Cancer-Associated-Fibroblast-Mediated Paracrine and Autocrine SDF-1/CXCR4 Signaling Promotes Stemness and Aggressiveness of Colorectal Cancers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation, Extraction, and Identification of CAFs and NFs
2.2. RNA Extraction, Quality Assessment, and Microarray Analysis
2.3. Cell Cultures and 3D Organotypic Culture System
2.4. Viral Production and Infection of Target Cells
2.5. In Vitro Migration and Invasion Assay
2.6. Western Blotting
2.7. Quantitative Real-Time PCR
2.8. Enzyme-Linked Immunosorbent Assay
2.9. Flow Cytometry
2.10. Xenograft Model
2.11. Sphere Culture
2.12. Clinical Tissue Collection with Immunohistochemical Staining
2.13. Statistical Analysis
3. Results
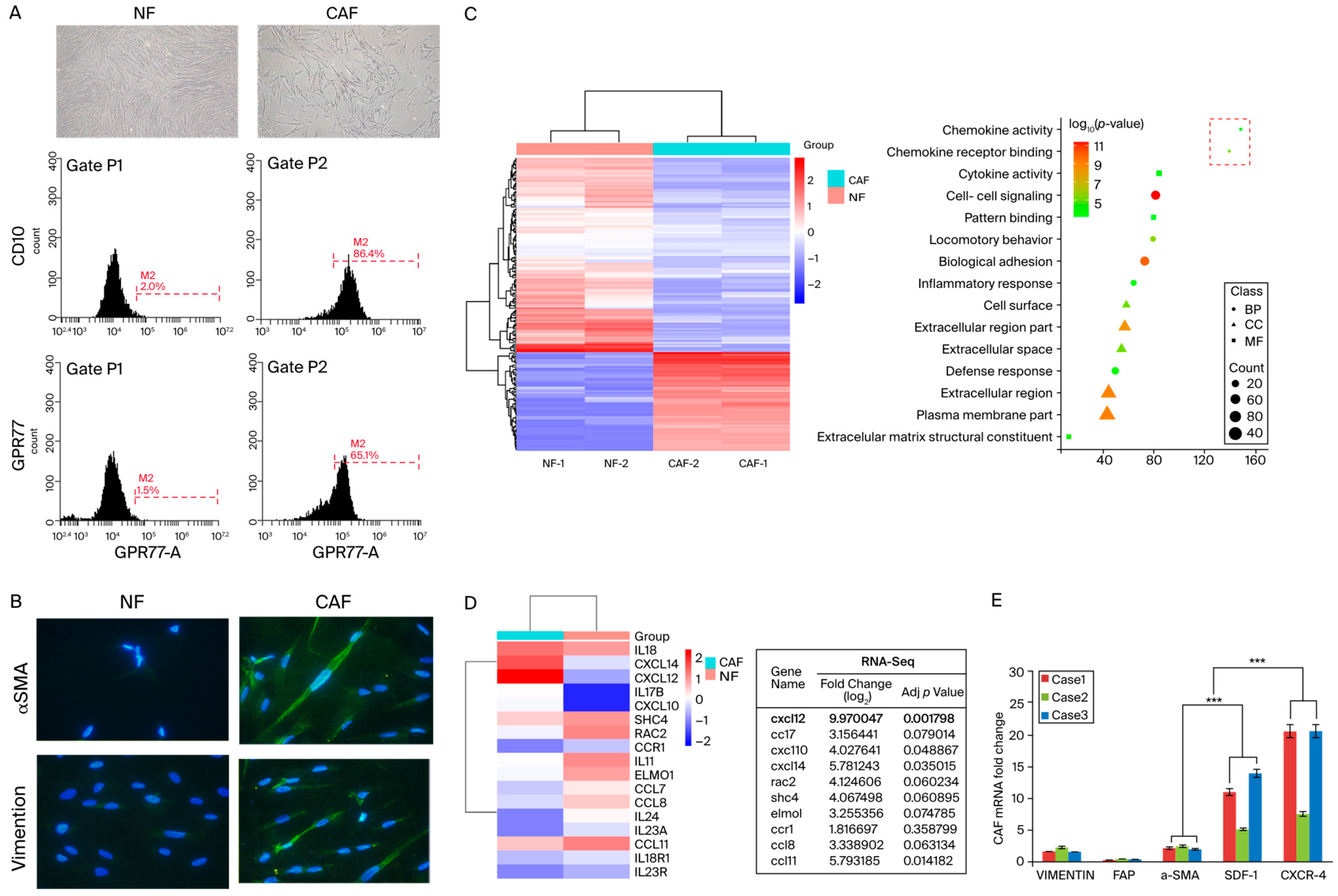
3.1. SDF-1 Was Markedly Increased in Cancer-Associated Fibroblasts
3.2. CAFs Induce Aggressive Phenotypes of CRC Cells
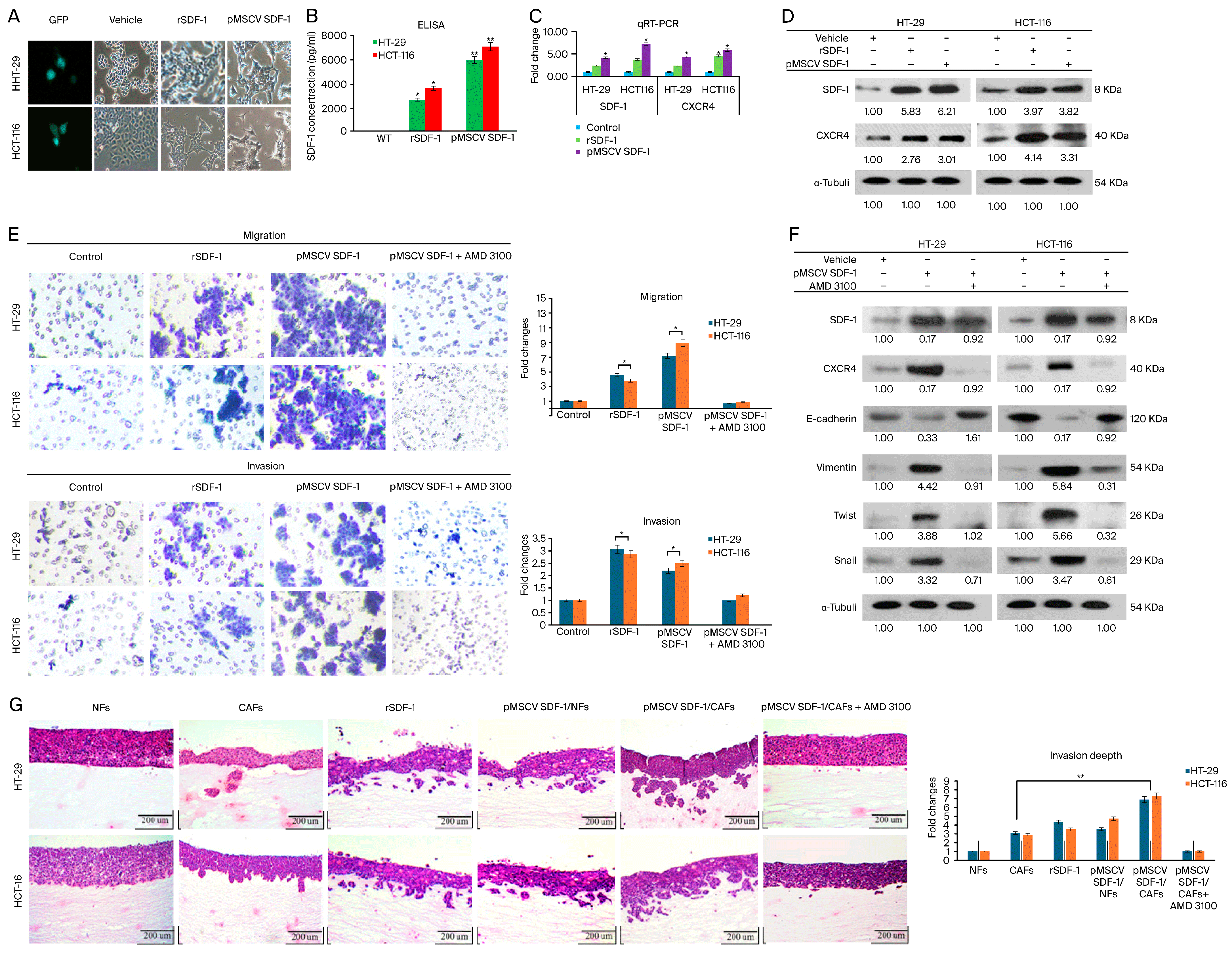
3.3. SDF-1 Expression in CAFs Promotes CRC Cell Migration and Invasion through Autocrine and Paracrine Signaling
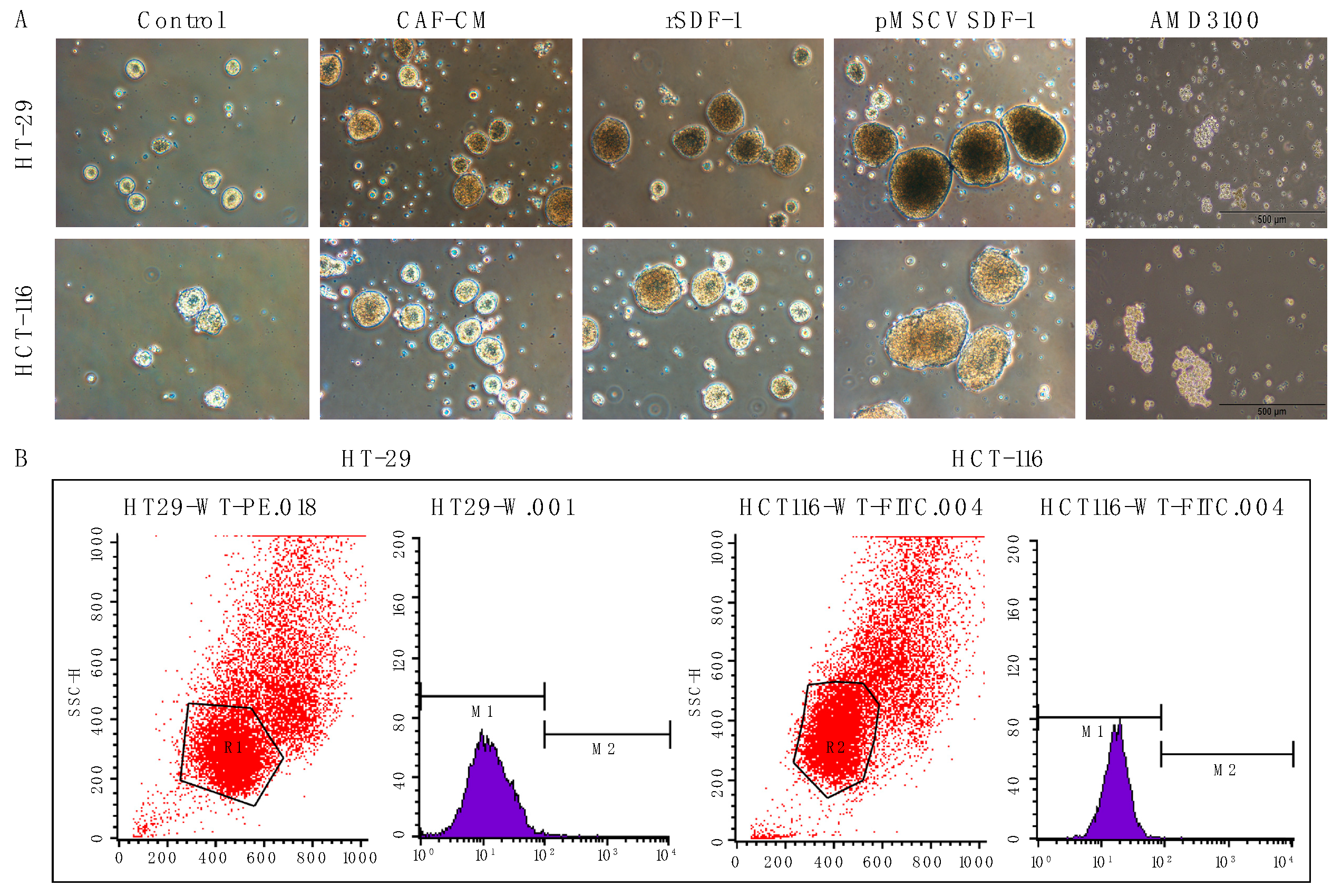
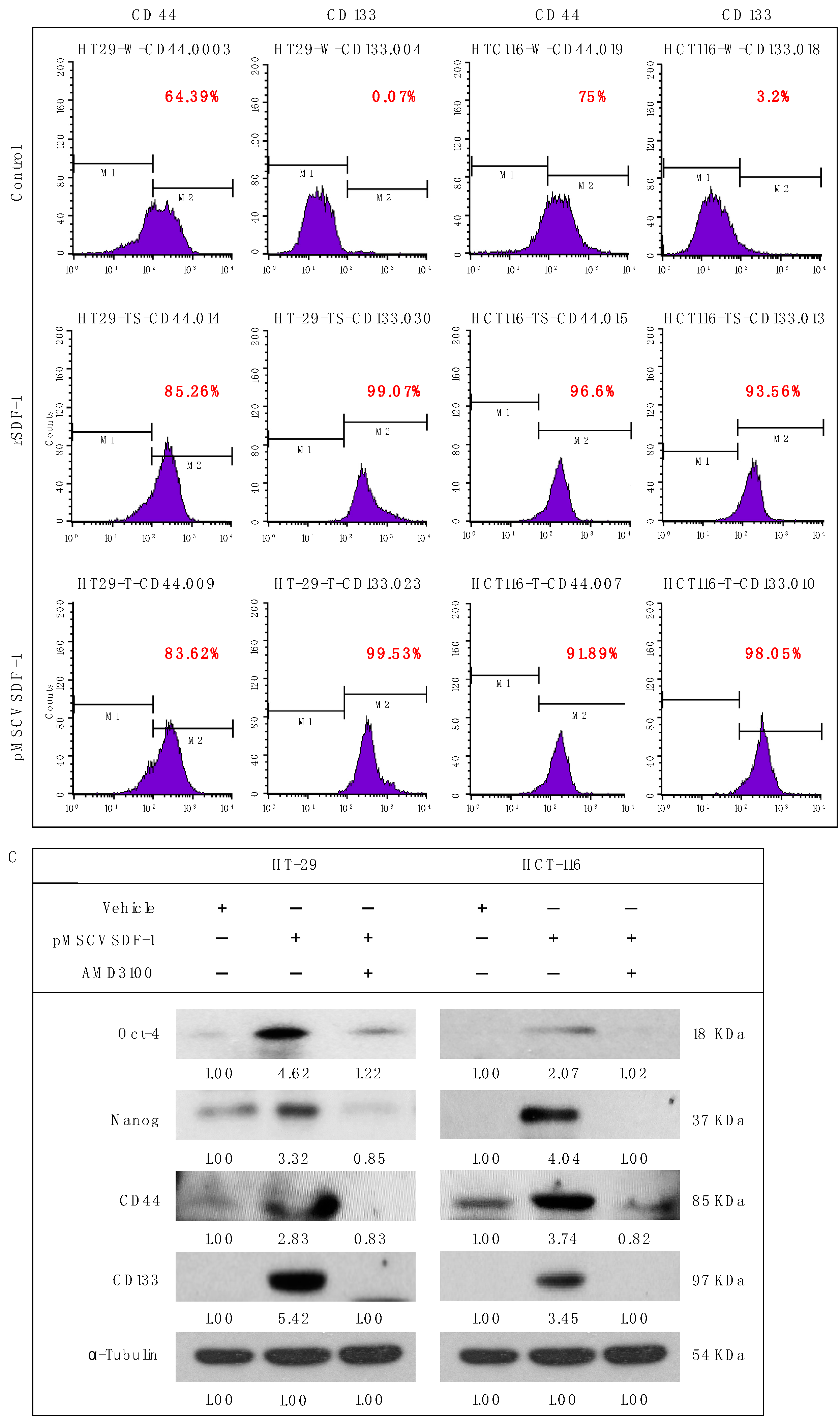
3.4. SDF-1 Enhances Spheroid Formation and Promotes Cancer Stemness in CRC Cells
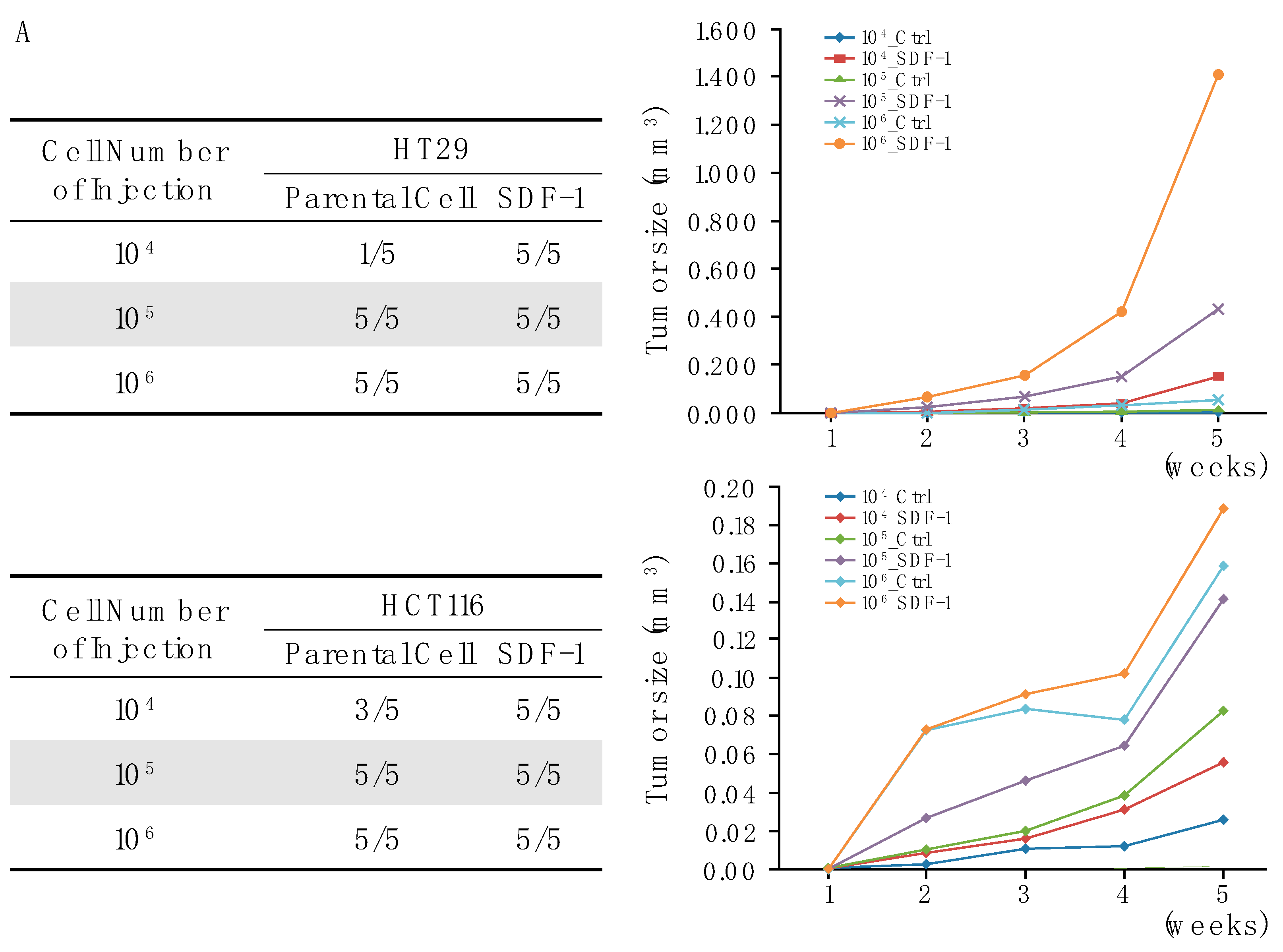
3.5. SDF-1-CXCR4 Signaling in Tumor Growth through Xenograft Models
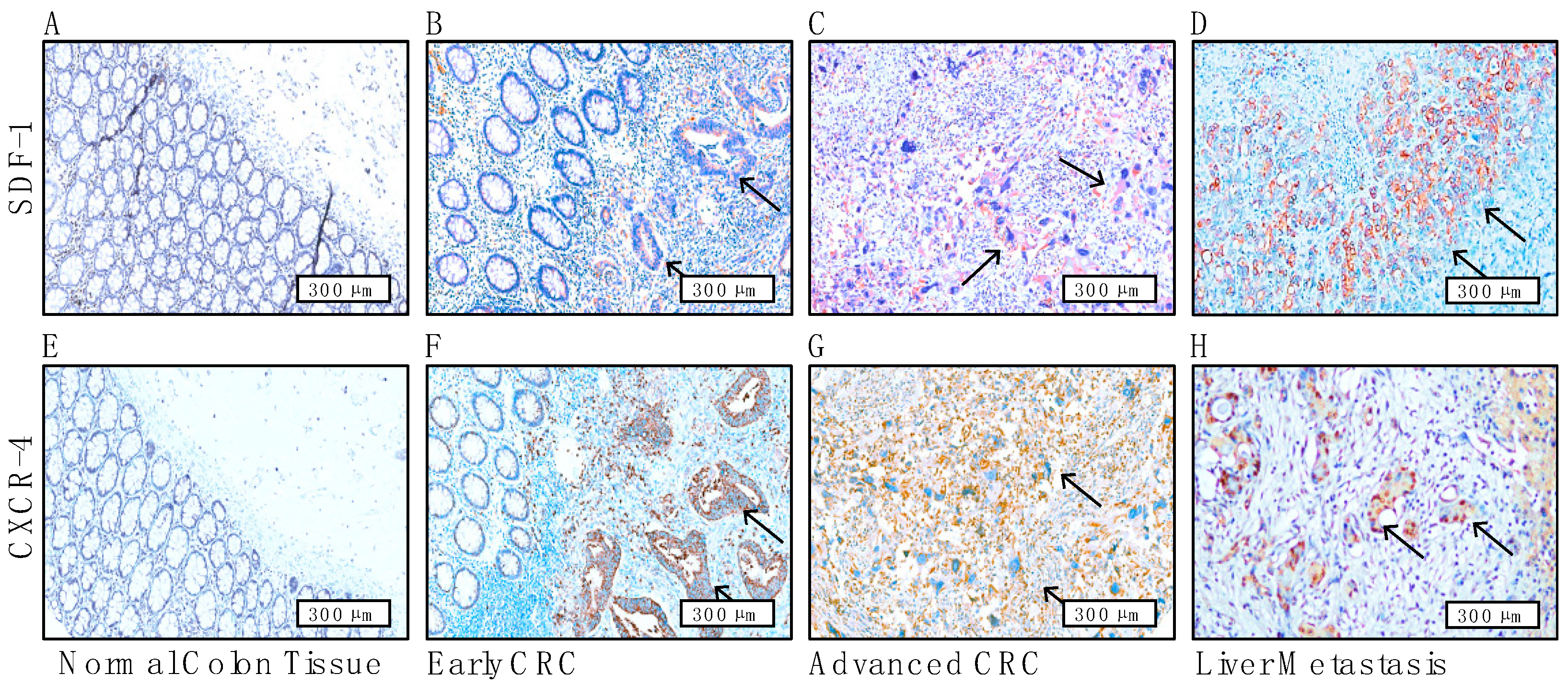
3.6. Immunohistochemical Staining Reveals the Prognostic Role of CAF-Mediated SDF-1/CXCR4 Signaling and Its Association with Liver Metastases in CRC Patients
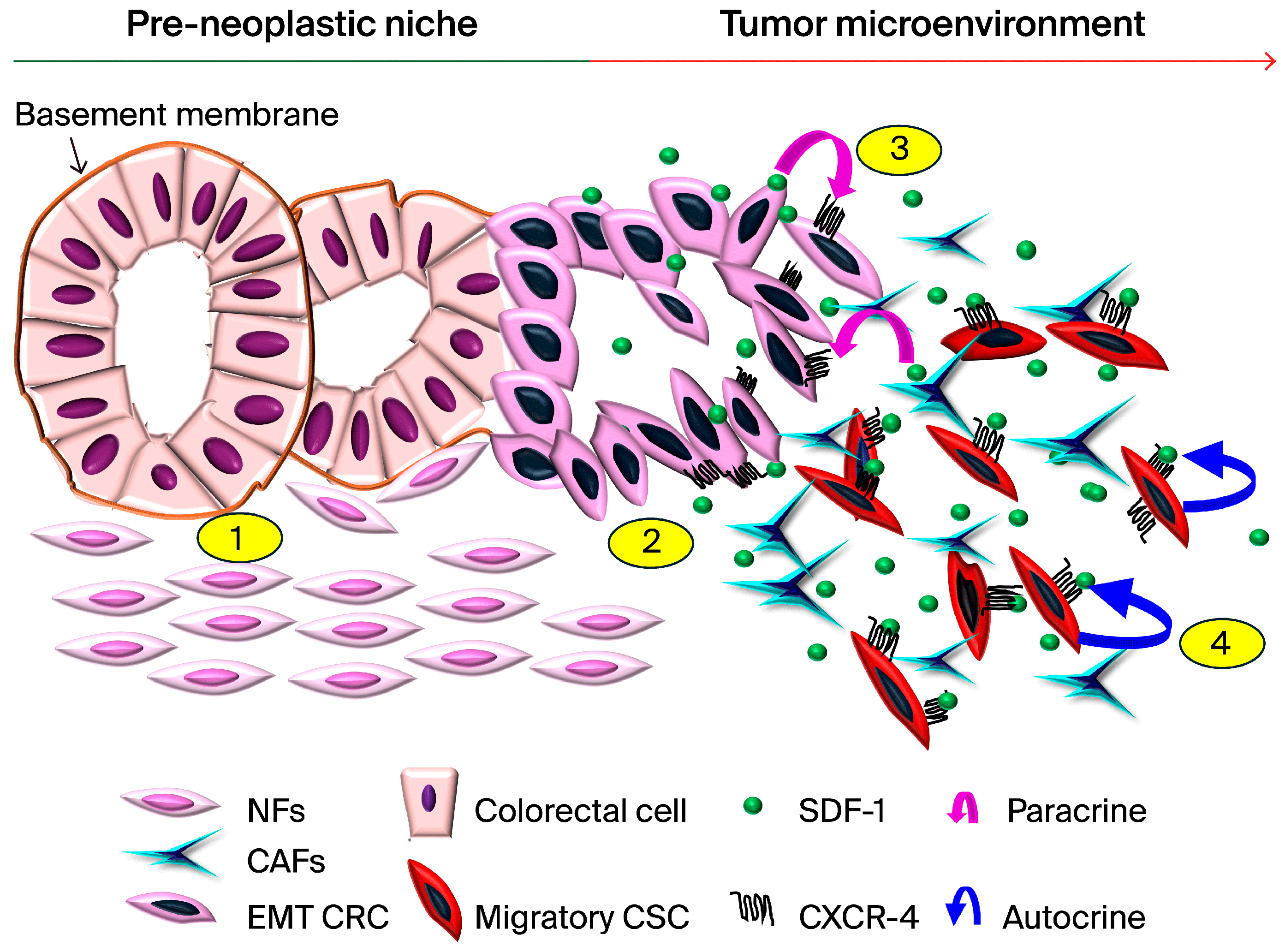
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2020, 70, 313. [Google Scholar] [CrossRef]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Janne, P.A.; Mayer, R.J. Chemoprevention of colorectal cancer. N. Engl. J. Med. 2000, 342, 1960–1968. [Google Scholar] [CrossRef]
- Chen, F.; Zhuang, X.; Lin, L.; Yu, P.; Wang, Y.; Shi, Y.; Hu, G.; Sun, Y. New horizons in tumor microenvironment biology: Challenges and opportunities. BMC Med. 2015, 13, 45. [Google Scholar] [CrossRef]
- Castells, M.; Thibault, B.; Delord, J.P.; Couderc, B. Implication of tumor microenvironment in chemoresistance: Tumor-associated stromal cells protect tumor cells from cell death. Int. J. Mol. Sci. 2012, 13, 9545–9571. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Catalano, V.; Turdo, A.; Di Franco, S.; Dieli, F.; Todaro, M.; Stassi, G. Tumor and its microenvironment: A synergistic interplay. Semin. Cancer Biol. 2013, 23, 522–532. [Google Scholar] [CrossRef]
- Isella, C.; Terrasi, A.; Bellomo, S.E.; Petti, C.; Galatola, G.; Muratore, A.; Mellano, A.; Senetta, R.; Cassenti, A.; Sonetto, C.; et al. Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 2015, 47, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Korkaya, H.; Liu, S.; Wicha, M.S. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J. Clin. Investig. 2011, 121, 3804–3809. [Google Scholar] [CrossRef]
- Parsonage, G.; Filer, A.D.; Haworth, O.; Nash, G.B.; Rainger, G.E.; Salmon, M.; Buckley, C.D. A stromal address code defined by fibroblasts. Trends Immunol. 2005, 26, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Tomasek, J.J.; Gabbiani, G.; Hinz, B.; Chaponnier, C.; Brown, R.A. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat. Rev. Mol. Cell Biol. 2002, 3, 349–363. [Google Scholar] [CrossRef]
- Watt, F.M.; Fujiwara, H. Cell-extracellular matrix interactions in normal and diseased skin. Cold Spring Harb. Perspect. Biol. 2011, 3, a005124. [Google Scholar] [CrossRef] [PubMed]
- Servais, C.; Erez, N. From sentinel cells to inflammatory culprits: Cancer-associated fibroblasts in tumour-related inflammation. J. Pathol. 2013, 229, 198–207. [Google Scholar] [CrossRef] [PubMed]
- LeBleu, V.S.; Kalluri, R. A peek into cancer-associated fibroblasts: Origins, functions and translational impact. Dis. Model. Mech. 2018, 11, dmm029447. [Google Scholar] [CrossRef] [PubMed]
- Gascard, P.; Tlsty, T.D. Carcinoma-associated fibroblasts: Orchestrating the composition of malignancy. Genes. Dev. 2016, 30, 1002–1019. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, N.A.; Sanchez-Salas, R.; Peske, J.D.; Vano, Y.; Becht, E.; Petitprez, F.; Validire, P.; Ingels, A.; Cathelineau, X.; Fridman, W.H.; et al. The clinical role of the TME in solid cancer. Br. J. Cancer 2019, 120, 45–53. [Google Scholar] [CrossRef]
- Zhuang, J.; Lu, Q.; Shen, B.; Huang, X.; Shen, L.; Zheng, X.; Huang, R.; Yan, J.; Guo, H. TGFbeta1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci. Rep. 2015, 5, 11924. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xiao, C.H.; Tan, L.D.; Wang, Q.S.; Li, X.Q.; Feng, Y.M. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-beta signalling. Br. J. Cancer 2014, 110, 724–732. [Google Scholar] [CrossRef]
- Xuefeng, X.; Hou, M.X.; Yang, Z.W.; Agudamu, A.; Wang, F.; Su, X.L.; Li, X.; Shi, L.; Terigele, T.; Bao, L.L.; et al. Epithelial-mesenchymal transition and metastasis of colon cancer cells induced by the FAK pathway in cancer-associated fibroblasts. J. Int. Med. Res. 2020, 48, 300060520931242. [Google Scholar] [CrossRef]
- You, J.; Li, M.; Cao, L.M.; Gu, Q.H.; Deng, P.B.; Tan, Y.; Hu, C.P. Snail1-dependent cancer-associated fibroblasts induce epithelial-mesenchymal transition in lung cancer cells via exosomes. QJM 2019, 112, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Calon, A.; Lonardo, E.; Berenguer-Llergo, A.; Espinet, E.; Hernando-Momblona, X.; Iglesias, M.; Sevillano, M.; Palomo-Ponce, S.; Tauriello, D.V.; Byrom, D.; et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015, 47, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, R.; Pietras, K. Heterogeneity of cancer-associated fibroblasts: Opportunities for precision medicine. Cancer Sci. 2020, 111, 2708–2717. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Huang, W.Y.; Lee, T.Y.; Chang, Y.M.; Chen, S.F.; Lin, Y.S.; Nieh, S. Interleukin-33-Enhanced CXCR4 Signaling Circuit Mediated by Carcinoma-Associated Fibroblasts Promotes Invasiveness of Head and Neck Cancer. Cancers 2021, 13, 3442. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Kipps, T.J. CXCR4: A key receptor in the crosstalk between tumor cells and their microenvironment. Blood 2006, 107, 1761–1767. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, O.; Bouchard, A.; Baccarelli, A.; Deschenes, J.; Sauter, G.; Simon, R.; Bianchi, R.; Basik, M. The role of CXCR4 receptor expression in breast cancer: A large tissue microarray study. Breast Cancer Res. Treat. 2006, 97, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.C.; Makena, P.S.; Gorantla, V.; Sinclair, S.E.; Waters, C.M. CXCR4 regulates migration of lung alveolar epithelial cells through activation of Rac1 and matrix metalloproteinase-2. Am. J. Physiol. Lung Cell Mol. Physiol. 2012, 302, L846–L856. [Google Scholar] [CrossRef] [PubMed]
- Pauls, E.; Ruiz, A.; Badia, R.; Permanyer, M.; Gubern, A.; Riveira-Munoz, E.; Torres-Torronteras, J.; Alvarez, M.; Mothe, B.; Brander, C.; et al. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J. Immunol. 2014, 193, 1988–1997. [Google Scholar] [CrossRef]
- Orimo, A.; Gupta, P.B.; Sgroi, D.C.; Arenzana-Seisdedos, F.; Delaunay, T.; Naeem, R.; Carey, V.J.; Richardson, A.L.; Weinberg, R.A. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005, 121, 335–348. [Google Scholar] [CrossRef]
- Amara, S.; Chaar, I.; Khiari, M.; Ounissi, D.; Weslati, M.; Boughriba, R.; Hmida, A.B.; Bouraoui, S. Stromal cell derived factor-1 and CXCR4 expression in colorectal cancer promote liver metastasis. Cancer Biomark. 2015, 15, 869–879. [Google Scholar] [CrossRef]
- Kim, J.; Takeuchi, H.; Lam, S.T.; Turner, R.R.; Wang, H.J.; Kuo, C.; Foshag, L.; Bilchik, A.J.; Hoon, D.S. Chemokine receptor CXCR4 expression in colorectal cancer patients increases the risk for recurrence and for poor survival. J. Clin. Oncol. 2005, 23, 2744–2753. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.; Carmical, J.R.; Ives, K.L.; Wood, T.G.; Aronson, J.F.; Gomez, G.A.; Djukom, C.D.; Hellmich, M.R. CD133+ colon cancer cells are more interactive with the tumor microenvironment than CD133- cells. Lab. Investig. 2012, 92, 420–436. [Google Scholar] [CrossRef] [PubMed]
- Saigusa, S.; Toiyama, Y.; Tanaka, K.; Yokoe, T.; Okugawa, Y.; Fujikawa, H.; Matsusita, K.; Kawamura, M.; Inoue, Y.; Miki, C.; et al. Cancer-associated fibroblasts correlate with poor prognosis in rectal cancer after chemoradiotherapy. Int. J. Oncol. 2011, 38, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Akishima-Fukasawa, Y.; Nakanishi, Y.; Ino, Y.; Moriya, Y.; Kanai, Y.; Hirohashi, S. Prognostic significance of CXCL12 expression in patients with colorectal carcinoma. Am. J. Clin. Pathol. 2009, 132, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.F.; Nieh, S.; Jao, S.W.; Wu, M.Z.; Liu, C.L.; Chang, Y.C.; Lin, Y.S. The paracrine effect of cancer-associated fibroblast-induced interleukin-33 regulates the invasiveness of head and neck squamous cell carcinoma. J. Pathol. 2013, 231, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.Y.; Lin, Y.S.; Lin, Y.C.; Nieh, S.; Chang, Y.M.; Lee, T.Y.; Chen, S.F.; Yang, K.D. Cancer-Associated Fibroblasts Promote Tumor Aggressiveness in Head and Neck Cancer through Chemokine Ligand 11 and C-C Motif Chemokine Receptor 3 Signaling Circuit. Cancers 2022, 14, 3141. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.F.; Chang, Y.C.; Nieh, S.; Liu, C.L.; Yang, C.Y.; Lin, Y.S. Nonadhesive culture system as a model of rapid sphere formation with cancer stem cell properties. PLoS ONE 2012, 7, e31864. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Zhang, Y.; Li, Y.; Wei, L. Crosstalk between colorectal cancer cells and cancer-associated fibroblasts in the tumor microenvironment mediated by exosomal noncoding RNAs. Front. Immunol. 2023, 14, 1161628. [Google Scholar] [CrossRef]
- Chhabra, Y.; Weeraratna, A.T. Fibroblasts in cancer: Unity in heterogeneity. Cell 2023, 186, 1580–1609. [Google Scholar] [CrossRef]
- Han, C.; Liu, T.; Yin, R. Biomarkers for cancer-associated fibroblasts. Biomark. Res. 2020, 8, 64. [Google Scholar] [CrossRef]
- Mhaidly, R.; Mechta-Grigoriou, F. Fibroblast heterogeneity in tumor micro-environment: Role in immunosuppression and new therapies. Semin. Immunol. 2020, 48, 101417. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Liu, J.; Qian, H.; Zhuang, Q. Cancer-associated fibroblasts: From basic science to anticancer therapy. Exp. Mol. Med. 2023, 55, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, D.P.; Quesnel, A.; Duran, C.L.; Filippou, P.S.; Karagiannis, G.S. An emerging paradigm of CXCL12 involvement in the metastatic cascade. Cytokine Growth Factor. Rev. 2024, 75, 12–30. [Google Scholar] [CrossRef] [PubMed]
- Mezzapelle, R.; Leo, M.; Caprioglio, F.; Colley, L.S.; Lamarca, A.; Sabatino, L.; Colantuoni, V.; Crippa, M.P.; Bianchi, M.E. CXCR4/CXCL12 Activities in the Tumor Microenvironment and Implications for Tumor Immunotherapy. Cancers 2022, 14, 2314. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Wang, Z.; Liu, B.; Lu, X.; Xiong, Y.; Shi, J.; Li, P.; Chen, J.; Zhang, Z.; Chen, M.; et al. A feed-forward loop between nuclear translocation of CXCR4 and HIF-1alpha promotes renal cell carcinoma metastasis. Oncogene 2019, 38, 881–895. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Z.; Yang, B.; Yang, Q.; Wang, L.; Sun, Y. CXCR4 nuclear localization follows binding of its ligand SDF-1 and occurs in metastatic but not primary renal cell carcinoma. Oncol. Rep. 2009, 22, 1333–1339. [Google Scholar] [CrossRef] [PubMed]
- Na, I.K.; Scheibenbogen, C.; Adam, C.; Stroux, A.; Ghadjar, P.; Thiel, E.; Keilholz, U.; Coupland, S.E. Nuclear expression of CXCR4 in tumor cells of non-small cell lung cancer is correlated with lymph node metastasis. Hum. Pathol. 2008, 39, 1751–1755. [Google Scholar] [CrossRef] [PubMed]
- Shim, B.; Jin, M.S.; Moon, J.H.; Park, I.A.; Ryu, H.S. High Cytoplasmic CXCR4 Expression Predicts Prolonged Survival in Triple-Negative Breast Cancer Patients Treated with Adjuvant Chemotherapy. J. Pathol. Transl. Med. 2018, 52, 369–377. [Google Scholar] [CrossRef]
- Nikkhoo, B.; Jalili, A.; Fakhari, S.; Sheikhesmaili, F.; Fathi, F.; Rooshani, D.; Hoseinpour Feizi, M.A.; Nikzaban, M. Nuclear pattern of CXCR4 expression is associated with a better overall survival in patients with gastric cancer. J. Oncol. 2014, 2014, 808012. [Google Scholar] [CrossRef]
- Wang, B.; Wang, W.; Niu, W.; Liu, E.; Liu, X.; Wang, J.; Peng, C.; Liu, S.; Xu, L.; Wang, L.; et al. SDF-1/CXCR4 axis promotes directional migration of colorectal cancer cells through upregulation of integrin alphavbeta6. Carcinogenesis 2014, 35, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.Y.; Makino, K.; Xia, W.; Matin, A.; Wen, Y.; Kwong, K.Y.; Bourguignon, L.; Hung, M.C. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 2001, 3, 802–808. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yang, Y.; Liu, S.; Tao, T.; Cai, J.; Wu, J.; Guan, H.; Zhu, X.; He, Z.; Li, J.; et al. EGF-induced nuclear localization of SHCBP1 activates beta-catenin signaling and promotes cancer progression. Oncogene 2019, 38, 747–764. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.J.; Zhao, J.; Sun, Y.; Hu, W.; Wu, Y.L.; Cen, B.; Wu, G.X.; Pei, G. beta-arrestin differentially regulates the chemokine receptor CXCR4-mediated signaling and receptor internalization, and this implicates multiple interaction sites between beta-arrestin and CXCR4. J. Biol. Chem. 2000, 275, 2479–2485. [Google Scholar] [CrossRef]
- Holland, J.D.; Kochetkova, M.; Akekawatchai, C.; Dottore, M.; Lopez, A.; McColl, S.R. Differential functional activation of chemokine receptor CXCR4 is mediated by G proteins in breast cancer cells. Cancer Res. 2006, 66, 4117–4124. [Google Scholar] [CrossRef] [PubMed]
- Groth, C.; Hu, X.; Weber, R.; Fleming, V.; Altevogt, P.; Utikal, J.; Umansky, V. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br. J. Cancer 2019, 120, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Feng, W.; Wang, H.; Wang, L.; Yang, X.; Yang, F.; Zhang, Y.; Liu, X.; Zhang, D.; Ren, Q.; et al. Blocking migration of regulatory T cells to leukemic hematopoietic microenvironment delays disease progression in mouse leukemia model. Cancer Lett. 2020, 469, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Li, J.; Zhang, J.; Wang, L.; Zhang, Q.; Ge, J.; Guo, Y.; Wang, B.; Huang, Y.; Yang, T.; et al. SDF-1/CXCR4 axis facilitates myeloid-derived suppressor cells accumulation in osteosarcoma microenvironment and blunts the response to anti-PD-1 therapy. Int. Immunopharmacol. 2019, 75, 105818. [Google Scholar] [CrossRef]
- Lanitis, E.; Dangaj, D.; Irving, M.; Coukos, G. Mechanisms regulating T-cell infiltration and activity in solid tumors. Ann. Oncol. 2017, 28, xii18–xii32. [Google Scholar] [CrossRef]
- D’Alterio, C.; Buoncervello, M.; Ierano, C.; Napolitano, M.; Portella, L.; Rea, G.; Barbieri, A.; Luciano, A.; Scognamiglio, G.; Tatangelo, F.; et al. Targeting CXCR4 potentiates anti-PD-1 efficacy modifying the tumor microenvironment and inhibiting neoplastic PD-1. J. Exp. Clin. Cancer Res. 2019, 38, 432. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.-Y.; Yang, S.-H.; Chang, P.-Y.; Chen, S.-F.; Nieh, S.; Huang, W.-Y.; Lin, Y.-C.; Lee, O.K.-S. Cancer-Associated-Fibroblast-Mediated Paracrine and Autocrine SDF-1/CXCR4 Signaling Promotes Stemness and Aggressiveness of Colorectal Cancers. Cells 2024, 13, 1334. https://doi.org/10.3390/cells13161334
Chen C-Y, Yang S-H, Chang P-Y, Chen S-F, Nieh S, Huang W-Y, Lin Y-C, Lee OK-S. Cancer-Associated-Fibroblast-Mediated Paracrine and Autocrine SDF-1/CXCR4 Signaling Promotes Stemness and Aggressiveness of Colorectal Cancers. Cells. 2024; 13(16):1334. https://doi.org/10.3390/cells13161334
Chicago/Turabian StyleChen, Chao-Yang, Shih-Hsien Yang, Ping-Ying Chang, Su-Feng Chen, Shin Nieh, Wen-Yen Huang, Yu-Chun Lin, and Oscar Kuang-Sheng Lee. 2024. "Cancer-Associated-Fibroblast-Mediated Paracrine and Autocrine SDF-1/CXCR4 Signaling Promotes Stemness and Aggressiveness of Colorectal Cancers" Cells 13, no. 16: 1334. https://doi.org/10.3390/cells13161334
APA StyleChen, C.-Y., Yang, S.-H., Chang, P.-Y., Chen, S.-F., Nieh, S., Huang, W.-Y., Lin, Y.-C., & Lee, O. K.-S. (2024). Cancer-Associated-Fibroblast-Mediated Paracrine and Autocrine SDF-1/CXCR4 Signaling Promotes Stemness and Aggressiveness of Colorectal Cancers. Cells, 13(16), 1334. https://doi.org/10.3390/cells13161334





