The Tumor Suppressor Par-4 Regulates Adipogenesis by Transcriptional Repression of PPARγ
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Culture
2.3. Constructs and RNA-Interference
2.4. In Vitro Adipogenesis
2.5. Antibodies
2.6. Western Blot Analysis
2.7. Real-Time qPCR
2.8. Oil Red O Staining of Cultured Cells
2.9. Luciferase Reporter Assays
2.10. Promoter Subcloning
2.11. Chromatin Immunoprecipitation (ChIP)
2.12. Statistical Analysis
3. Results
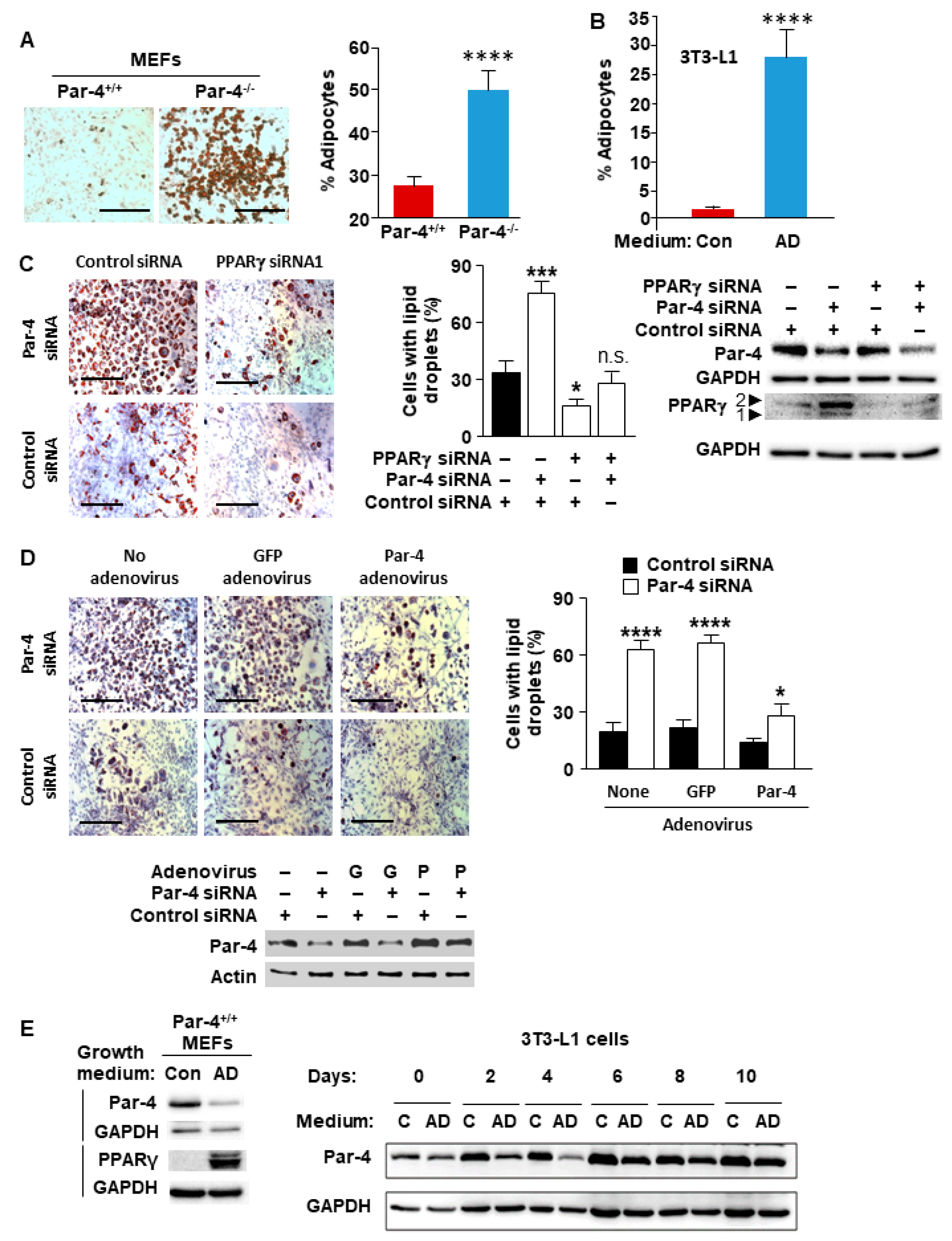
3.1. Par-4 Inhibits Adipogenesis In Vitro
3.2. Par-4 Overexpression Inhibits Adipogenesis
3.3. Adipogenesis Downregulates Par-4 Expression
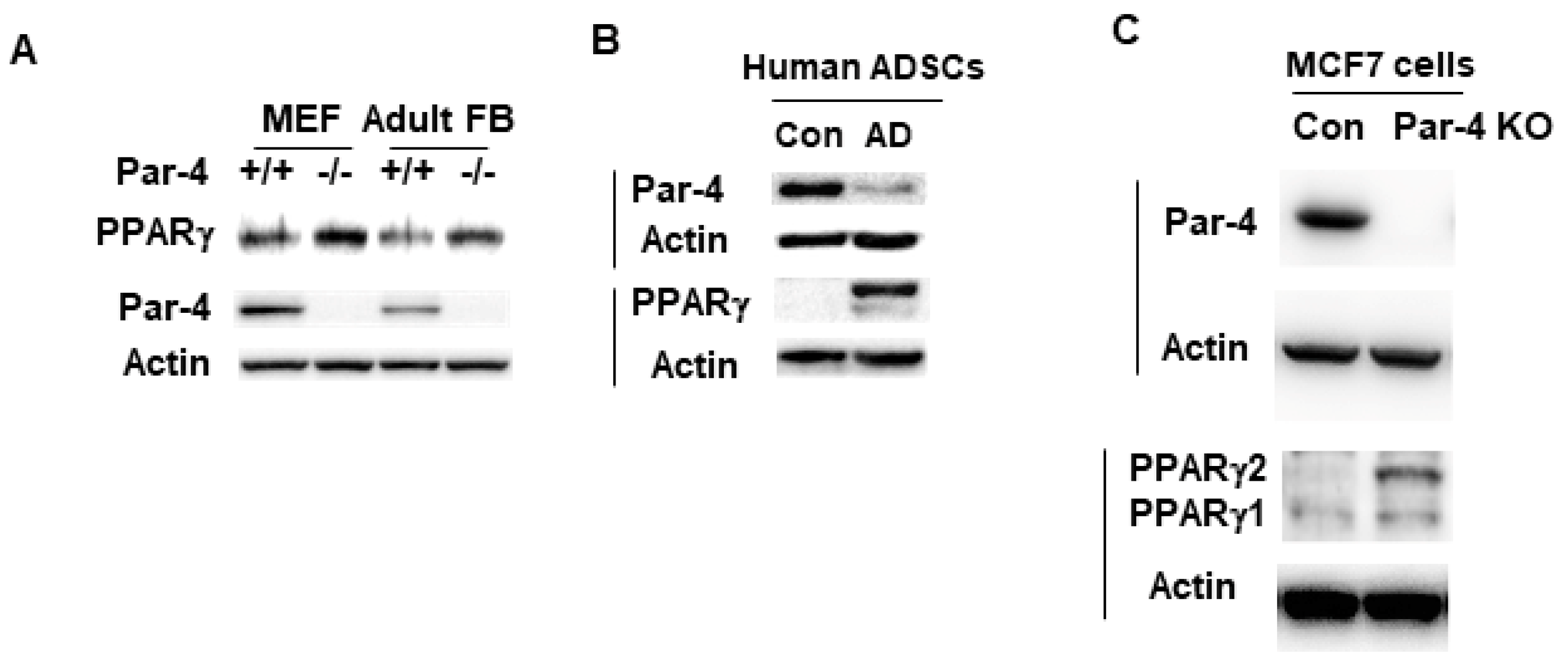
3.4. PPARγ Expression Is Inversely Associated with Par-4 Status
3.5. Par-4 Transcriptionally Inhibits PPARγ
3.6. Par-4 Overexpression Inhibits PPARγ1 and γ2 Isoforms
3.7. Par-4 Inhibits Transcriptional Activity of the PPARγ2 Promoter
3.8. Nuclear Localization of Par-4 Is Required for Regulation of the PPARγ2 Promoter
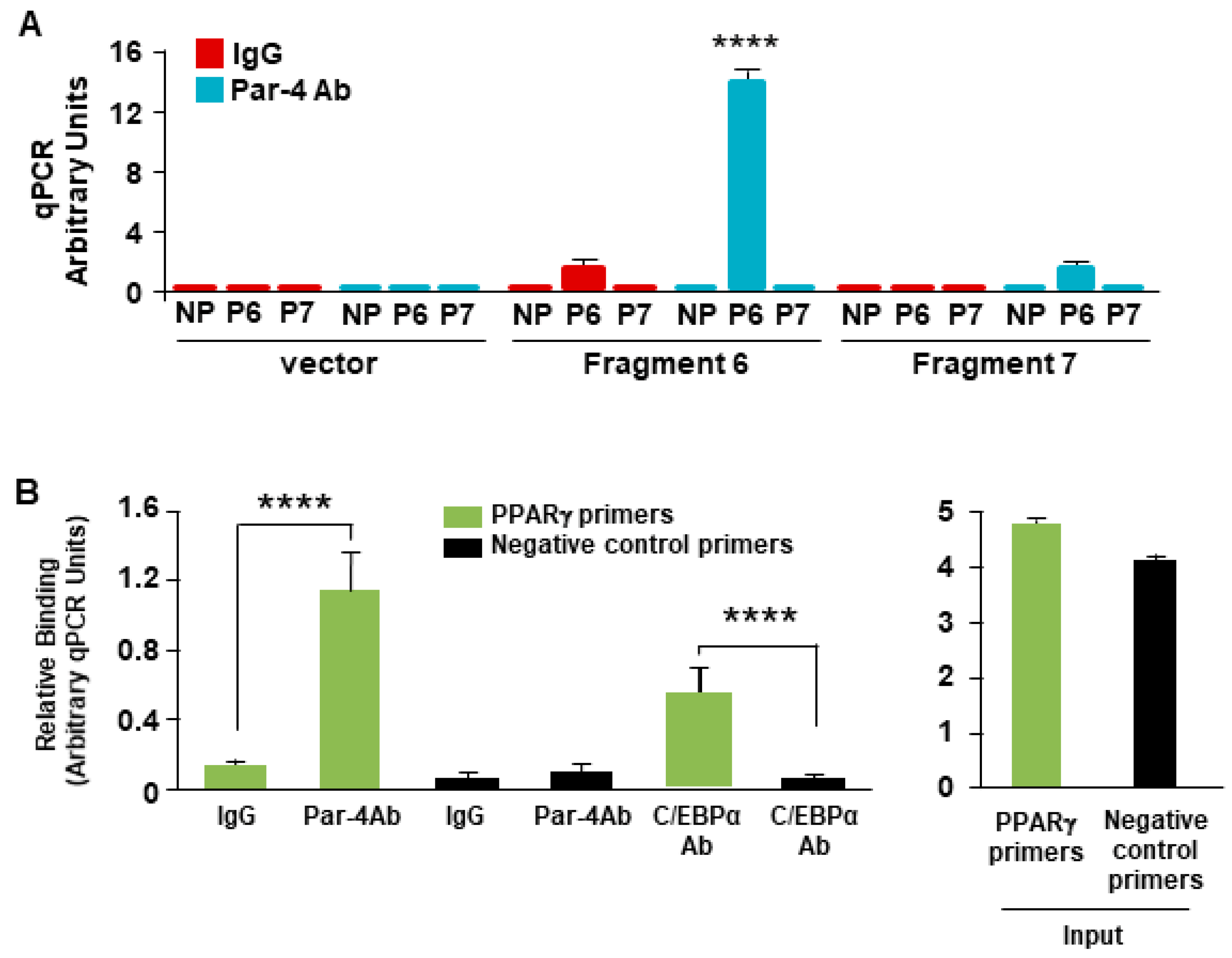
3.9. Par-4 Binds the PPARγ2 Promoter
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- El-Guendy, N.; Zhao, Y.; Gurumurthy, S.; Burikhanov, R.; Rangnekar, V.M. Identification of a unique core domain of par-4 sufficient for selective apoptosis induction in cancer cells. Molec. Cell Biol. 2003, 23, 5516–5525. [Google Scholar] [CrossRef] [PubMed]
- Cheratta, A.R.; Thayyullathil, F.; Pallichankandy, S.; Subburayan, K.; Alakkal, A.; Galadari, S. Prostate apoptosis response-4 and tumor suppression: It’s not just about apoptosis anymore. Cell Death Dis. 2021, 12, 47. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.; Krishnan, S.; Ananth, S.; Sells, S.F.; Shi, Y.; Walther, M.M.; Linehan, W.M.; Sukhatme, V.P.; Weinstein, M.H.; Rangnekar, V.M. Decreased expression of the pro-apoptotic protein Par-4 in renal cell carcinoma. Oncogene 1999, 18, 1205–1208. [Google Scholar] [CrossRef]
- Goswami, A.; Burikhanov, R.; de Thonel, A.; Fujita, N.; Goswami, M.; Zhao, Y.; Eriksson, J.E.; Tsuruo, T.; Rangnekar, V.M. Binding and phosphorylation of par-4 by akt is essential for cancer cell survival. Molec. Cell 2005, 20, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Hebbar, N.; Wang, C.; Rangnekar, V.M. Mechanisms of apoptosis by the tumor suppressor Par-4. J. Cell Physol. 2012, 227, 3715–3721. [Google Scholar] [CrossRef]
- Moreno-Bueno, G.; Fernandez-Marcos, P.J.; Collado, M.; Tendero, M.J.; Rodriguez-Pinilla, S.M.; Garcia-Cao, I.; Hardisson, D.; Diaz-Meco, M.T.; Moscat, J.; Serrano, M.; et al. Inactivation of the candidate tumor suppressor par-4 in endometrial cancer. Cancer Res. 2007, 67, 1927–1934. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Cao, I.; Lafuente, M.J.; Criado, L.M.; Diaz-Meco, M.T.; Serrano, M.; Moscat, J. Genetic inactivation of Par4 results in hyperactivation of NF-kappaB and impairment of JNK and p38. EMBO Rep. 2003, 4, 307–312. [Google Scholar] [CrossRef][Green Version]
- Contract, D.; Mackley, H.; Irby, R.B. Par-4 sensitizes human colon cancer cells to chemoradiotherapy. J. Cancer Ther. 2011, 2, 542–547. [Google Scholar] [CrossRef][Green Version]
- Alvarez, J.V.; Pan, T.-C.; Ruth, J.; Feng, Y.; Zhou, A.; Pant, D.; Grimley, J.S.; Wandless, T.J.; Demichele, A.; Chodosh, L.A. Par-4 downregulation promotes breast cancer recurrence by preventing multinucleation following targeted therapy. Cancer Cell 2013, 24, 30–44. [Google Scholar] [CrossRef]
- Pereira, M.C.; de Bessa-Garcia, S.A.; Burikhanov, R.; Pavanelli, A.C.; Antunes, L.; Rangnekar, V.M.; Nagai, M.A. Prostate apoptosis response-4 is involved in the apoptosis response to docetaxel in MCF-7 breast cancer cells. Int. J. Oncol. 2013, 43, 531–538. [Google Scholar] [CrossRef]
- Zhao, Y.; Burikhanov, R.; Qiu, S.; Lele, S.M.; Jennings, C.D.; Bondada, S.; Spear, B.; Rangnekar, V.M. Cancer resistance in transgenic mice expressing the SAC module of Par-4. Cancer Res. 2007, 67, 9276–9285. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Burikhanov, R.; Brandon, J.; Qiu, S.; Shelton, B.J.; Spear, B.; Bondada, S.; Bryson, S.; Rangnekar, V.M. Systemic Par-4 inhibits non-autochthonous tumor growth. Cancer Biol. Ther. 2011, 12, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Burikhanov, R.; Zhao, Y.; Goswami, A.; Qiu, S.; Schwarze, S.R.; Rangnekar, V.M. The tumor suppressor Par-4 activates an extrinsic pathway for apoptosis. Cell 2009, 138, 377–388. [Google Scholar] [CrossRef]
- Gurumurthy, S.; Goswami, A.; Vasudevan, K.M.; Rangnekar, V.M. Phosphorylation of Par-4 by protein kinase A is critical for apoptosis. Mol. Cell Biol. 2005, 25, 1146–1161. [Google Scholar] [CrossRef]
- Johnstone, R.W.; See, R.H.; Sells, S.F.; Wang, J.; Muthukkumar, S.; Englert, C.; Haber, D.A.; Licht, J.D.; Sugrue, S.P.; Roberts, T.; et al. A novel repressor, par-4, modulates transcription and growth suppression functions of the Wilms’ tumor suppressor WT1. Mol. Cell Biol. 1996, 16, 6945–6956. [Google Scholar] [CrossRef]
- Cheema, S.K.; Mishra, S.K.; Rangnekar, V.M.; Tari, A.M.; Kumar, R.; Lopez-Berestein, G. Par-4 transcriptionally regulates Bcl-2 through a WT1-binding site on the bcl-2 promoter. J. Biol. Chem. 2003, 278, 19995–20005. [Google Scholar] [CrossRef]
- Lu, C.; Li, J.-Y.; Ge, Z.; Zhang, L.; Zhou, G.-P. Par-4/THAP1 complex and Notch3 competitively regulated pre-mRNA splicing of CCAR1 and affected inversely the survival of T-cell acute lymphoblastic leukemia cells. Oncogene 2013, 32, 5602–5613. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, X.; Zhang, J.; Lam, E.K.Y.; Shin, V.Y.; Cheng, A.S.L.; Yu, J.; Chan, F.K.L.; Sung, J.J.Y.; Jin, H.C. Warburg effect revisited: An epigenetic link between glycolysis and gastric carcinogenesis. Oncogene 2010, 29, 442–450. [Google Scholar] [CrossRef]
- Faubert, B.; Boily, G.; Izreig, S.; Griss, T.; Samborska, B.; Dong, Z.; Dupuy, F.; Chambers, C.; Fuerth, B.J.; Viollet, B.; et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013, 17, 113–124. [Google Scholar] [CrossRef]
- Liang, Y.; Liu, J.; Feng, Z. The regulation of cellular metabolism by tumor suppressor p53. Cell Biosci. 2013, 3, 9. [Google Scholar] [CrossRef]
- Araujo, N.; Sledziona, J.; Noothi, S.K.; Burikhanov, R.; Hebbar, N.; Ganguly, S.; Shrestha-Bhattarai, T.; Zhu, B.; Katz, W.S.; Zhang, Y.; et al. Tumor suppressor Par-4 regulates complement factor C3 and obesity. Front. Oncol. 2022, 12, 860446. [Google Scholar] [CrossRef] [PubMed]
- Yahagi, N.; Shimano, H.; Matsuzaka, T.; Najima, Y.; Sekiya, M.; Nakagawa, Y.; Ide, T.; Tomita, S.; Okazaki, H.; Tamura, Y.; et al. p53 Activation in adipocytes of obese mice. J. Biol. Chem. 2003, 278, 25395–25400. [Google Scholar] [CrossRef] [PubMed]
- Molchadsky, A.; Ezra, O.; Amendola, P.G.; Krantz, D.; Kogan-Sakin, I.; Buganim, Y.; Rivlin, N.; Goldfinger, N.; Folgiero, V.; Falcioni, R.; et al. p53 is required for brown adipogenic differentiation and has a protective role against diet-induced obesity. Cell Death Differ. 2013, 20, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, X.; Gao, X.; Mei, Y.; Wu, M. A new role of p53 in regulating lipid metabolism. J. Mol. Cell Biol. 2013, 5, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Hegele, R.A. Lessons from human mutations in PPARγ. Int. J. Obes. 2005, 29, S31–S35. [Google Scholar] [CrossRef][Green Version]
- Zhu, Y.; Qi, C.; Korenbergt, J.R.; Chent, X.-N.; Noyat, D.; Sambasiva Rao, M.; Reddyt, J.K.; Tolbert, N.E. Structural organization of mouse peroxisome proliferator- activated receptor y (mPPARy) gene: Alternative promoter use and different splicing yield two mPPARy isoforms (peroxisome proliferation/nuclear receptor superfamily/fatty acid P-oxidation). Proc. Natl. Acad. Sci. USA 1995, 92, 7921–7925. [Google Scholar] [CrossRef]
- Mukherjee, R.; Jow, L.; Croston, G.E.; Paterniti, J.R. Identification, Characterization, and Tissue Distribution of Human Peroxisome Proliferator-activated Receptor (PPAR) Isoforms PPARγ2 versus PPARγ1 and activation with retinoid x receptor agonists and antagonists. J. Biol. Chem. 1997, 272, 8071–8076. [Google Scholar] [CrossRef]
- Schadinger, S.E.; Bucher, N.L.R.; Schreiber, B.M.; Farmer, S.R. PPAR 2 regulates lipogenesis and lipid accumulation in steatotic hepatocytes. AJP Endocrinol. Metab. 2005, 288, E1195–E1205. [Google Scholar] [CrossRef]
- Lee, Y.K.; Park, J.E.; Lee, M.; Hardwick, J.P. Hepatic lipid homeostasis by peroxisome proliferator-activated receptor gamma 2. Liver Res. 2018, 2, 209–215. [Google Scholar] [CrossRef]
- Tontonoz, P.; Hu, E.; Spiegelman, B.M. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 2014, 79, 1147–1156. [Google Scholar] [CrossRef]
- Ruiz-Ojeda, F.J.; Rupérez, A.I.; Gomez-Llorente, C.; Gil, A.; Aguilera, C.M. Cell models and their application for studying adipogenic differentiation in relation to obesity: A review. Int. J. Mol. Sci. 2016, 17, 1040. [Google Scholar] [CrossRef] [PubMed]
- Cristancho, A.G.; Mitchell, L.A. Forming functional fat: A growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 2011, 12, 722–734. [Google Scholar] [CrossRef]
- JASPAR 2018. Available online: http://jaspar.genereg.net/analysis (accessed on 3 July 2018).
- Dastagir, K.; Reimers, K.; Lazaridis, A.; Jahn, S.; Maurer, V.; Strauß, S.; Dastagir, N.; Radtke, C.; Kampmann, A.; Bucan, V.; et al. Murine embryonic fibroblast cell lines differentiate into three mesenchymal lineages to different extents: New models to investigate differentiation processes. Cell Reprog. 2014, 16, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Al-Sayegh, M.; Ali, H.; Jamal, M.H.; El-Gindi, M.; Chanyong, T.; Al-Awadi, K.; Abu-Farha, M. Mouse embryonic fibroblast adipogenic potential: A comprehensive transcriptome analysis. Adipocyte 2021, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Eeda, V.; Undi, R.B.; Mann, S.; Stout, M.; Lim, H.-Y.; Wang, W. A novel peroxisome proliferator-activated receptor gamma ligand improves insulin sensitivity and promotes browning of white adipose tissue in obese mice. Mol. Metab. 2021, 54, 101363. [Google Scholar] [CrossRef]
- Lauschke, V.M.; Hagberg, C.E. Next-generation human adipose tissue culture methods. Curr. Opin. Genet. Dev. 2023, 80, 102057. [Google Scholar] [CrossRef]
- Kleinert, M.; Clemmensen, C.; Hofmann, S.M.; Moore, M.C.; Renner, S.; Woods, S.C.; Huypens, P.; Beckers, J.; de Angelis, M.H.; Schürmann, A.; et al. Animal models of obesity and diabetes mellitus. Nat. Rev. Endocrinol. 2018, 14, 140–162. [Google Scholar] [CrossRef] [PubMed]
- Martins, T.; Castro-Ribeiro, C.; Lemos, S.; Ferreira, T.; Nascimento-Gonçalves, E.; Rosa, E.; Oliveira, P.A.; Antunes, L.M. Murine models of obesity. Obesities 2022, 2, 127–147. [Google Scholar] [CrossRef]
- Barradas, M.; Monjas, A.; Diaz-Meco, M.T.; Serrano, M.; Moscat, J. The downregulation of the pro-apoptotic protein Par-4 is critical for Ras-induced survival and tumor progression. EMBO J. 1999, 18, 6362–6369. [Google Scholar] [CrossRef]
- Kukoc-Zivojnov, N.; Puccetti, E.; Chow, K.U.; Bergmann, M.; Ruthardt, M.; Hoelzer, D.; Mitrou, P.S.; Weidmann, E.; Boehrer, S. Prostate apoptosis response gene-4 (par-4) abrogates the survival function of p185(BCR-ABL) in hematopoietic cells. Exp. Hematol. 2004, 32, 49–56. [Google Scholar] [CrossRef]
- Diaz-Meco, M.T.; Abu-Baker, S. The Par-4/PTEN connection in tumor suppression. Cell Cycle 2009, 8, 2518–2522. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nagai, M.A.; Gerhard, R.; Salaorni, S.; Fregnani, J.H.; Nonogaki, S.; Netto, M.M.; Soares, F.A. Down-regulation of the candidate tumor suppressor gene PAR-4 is associated with poor prognosis in breast cancer. Int. J. Oncol. 2010, 37, 41–49. [Google Scholar] [CrossRef]
- Chaudhry, P.; Singh, M.; Parent, S.; Asselin, E. Prostate apoptosis response 4 (Par-4), a novel substrate of caspase-3 during apoptosis activation. Mol. Cell Biol. 2012, 32, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Meynier, S.; Kramer, M.; Ribaux, P.; Tille, J.C.; Delie, F.; Petignat, P.; Cohen, M. Role of PAR-4 in ovarian cancer. Oncotarget 2015, 6, 22641–22652. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Ma, F.; Zhou, H.; Ding, R.; Lu, B.; Zou, L.; Li, J.; Lu, R. Inhibitory effect of Par-4 combined with cisplatin on human Wilms’ tumor cells. Tumor Biol. 2017, 39, 1010428317716689. [Google Scholar] [CrossRef]
- Mabe, N.W.; Fox, D.B.; Lupo, R.; Decker, A.E.; Phelps, S.N.; Thompson, J.W.; Alvarez, J.V. Epigenetic silencing of tumor suppressor Par-4 promotes chemoresistance in recurrent breast cancer. J. Clin. Investig. 2018, 128, 4413–4428. [Google Scholar] [CrossRef]
- Clark, A.M.; Ponniah, K.; Warden, M.S.; Raitt, E.M.; Smith, B.G.; Pascal, S.M. Tetramer formation by the caspase-activated fragment of the Par-4 tumor suppressor. FEBS J. 2019, 286, 4060–4073. [Google Scholar] [CrossRef]
- Katoch, A.; Jamwal, V.L.; Faheem, M.M.; Kumar, S.; Senapati, S.; Yadav, G.; Gandhi, S.G.; Goswami, A. Overlapping targets exist between the Par-4 and miR-200c axis which regulate EMT and proliferation of pancreatic cancer cells. Transl. Oncol. 2021, 14, 100879. [Google Scholar] [CrossRef]
- Sonawane, V.; Ghosalkar, J.; Achrekar, S.; Joshi, K. Ketorolac modulates Rac-1/HIF-1alpha/DDX3/beta-catenin signalling via a tumor suppressor prostate apoptosis response-4 (Par-4) in renal cell carcinoma. Sci. Rep. 2023, 13, 5659. [Google Scholar] [CrossRef]
- Pandey, S.; Raut, K.K.; Clark, A.M.; Baudin, A.; Djemr, I.L.; Libich, D.S.; Ponniah, K.; Pascal, S.M. Enhancing the conformational stability of the cl-Par-4 tumor suppressor via site-directed mutagenesis. Biomolecules 2023, 13, 667. [Google Scholar] [CrossRef]
- Raut, K.K.; Pandey, S.; Kharel, G.; Pascal, S.M. Evidence of direct interaction between cisplatin and the caspase-cleaved prostate apoptosis response-4 tumor suppressor. Protein Sci. 2024, 33, e4867. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Abderrahman, B.; Chai, T.S.; Yerrum, S.; Jordan, V.C. Targeting peroxisome proliferator-activated receptor γ to increase estrogen-induced apoptosis in estrogen-deprived breast cancer cells. Mol. Cancer Ther. 2018, 17, 2732–2745. [Google Scholar] [CrossRef] [PubMed]
- Augimeri, G.; Giordano, C.; Gelsomino, L.; Plastina, P.; Barone, I.; Catalano, S.; Andò, S.; Bonofiglio, D. The role of PPARγ ligands in breast cancer: From basic research to clinical studies. Cancers 2020, 12, 2623. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Quiles, M.; Broekema, M.F.; Kalkhoven, E. PPARgamma in metabolism, immunity, and cancer: Unified and diverse mechanisms of action. Front. Endocrinol. 2021, 12, 624112. [Google Scholar] [CrossRef]
- Wagner, N.; Wagner, K.D. Peroxisome proliferator-activated receptors and the hallmarks of cancer. Cells 2022, 11, 2432. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pan, Y.; Zhao, X.; Wu, S.; Li, F.; Wang, Y.; Liu, B.; Zhang, Y.; Gao, X.; Wang, Y.; et al. Peroxisome proliferator-activated receptors: A key link between lipid metabolism and cancer progression. Clin. Nutr. 2024, 43, 332–345. [Google Scholar] [CrossRef]

| Target | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Mouse Par-4 | AGAATGAAGCTGCGACCCTC | ATCTTCTGGGGCACTGGTTG |
| Mouse PPARγ1 | GTCTCGGTTGAGGGGAC | TGTCAACCATGGTAATTTCAGT |
| Mouse GAPDH | AAATGGTGAAGGTCGGTGTG | TGAATTTGCCGTGAGTGGAG |
| Fragment | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| 1 | GAGTGGTACCGTAAGCAACATTTATTG | GAGCCTCGAGAACAGCATAAAACAGAG |
| 2 | GAGTGGTACCGTAAGCAACATTTATTG | GAGCCTCGAGTTTAACAAGAATTCTTA |
| 3 | GAGTGGTACCTTTTACATTCTAGACAC | GAGCCTCGAGAACAGCATAAAACAGAG |
| 4 | GAGTGGTACCTTTTACATTCTAGACAC | GAGCCTCGAGGGTCTAAATATCAGTCA |
| 5 | GAGTGGTACCCATCATTTGGACTACTG | GAGCCTCGAGGCCTTTGCCCTTTTTGG |
| 6 | GAGTGGTACCGCTCTTTTAAAGTCCAC | GAGCCTCGAGAGGTCCAAAATGTTACT |
| 7 | GAGTGGTACCGATAGATAAACAAATTT | GAGCCTCGAGGTACAGTAGTTGGAATT |
| 7 + 8 | GAGTGGTACCGATAGATAAACAAATTT | GAGCCTCGAGAACAGCATAAAACAGAG |
| Primer Pair Designation | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| 6 | GCTCTTTTAAAGTCCACAAGTCACTG | GGAAAACTCTGGCTTCTTGCTTAA |
| 7 | ATGTGTGATTAGGAGTTTCAACCAAA | GAATTACCAGAGCAGAGATTGTTCA |
| Mouse Negative Control Primer Set 2 | Proprietary Sequence | Proprietary Sequence |
| Mouse Positive Control Primer Set (GAPDH) | Proprietary Sequence | Proprietary Sequence |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sledziona, J.; Burikhanov, R.; Araujo, N.; Jiang, J.; Hebbar, N.; Rangnekar, V.M. The Tumor Suppressor Par-4 Regulates Adipogenesis by Transcriptional Repression of PPARγ. Cells 2024, 13, 1495. https://doi.org/10.3390/cells13171495
Sledziona J, Burikhanov R, Araujo N, Jiang J, Hebbar N, Rangnekar VM. The Tumor Suppressor Par-4 Regulates Adipogenesis by Transcriptional Repression of PPARγ. Cells. 2024; 13(17):1495. https://doi.org/10.3390/cells13171495
Chicago/Turabian StyleSledziona, James, Ravshan Burikhanov, Nathalia Araujo, Jieyun Jiang, Nikhil Hebbar, and Vivek M. Rangnekar. 2024. "The Tumor Suppressor Par-4 Regulates Adipogenesis by Transcriptional Repression of PPARγ" Cells 13, no. 17: 1495. https://doi.org/10.3390/cells13171495
APA StyleSledziona, J., Burikhanov, R., Araujo, N., Jiang, J., Hebbar, N., & Rangnekar, V. M. (2024). The Tumor Suppressor Par-4 Regulates Adipogenesis by Transcriptional Repression of PPARγ. Cells, 13(17), 1495. https://doi.org/10.3390/cells13171495








