Curcumin Inhibits TORC1 and Prolongs the Lifespan of Cells with Mitochondrial Dysfunction
Abstract
1. Introduction
2. Results
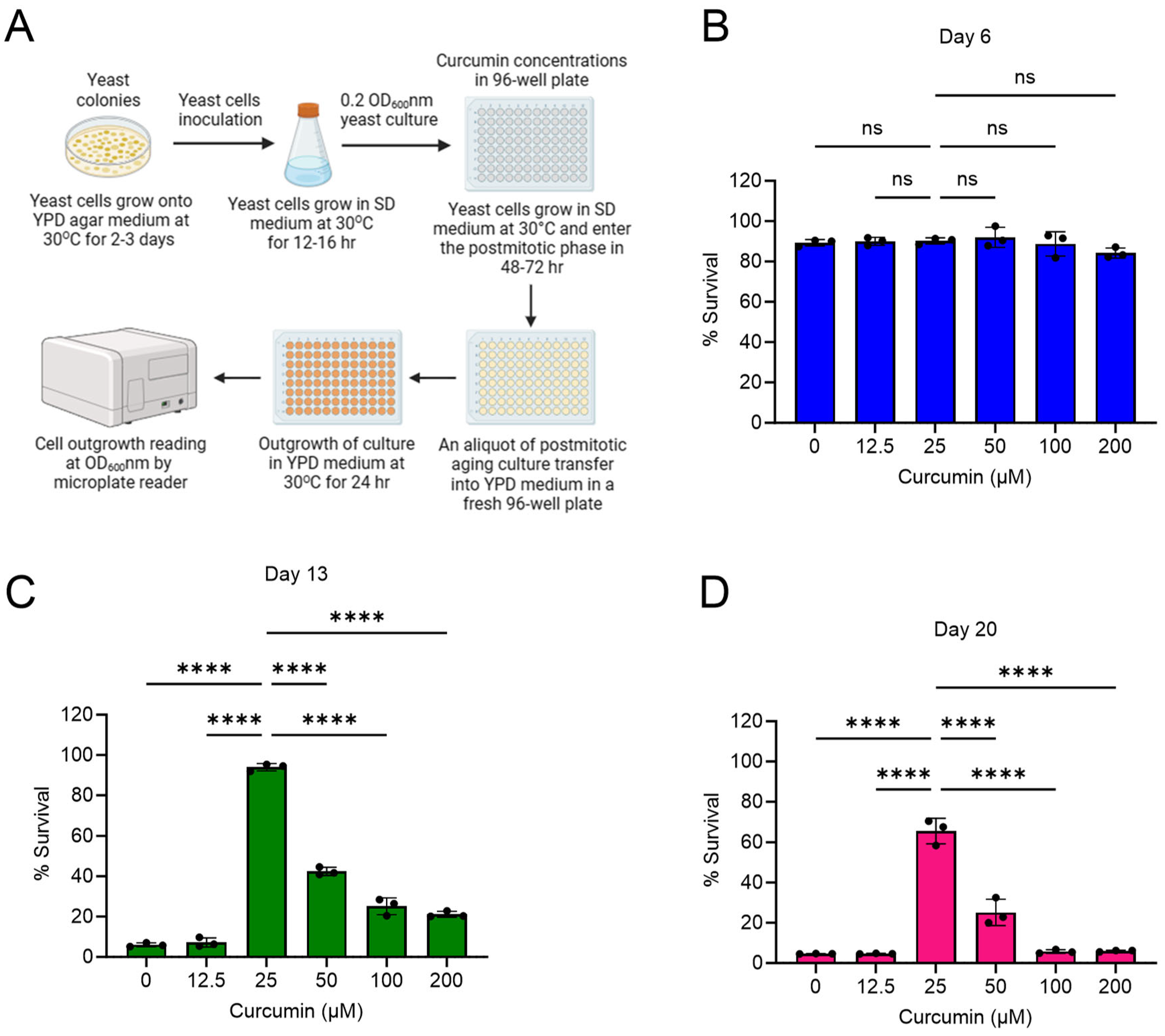
2.1. Curcumin Extends Lifespan of Postmitotic Cells during Chronological Aging
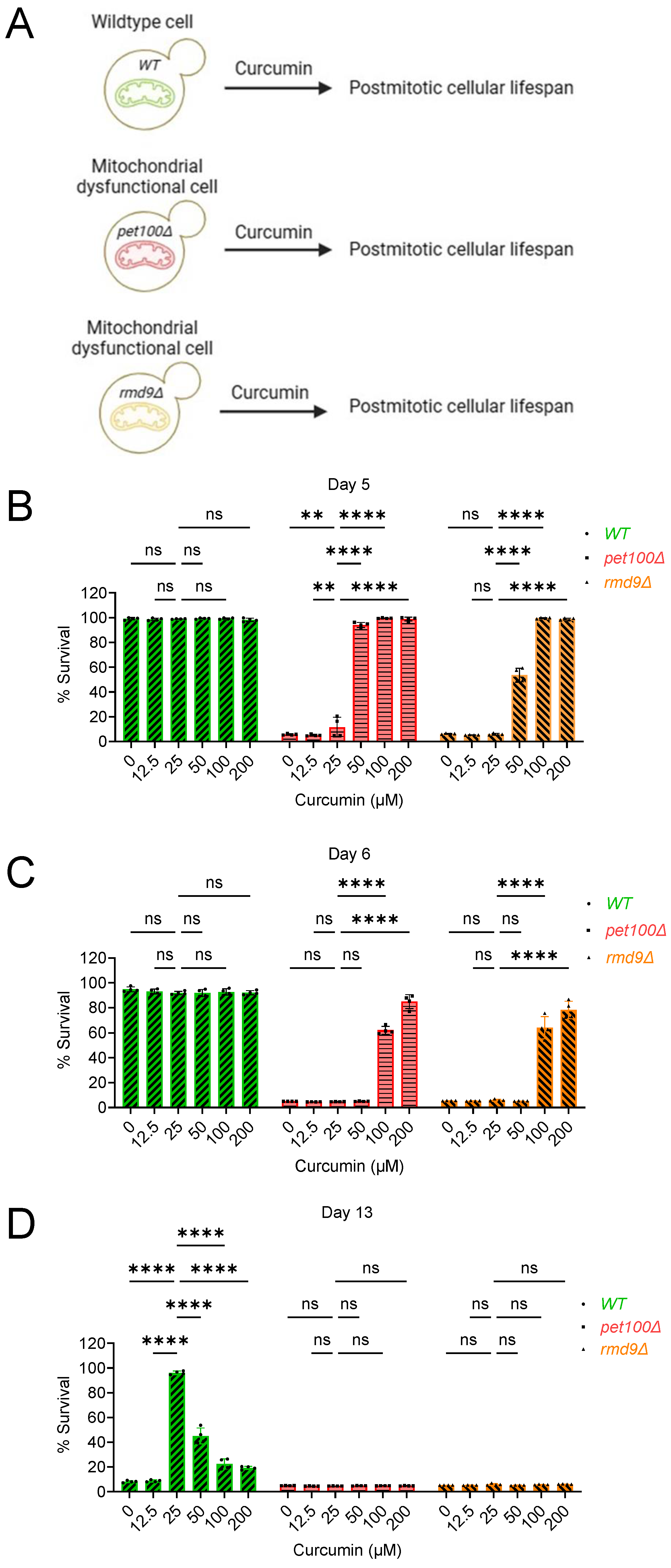
2.2. Curcumin Increases Postmitotic Cellular Lifespan of Mitochondrial Dysfunctional Cells during Chronological Aging in Yeast
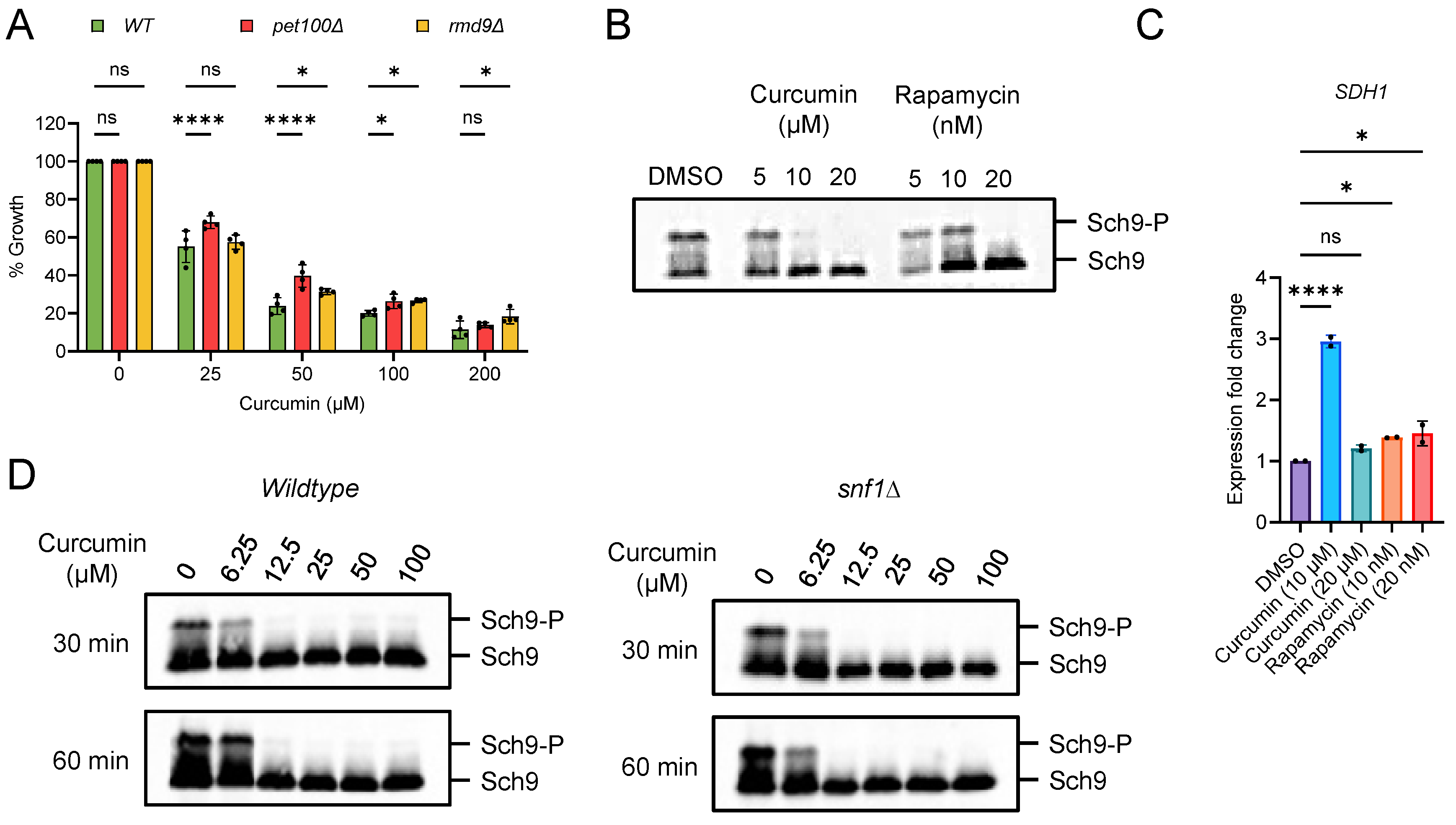
2.3. Curcumin Inhibits TORC1 and Prevents Accelerated Aging in Postmitotic Cells with Mitochondrial Dysfunction
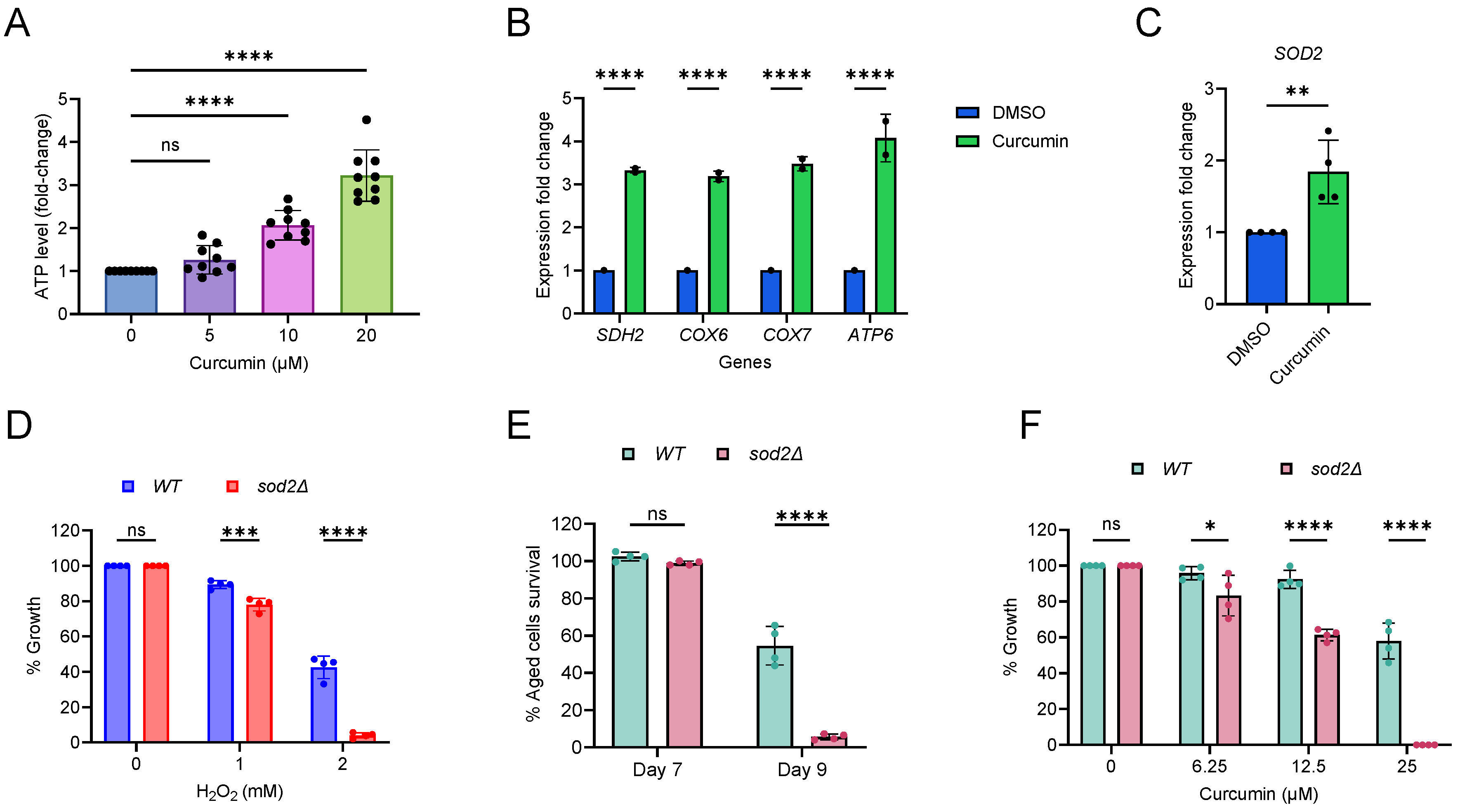
2.4. Curcumin Increases Mitochondrial Function and Oxidative Stress
3. Discussion
4. Methods
4.1. Yeast Strains and Culture Conditions
4.2. Chemical Treatments of Cell Cultures
4.3. Analysis of Postmitotic Cellular Lifespan during Chronological Aging in Yeast
4.4. Measurement of ATP Level
4.5. RNA Extraction and qRT-PCR
4.6. Oxidative Stress Growth Sensitivity Assay
4.7. Analysis of TORC1 Activity
4.8. Data Quantification and Statistical Interpretation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kennedy, B.K.; Berger, S.L.; Brunet, A.; Campisi, J.; Cuervo, A.M.; Epel, E.S.; Franceschi, C.; Lithgow, G.J.; Morimoto, R.I.; Pessin, J.E.; et al. Geroscience: Linking aging to chronic disease. Cell 2014, 159, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Tchkonia, T.; Kirkland, J.L. Aging, Cell Senescence, and Chronic Disease: Emerging Therapeutic Strategies. JAMA—J. Am. Med. Assoc. 2018, 320, 1319–1320. [Google Scholar] [CrossRef]
- Howlett, S.E.; Rutenberg, A.D.; Rockwood, K. The degree of frailty as a translational measure of health in aging. Nat. Aging 2021, 1, 651–665. [Google Scholar] [CrossRef]
- Litomericky, S. Ageing and Health; WHO: Geneva, Switzerland, 2022; Volume 50, pp. 584–588. Available online: https://www.who.int/news-room/fact-sheets/detail/ageing-and-health (accessed on 6 June 2023).
- Lima, T.; Li, T.Y.; Mottis, A.; Auwerx, J. Pleiotropic effects of mitochondria in aging. Nat. Aging 2022, 2, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Amorim, J.A.; Coppotelli, G.; Rolo, A.P.; Palmeira, C.M.; Ross, J.M.; Sinclair, D.A. Mitochondrial and metabolic dysfunction in ageing and age-related diseases. Nat. Rev. Endocrinol. 2022, 18, 243–258. [Google Scholar] [CrossRef]
- Sun, N.; Youle, R.J.; Finkel, T. The Mitochondrial Basis of Aging. Mol. Cell 2016, 61, 654–666. [Google Scholar] [CrossRef]
- Bratic, A.; Larsson, N.G. The role of mitochondria in aging. J. Clin. Investig. 2013, 123, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Monzel, A.S.; Enríquez, J.A.; Picard, M. Multifaceted mitochondria: Moving mitochondrial science beyond function and dysfunction. Nat. Metab. 2023, 5, 546–562. [Google Scholar] [CrossRef]
- Bonora, M.; Wieckowski, M.R.; Sinclair, D.A.; Kroemer, G.; Pinton, P.; Galluzzi, L. Targeting mitochondria for cardiovascular disorders: Therapeutic potential and obstacles. Nat. Rev. Cardiol. 2019, 16, 33–55. [Google Scholar] [CrossRef]
- Xie, S.; Xu, S.C.; Deng, W.; Tang, Q. Metabolic landscape in cardiac aging: Insights into molecular biology and therapeutic implications. Signal Transduct. Target. Ther. 2023, 8, 114. [Google Scholar] [CrossRef]
- Abdellatif, M.; Rainer, P.P.; Sedej, S.; Kroemer, G. Hallmarks of Cardiovascular Ageing. Nat. Rev. Cardiol. 2023, 20, 754–777. [Google Scholar] [CrossRef] [PubMed]
- Cunnane, S.C.; Trushina, E.; Morland, C.; Prigione, A.; Casadesus, G.; Andrews, Z.B.; Beal, M.F.; Bergersen, L.H.; Brinton, R.D.; de la Monte, S.; et al. Brain energy rescue: An emerging therapeutic concept for neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 2020, 19, 609–633. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Sligar, J.; DeBruin, D.A.; Saner, N.J.; Philp, A.M.; Philp, A. The importance of mitochondrial quality control for maintaining skeletal muscle function across health span. Am. J. Physiol. Cell Physiol. 2022, 322, C461–C467. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.L.; Robinson, M.M.; Nair, S.K. Skeletal muscle aging and the mitochondrion. Trends Endocrinol. Metab. 2013, 24, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.G.; Klein, R.; Cheng, C.Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef] [PubMed]
- Ferrington, D.A.; Ebeling, M.C.; Kapphahn, R.J.; Terluk, M.R.; Fisher, C.R.; Polanco, J.R.; Roehrich, H.; Leary, M.M.; Geng, Z.; Dutton, J.R.; et al. Altered bioenergetics and enhanced resistance to oxidative stress in human retinal pigment epithelial cells from donors with age-related macular degeneration. Redox Biol. 2017, 13, 255–265. [Google Scholar] [CrossRef]
- Feher, J.; Kovacs, I.; Artico, M.; Cavallotti, C.; Papale, A.; Balacco Gabrieli, C. Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiol. Aging 2006, 27, 983–993. [Google Scholar] [CrossRef]
- Partridge, L.; Fuentealba, M.; Kennedy, B.K. The quest to slow ageing through drug discovery. Nat. Rev. Drug Discov. 2020, 19, 513–532. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Lushchak, O.; Strilbytska, O.; Koliada, A.; Zayachkivska, A.; Burdyliuk, N.; Yurkevych, I.; Storey, K.B.; Vaiserman, A. Nanodelivery of phytobioactive compounds for treating aging-associated disorders. GeroScience 2020, 42, 117–139. [Google Scholar] [CrossRef]
- Tyagi, A.K.; Prasad, S.; Yuan, W.; Li, S.; Aggarwal, B.B. Identification of a novel compound (β-sesquiphellandrene) from turmeric (Curcuma longa) with anticancer potential: Comparison with curcumin. Investig. New Drugs 2015, 33, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Tayyem, R.F.; Heath, D.D.; Al-Delaimy, W.K.; Rock, C.L. Curcumin content of turmeric and curry powders. Nutr. Cancer 2006, 55, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Vaiserman, A.; Koliada, A.; Zayachkivska, A.; Lushchak, O. Curcumin: A therapeutic potential in ageing-related disorders. PharmaNutrition 2020, 4, 100226. [Google Scholar] [CrossRef]
- Longo, V.D.; Shadel, G.S.; Kaeberlein, M.; Kennedy, B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012, 16, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, M.; Burtner, C.R.; Kennedy, B.K. Recent developments in yeast aging. PLoS Genet. 2007, 3, e84. [Google Scholar] [CrossRef]
- Zimmermann, A.; Hofer, S.; Pendl, T.; Kainz, K.; Madeo, F.; Carmona-Gutierrez, D. Yeast as a tool to identify anti-aging compounds. FEMS Yeast Res. 2018, 18, foy020. [Google Scholar] [CrossRef]
- Alfatah, M.; Eisenhaber, F. The PICLS high-throughput screening method for agents extending cellular longevity identifies 2,5-anhydro-D-mannitol as novel anti-aging compound. GeroScience 2022, 45, 141–158. [Google Scholar] [CrossRef]
- Sinha, S.; Nge, C.E.; Leong, C.Y.; Ng, V.; Crasta, S.; Alfatah, M.; Goh, F.; Low, K.N.; Zhang, H.; Arumugam, P.; et al. Genomics-driven discovery of a biosynthetic gene cluster required for the synthesis of BII-Rafflesfungin from the fungus Phoma sp. F3723. BMC Genom. 2019, 20, 374. [Google Scholar] [CrossRef]
- Alfatah, M.; Wong, J.H.; Nge, C.E.; Kong, K.W.; Low, K.N.; Leong, C.Y.; Crasta, S.; Munusamy, M.; Chang, A.M.L.; Hoon, S.; et al. Hypoculoside, a sphingoid base-like compound from Acremonium disrupts the membrane integrity of yeast cells. Sci. Rep. 2019, 9, 710. [Google Scholar] [CrossRef] [PubMed]
- Soh, J.W.; Marowsky, N.; Nichols, T.J.; Rahman, A.M.; Miah, T.; Sarao, P.; Khasawneh, R.; Unnikrishnan, A.; Heydari, A.R.; Silver, R.B.; et al. Curcumin is an early-acting stage-specific inducer of extended functional longevity in Drosophila. Exp. Gerontol. 2013, 48, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Liao, V.H.C.; Yu, C.W.; Chu, Y.J.; Li, W.H.; Hsieh, Y.C.; Wang, T.T. Curcumin-mediated lifespan extension in Caenorhabditis elegans. Mech. Ageing Dev. 2011, 132, 480–487. [Google Scholar] [CrossRef]
- Dehghan, E.; Zhang, Y.; Saremi, B.; Yadavali, S.; Hakimi, A.; Dehghani, M.; Goodarzi, M.; Tu, X.; Robertson, S.; Lin, R.; et al. Hydralazine induces stress resistance and extends C. elegans lifespan by activating the NRF2/SKN-1 signalling pathway. Nat. Commun. 2017, 8, 2223. [Google Scholar] [CrossRef] [PubMed]
- Stępień, K.; Wojdyła, D.; Nowak, K.; Mołoń, M. Impact of curcumin on replicative and chronological aging in the Saccharomyces cerevisiae yeast. Biogerontology 2020, 21, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Alfatah, M.; Jia, J.L.J.; Zhang, Y.; Naaz, A.; Cheng, T.Y.N.; Yogasundaram, S.; Faidzinn, N.A.; Jing, J.L.; Eisenhaber, B.; Eisenhaber, F. Uncharacterized yeast gene YBR238C, an effector of TORC1 signaling in a mitochondrial feedback loop, accelerates cellular aging via HAP4- and RMD9-dependent mechanisms. eLife 2023, 12, RP92178. [Google Scholar] [CrossRef]
- Merz, S.; Westermann, B. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 2009, 10, 95. [Google Scholar] [CrossRef]
- Nouet, C.; Bourens, M.; Hlavacek, O.; Marsy, S.; Lemaire, C.; Ve Dujardin, G. Rmd9p Controls the Processing/Stability of Mitochondrial mRNAs and Its Overexpression Compensates for a Partial Deficiency of Oxa1p in Saccharomyces cerevisiae. Genetics 2007, 175, 1105–1115. [Google Scholar] [CrossRef]
- Hillen, H.S.; Markov, D.A.; Wojtas, I.D.; Hofmann, K.B.; Lidschreiber, M.; Cowan, A.T.; Jones, J.L.; Temiakov, D.; Cramer, P.; Anikin, M. The pentatricopeptide repeat protein Rmd9 recognizes the dodecameric element in the 3′-UTRs of yeast mitochondrial mRNAs. Proc. Natl. Acad. Sci. USA 2021, 118, e200932911. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Hall, M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR signaling in growth, metabolism, and disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Hall, M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017, 36, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Alfatah, M.; Wong, J.H.; Krishnan, V.G.; Lee, Y.C.; Sin, Q.F.; Goh, C.J.H.; Kong, K.W.; Hoon, S.; Arumugam, P.; Lee, W.T.; et al. TORC1 regulates the transcriptional response to glucose and developmental cycle via the Tap42-Sit4-Rrd1/2 pathway in Saccharomyces cerevisiae. BMC Biol. 2021, 19, 859793. [Google Scholar] [CrossRef]
- Alfatah, M.; Cui, L.; Goh, C.J.H.; Cheng, T.Y.N.; Zhang, Y.; Naaz, A.; Wong, J.H.; Lewis, J.; Poh, W.J.; Arumugam, P. Metabolism of glucose activates TORC1 through multiple mechanisms in Saccharomyces cerevisiae. Cell Rep. 2023, 42, 113205. [Google Scholar] [CrossRef]
- González, A.; Hall, M.N.; Lin, S.C.; Hardie, D.G. AMPK and TOR: The Yin and Yang of Cellular Nutrient Sensing and Growth Control. Cell Metab. 2020, 31, 472–492. [Google Scholar] [CrossRef]
- Lin, S.C.; Hardie, D.G. AMPK: Sensing Glucose as well as Cellular Energy Status. Cell Metab. 2018, 27, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK Phosphorylation of Raptor Mediates a Metabolic Checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007, 8, 774–785. [Google Scholar] [CrossRef]
- Hedbacker, K.; Carlson, M. SNF1/AMPK pathways in yeast. Front. Biosci. 2008, 3, 2408–2420. [Google Scholar] [CrossRef]
- Hallett, J.E.H.; Luo, X.; Capaldi, A.P.; Hughes Hallett, J.E.; Luo, X.; Capaldi, A.P. Snf1/AMPK promotes the formation of Kog1/Raptor-bodies to increase the activation threshold of TORC1 in budding yeast. Elife 2015, 4, e09181. [Google Scholar] [CrossRef]
- Jing, J.L.; Ning, T.C.Y.; Natali, F.; Eisenhaber, F.; Alfatah, M. Iron Supplementation Delays Aging and Extends Cellular Lifespan through Potentiation of Mitochondrial Function. Cells 2022, 11, 862. [Google Scholar] [CrossRef]
- Balaban, R.S.; Nemoto, S.; Finkel, T. Mitochondria, oxidants, and aging. Cell 2005, 120, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Handy, D.E.; Loscalzo, J. Redox regulation of mitochondrial function. Antioxid. Redox Signal. 2012, 16, 1323–1367. [Google Scholar] [CrossRef] [PubMed]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef]
- Ng, T.P.; Nyunt, M.S.Z.; Gao, Q.; Gwee, X.; Chua, D.Q.L.; Yap, K.B. Curcumin-Rich Curry Consumption and Neurocognitive Function from 4.5-Year Follow-Up of Community-Dwelling Older Adults (Singapore Longitudinal Ageing Study). Nutrients 2022, 14, 1189. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.P.; Nyunt, S.Z.; Gao, Q.; Gwee, X.; Chua, D.Q.L.; Yap, K.B. Curcumin-rich curry consumption and life expectancy: Singapore longitudinal ageing study. GeroScience 2023, 46, 969–980. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- van Dijken, J.P.; Bauer, J.; Brambilla, L.; Duboc, P.; Francois, J.; Gancedo, C.; Giuseppin, M.; Heijnen, J.; Hoare, M.; Lange, H.; et al. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 2000, 26, 706–714. [Google Scholar] [CrossRef]
- Longtine, M.S.; McKenzie, A.; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Verma, H.K.; Shukla, P.; Alfatah, M.; Khare, A.K.; Upadhyay, U.; Ganesan, K.; Singh, J. High level constitutive expression of luciferase reporter by lsd90 promoter in fission yeast. PLoS ONE 2014, 9, e101201. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naaz, A.; Zhang, Y.; Faidzinn, N.A.; Yogasundaram, S.; Dorajoo, R.; Alfatah, M. Curcumin Inhibits TORC1 and Prolongs the Lifespan of Cells with Mitochondrial Dysfunction. Cells 2024, 13, 1470. https://doi.org/10.3390/cells13171470
Naaz A, Zhang Y, Faidzinn NA, Yogasundaram S, Dorajoo R, Alfatah M. Curcumin Inhibits TORC1 and Prolongs the Lifespan of Cells with Mitochondrial Dysfunction. Cells. 2024; 13(17):1470. https://doi.org/10.3390/cells13171470
Chicago/Turabian StyleNaaz, Arshia, Yizhong Zhang, Nashrul Afiq Faidzinn, Sonia Yogasundaram, Rajkumar Dorajoo, and Mohammad Alfatah. 2024. "Curcumin Inhibits TORC1 and Prolongs the Lifespan of Cells with Mitochondrial Dysfunction" Cells 13, no. 17: 1470. https://doi.org/10.3390/cells13171470
APA StyleNaaz, A., Zhang, Y., Faidzinn, N. A., Yogasundaram, S., Dorajoo, R., & Alfatah, M. (2024). Curcumin Inhibits TORC1 and Prolongs the Lifespan of Cells with Mitochondrial Dysfunction. Cells, 13(17), 1470. https://doi.org/10.3390/cells13171470







