cells. 2024; 13(16):1319. https://doi.org/10.3390/cells13161319
Chicago/Turabian StyleNazari, Mansoureh, Mojtaba Kordrostami, Ali Akbar Ghasemi-Soloklui, Julian J. Eaton-Rye, Pavel Pashkovskiy, Vladimir Kuznetsov, and Suleyman I. Allakhverdiev. 2024. "Enhancing Photosynthesis and Plant Productivity through Genetic Modification" Cells 13, no. 16: 1319. https://doi.org/10.3390/cells13161319







