Somatic MED12 Mutations in Myometrial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Case Selection
2.2. Histologic Evaluation Criteria
2.3. DNA Extraction and Sanger Sequencing
DNA Library Preparation and Duplex Deep Sequencing
2.4. Statistical Analysis
3. Results
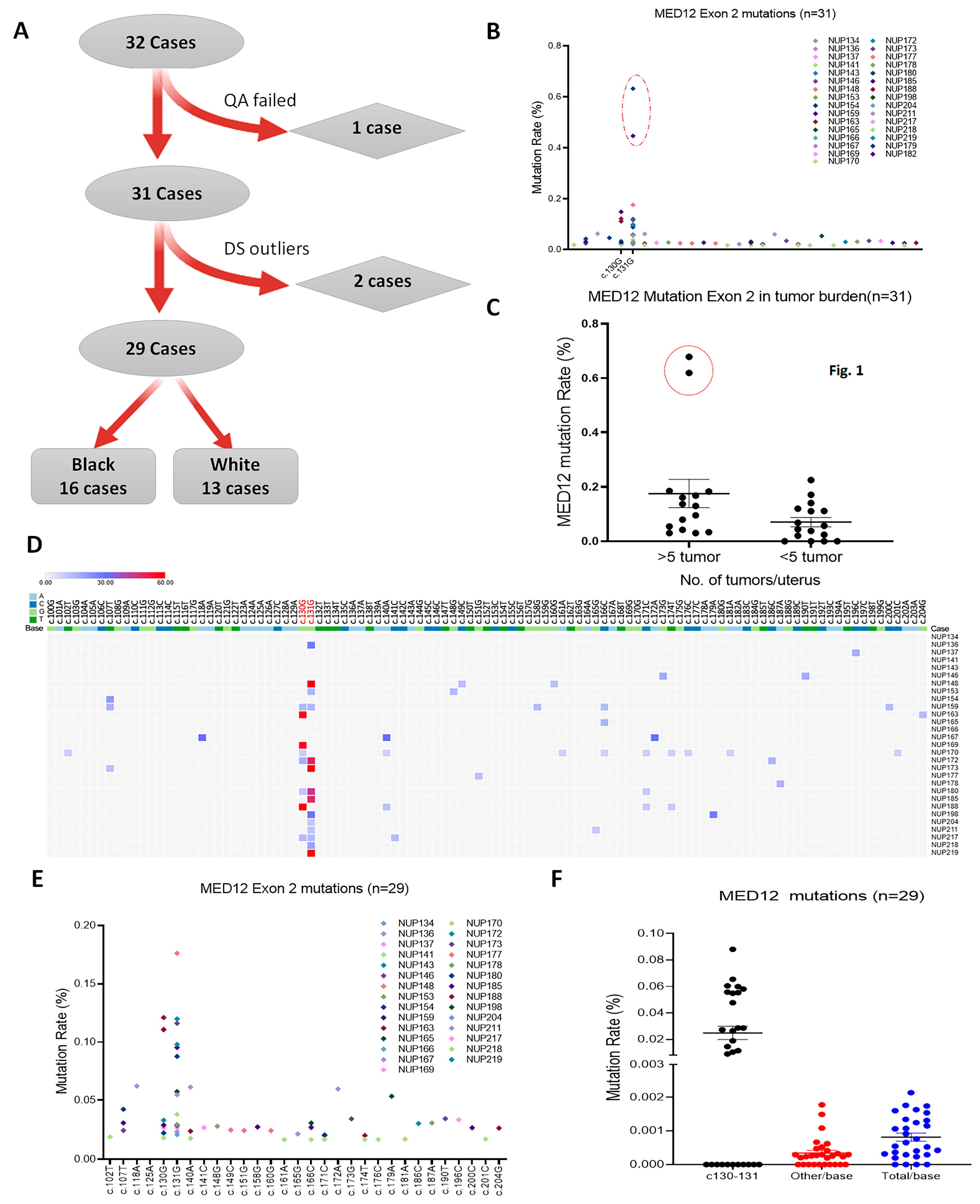
3.1. Case Selection and Sanger Sequencing Analysis of MED12 in LM
3.2. Duplex Deep Sequencing of Myometrial Tissues
3.3. MED12 Mutation Analysis in Tumor-Free Myometrium
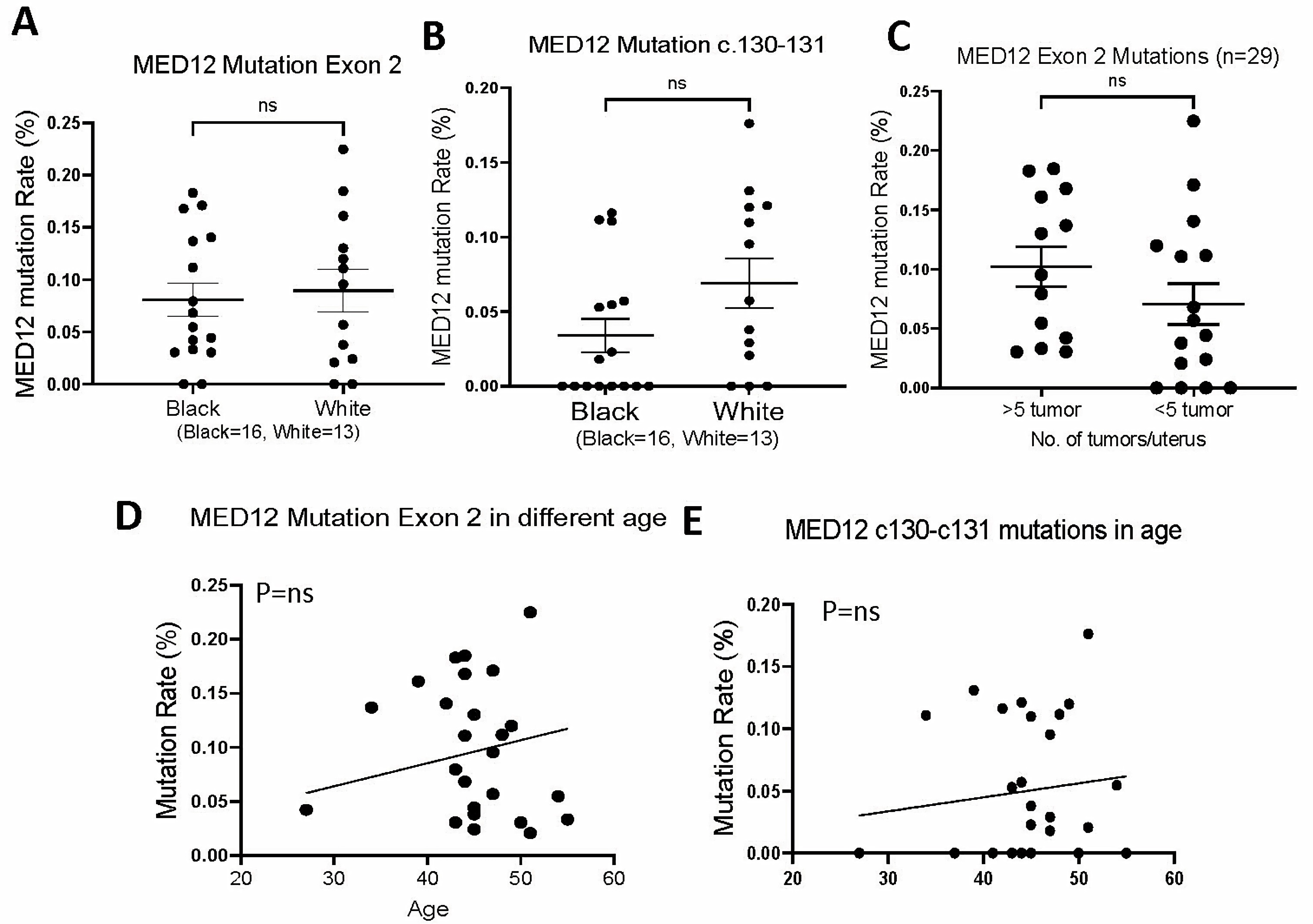
3.4. MED12 Mutations in Association with Pathological Parameters
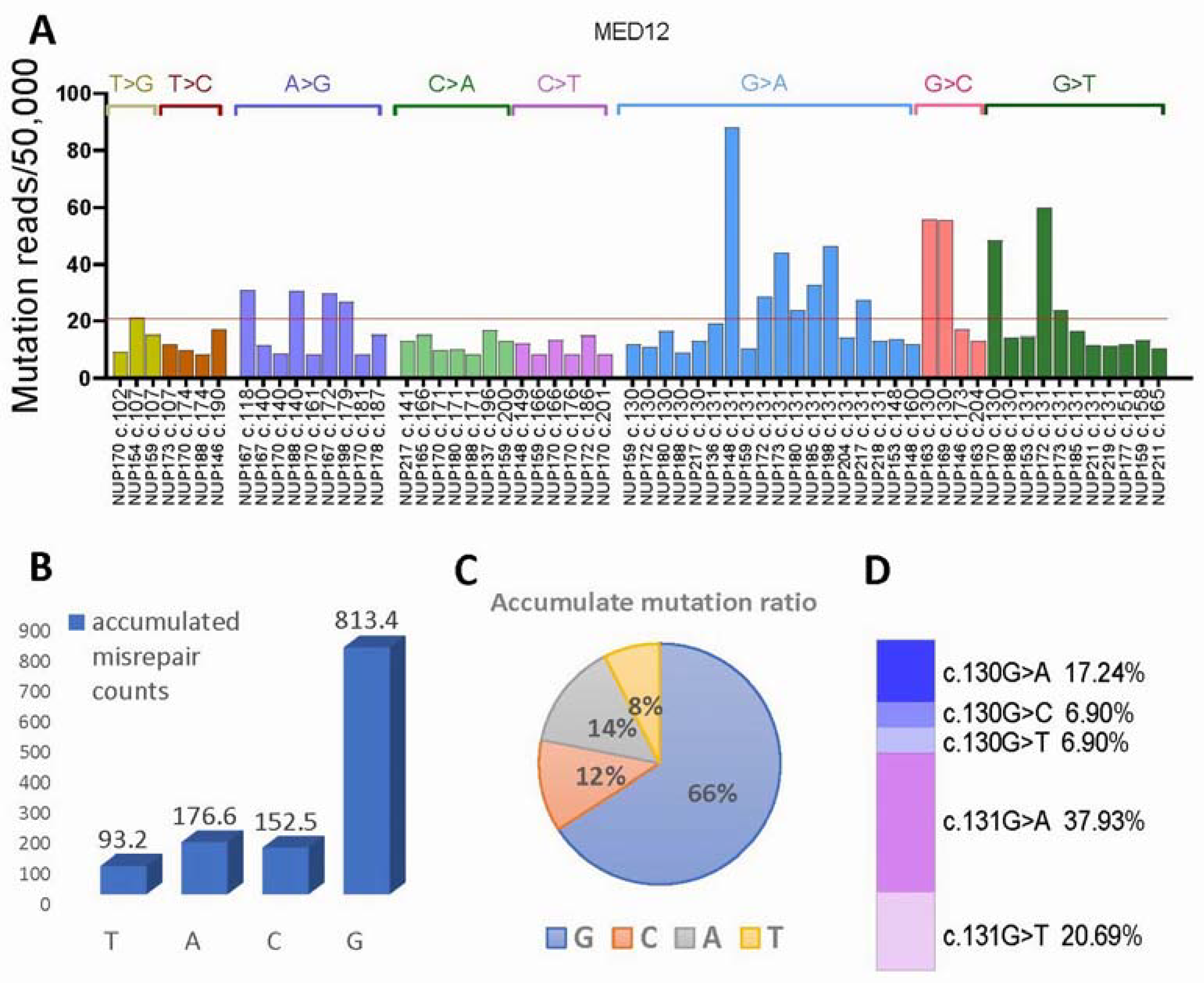
3.5. MED12 Mutational Signature Analysis
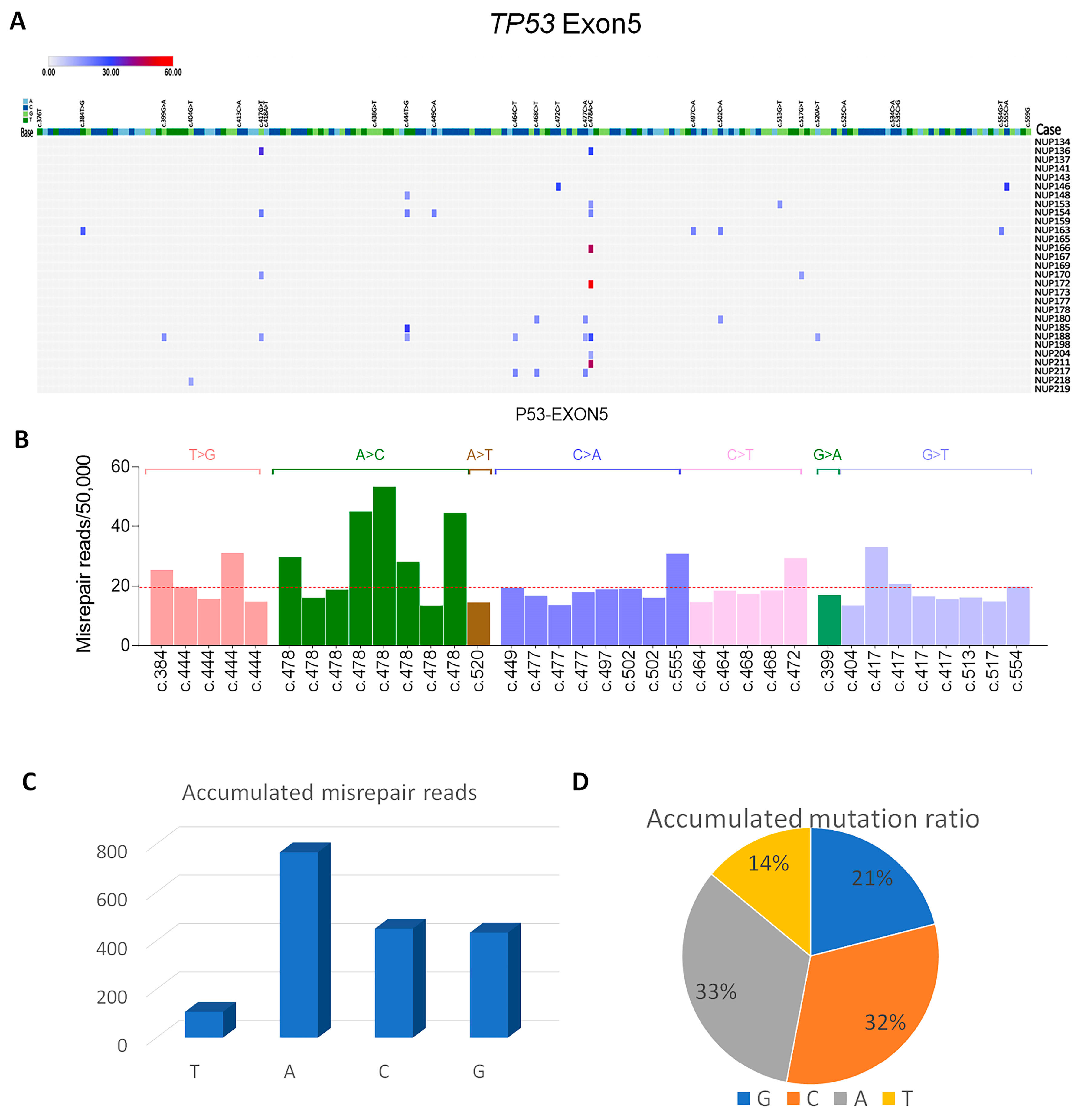
3.6. P53 Mutations in Exon 5 Detected by DDS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bulun, S.E. Uterine fibroids. N. Engl. J. Med. 2013, 369, 1344–1355. [Google Scholar] [CrossRef]
- Catherino, W.H.; Parrott, E.; Segars, J. Proceedings from theNational Institute of Child Health and Human Development conference on the Uterine Fibroid Research Update Workshop. Fertil. Steril. 2011, 95, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Makinen, N.; Mehine, M.; Tolvanen, J.; Kaasinen, E.; Li, Y.; Lehtonen, H.J.; Gentile, M.; Yan, J.; Enge, M.; Taipale, M.; et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 2011, 334, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Bertsch, E.; Qiang, W.; Zhang, Q.; Espona-Fiedler, M.; Druschitz, S.; Liu, Y.; Mittal, K.; Kong, B.; Kurita, T.; Wei, J.J. MED12 and HMGA2 mutations: Two independent genetic events in uterine leiomyoma and leiomyosarcoma. Mod. Pathol. 2014, 27, 1144–1153. [Google Scholar] [CrossRef]
- Heinonen, H.R.; Pasanen, A.; Heikinheimo, O.; Tanskanen, T.; Palin, K.; Tolvanen, J.; Vahteristo, P.; Sjoberg, J.; Pitkanen, E.; Butzow, R.; et al. Multiple clinical characteristics separate MED12-mutation-positive and -negative uterine leiomyomas. Sci. Rep. 2017, 7, 1015. [Google Scholar] [CrossRef] [PubMed]
- Bullerdiek, J.; Rommel, B. Factors targeting MED12 to drive tumorigenesis? F1000Research 2018, 7, 359. [Google Scholar] [CrossRef]
- Lim, W.K.; Ong, C.K.; Tan, J.; Thike, A.A.; Ng, C.C.; Rajasegaran, V.; Myint, S.S.; Nagarajan, S.; Nasir, N.D.; McPherson, J.R.; et al. Exome sequencing identifies highly recurrent MED12 somatic mutations in breast fibroadenoma. Nat. Genet. 2014, 46, 877–880. [Google Scholar] [CrossRef]
- Li, Y.; Xu, X.; Asif, H.; Feng, Y.; Kohrn, B.F.; Kennedy, S.R.; Kim, J.J.; Wei, J.J. Myometrial oxidative stress drives MED12 mutations in leiomyoma. Cell Biosci. 2022, 12, 111. [Google Scholar] [CrossRef]
- Fletcher, N.M.; Abusamaan, M.S.; Memaj, I.; Saed, M.G.; Al-Hendy, A.; Diamond, M.P.; Saed, G.M. Oxidative stress: A key regulator of leiomyoma cell survival. Fertil. Steril. 2017, 107, 1387–1394.e1. [Google Scholar] [CrossRef]
- Foksinski, M.; Kotzbach, R.; Szymanski, W.; Olinski, R. The level of typical biomarker of oxidative stress 8-hydroxy-2′-deoxyguanosine is higher in uterine myomas than in control tissues and correlates with the size of the tumor. Free Radic. Biol. Med. 2000, 29, 597–601. [Google Scholar] [CrossRef]
- Torres, M.J.; Kew, K.A.; Ryan, T.E.; Pennington, E.R.; Lin, C.T.; Buddo, K.A.; Fix, A.M.; Smith, C.A.; Gilliam, L.A.; Karvinen, S.; et al. 17β-Estradiol Directly Lowers Mitochondrial Membrane Microviscosity and Improves Bioenergetic Function in Skeletal Muscle. Cell Metab. 2018, 27, 167–179 e167. [Google Scholar] [CrossRef] [PubMed]
- Vidimar, V.; Gius, D.; Chakravarti, D.; Bulun, S.E.; Wei, J.J.; Kim, J.J. Dysfunctional MnSOD leads to redox dysregulation and activation of prosurvival AKT signaling in uterine leiomyomas. Sci. Adv. 2016, 2, e1601132. [Google Scholar] [CrossRef] [PubMed]
- Ziech, D.; Franco, R.; Pappa, A.; Panayiotidis, M.I. Reactive oxygen species (ROS)--induced genetic and epigenetic alterations in human carcinogenesis. Mutat. Res. 2011, 711, 167–173. [Google Scholar] [CrossRef]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef]
- Poetsch, A.R. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput. Struct. Biotechnol. J. 2020, 18, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, M.; Tsuzuki, T. Oxidative nucleotide damage: Consequences and prevention. Oncogene 2002, 21, 8895–8904. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.C.; Cahill, D.S.; Kasai, H.; Nishimura, S.; Loeb, L.A. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G-T and A-C substitutions. J. Biol. Chem. 1992, 267, 166–172. [Google Scholar] [CrossRef]
- Kucab, J.E.; Zou, X.; Morganella, S.; Joel, M.; Nanda, A.S.; Nagy, E.; Gomez, C.; Degasperi, A.; Harris, R.; Jackson, S.P.; et al. A Compendium of Mutational Signatures of Environmental Agents. Cell 2019, 177, 821–836 e816. [Google Scholar] [CrossRef]
- Chin, E.L.; da Silva, C.; Hegde, M. Assessment of clinical analytical sensitivity and specificity of next-generation sequencing for detection of simple and complex mutations. BMC Genet. 2013, 14, 6. [Google Scholar] [CrossRef]
- Goldman, D.; Domschke, K. Making sense of deep sequencing. Int. J. Neuropsychopharmacol. 2014, 17, 1717–1725. [Google Scholar] [CrossRef]
- Peng, Q.; Xu, C.; Kim, D.; Lewis, M.; DiCarlo, J.; Wang, Y. Targeted Single Primer Enrichment Sequencing with Single End Duplex-UMI. Sci. Rep. 2019, 9, 4810. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.W.; Kennedy, S.R.; Salk, J.J.; Fox, E.J.; Hiatt, J.B.; Loeb, L.A. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 14508–14513. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, S.R.; Schmitt, M.W.; Fox, E.J.; Kohrn, B.F.; Salk, J.J.; Ahn, E.H.; Prindle, M.J.; Kuong, K.J.; Shen, J.C.; Risques, R.A.; et al. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat. Protoc. 2014, 9, 2586–2606. [Google Scholar] [CrossRef]
- Dilliott, A.A.; Farhan, S.M.K.; Ghani, M.; Sato, C.; Liang, E.; Zhang, M.; McIntyre, A.D.; Cao, H.; Racacho, L.; Robinson, J.F.; et al. Targeted Next-generation Sequencing and Bioinformatics Pipeline to Evaluate Genetic Determinants of Constitutional Disease. J. Vis. Exp. 2018, 134, e57266. [Google Scholar] [CrossRef]
- Salk, J.J.; Loubet-Senear, K.; Maritschnegg, E.; Valentine, C.C.; Williams, L.N.; Higgins, J.E.; Horvat, R.; Vanderstichele, A.; Nachmanson, D.; Baker, K.T.; et al. Ultra-Sensitive TP53 Sequencing for Cancer Detection Reveals Progressive Clonal Selection in Normal Tissue over a Century of Human Lifespan. Cell Rep. 2019, 28, 132–144 e133. [Google Scholar] [CrossRef]
- Cole, A.J.; Dwight, T.; Gill, A.J.; Dickson, K.A.; Zhu, Y.; Clarkson, A.; Gard, G.B.; Maidens, J.; Valmadre, S.; Clifton-Bligh, R.; et al. Assessing mutant p53 in primary high-grade serous ovarian cancer using immunohistochemistry and massively parallel sequencing. Sci. Rep. 2016, 6, 26191. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015, 27, 15–26. [Google Scholar] [CrossRef]
- Goad, J.; Rudolph, J.; Zandigohar, M.; Tae, M.; Dai, Y.; Wei, J.J.; Bulun, S.E.; Chakravarti, D.; Rajkovic, A. Single-cell sequencing reveals novel cellular heterogeneity in uterine leiomyomas. Hum. Reprod. 2022, 37, 2334–2349. [Google Scholar] [CrossRef]
- Shynlova, O.; Oldenhof, A.; Dorogin, A.; Xu, Q.; Mu, J.; Nashman, N.; Lye, S.J. Myometrial apoptosis: Activation of the caspase cascade in the pregnant rat myometrium at midgestation. Biol. Reprod. 2006, 74, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.; Hockel, M.; Wree, A.; Leo, C.; Horn, L.C.; Vaupel, P. Lack of hypoxic response in uterine leiomyomas despite severe tissue hypoxia. Cancer Res. 2008, 68, 4719–4726. [Google Scholar] [CrossRef]
- Wu, B.; Chen, X.; He, B.; Liu, S.; Li, Y.; Wang, Q.; Gao, H.; Wang, S.; Liu, J.; Zhang, S.; et al. ROS are critical for endometrial breakdown via NF-kappaB-COX-2 signaling in a female mouse menstrual-like model. Endocrinology 2014, 155, 3638–3648. [Google Scholar] [CrossRef]
- Li, Y.; McNally, R.P.; Feng, Y.; Kim, J.J.; Wei, J.-J. Racial differences in transcriptomics and reactive oxygen species burden in myometrium and leiomyoma. Hum. Reprod. 2023, 38, 609–620. [Google Scholar] [CrossRef]
- Silbergeld, E.K.; Patrick, T.E. Environmental exposures, toxicologic mechanisms, and adverse pregnancy outcomes. Am. J. Obstet. Gynecol. 2005, 192, S11–S21. [Google Scholar] [CrossRef] [PubMed]
- Mas, A.; Prusinski, L.; Yang, Q.; Diaz-Gimeno, P.; Stone, L.; Diamond, M.P.; Simon, C.; Al-Hendy, A. Role of Stro1+/CD44+ stem cells in myometrial physiology and uterine remodeling during pregnancy. Biol. Reprod. 2017, 96, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.R.; Mohammed, S.A. Discrimination and racial disparities in health: Evidence and needed research. J. Behav. Med. 2009, 32, 20–47. [Google Scholar] [CrossRef]
- Wise, L.A.; Palmer, J.R.; Cozier, Y.C.; Hunt, M.O.; Stewart, E.A.; Rosenberg, L. Perceived racial discrimination and risk of uterine leiomyomata. Epidemiology 2007, 18, 747–757. [Google Scholar] [CrossRef]
- Geronimus, A.T.; Hicken, M.; Keene, D.; Bound, J. “Weathering” and age patterns of allostatic load scores among blacks and whites in the United States. Am. J. Public Health 2006, 96, 826–833. [Google Scholar] [CrossRef]
- Van Dyke, M.E.; Baumhofer, N.K.; Slopen, N.; Mujahid, M.S.; Clark, C.R.; Williams, D.R.; Lewis, T.T. Pervasive Discrimination and Allostatic Load in African American and White Adults. Psychosom. Med. 2020, 82, 316–323. [Google Scholar] [CrossRef] [PubMed]
| Black | White | Total | |
|---|---|---|---|
| No. of Cases | 16 | 13 | 29 |
| Age | 43.12 ± 1.79 | 45.54 ± 0.97 | 44.31 ± 1.08 |
| >5 | 56.25% (9/16) | 30.77% (4/13) | 44.83% (13/29) |
| <5 | 43.75% (7/16) | 69.23% (9/13) | 55.17% (16/29) |
| MED12 Mut in LM | 87.50% (14/16) | 69.23% (9/13) | 79.31% (23/29) |
| MED12 Mut in MM | 50.00% (8/16) | 76.92% (10/13) | 62.07% (18/29) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Asif, H.; Feng, Y.; Kim, J.J.; Wei, J.-J. Somatic MED12 Mutations in Myometrial Cells. Cells 2024, 13, 1432. https://doi.org/10.3390/cells13171432
Li Y, Asif H, Feng Y, Kim JJ, Wei J-J. Somatic MED12 Mutations in Myometrial Cells. Cells. 2024; 13(17):1432. https://doi.org/10.3390/cells13171432
Chicago/Turabian StyleLi, Yinuo, Huma Asif, Yue Feng, Julie J. Kim, and Jian-Jun Wei. 2024. "Somatic MED12 Mutations in Myometrial Cells" Cells 13, no. 17: 1432. https://doi.org/10.3390/cells13171432






