Ablation of the Integrin CD11b Mac-1 Limits Deleterious Responses to Traumatic Spinal Cord Injury and Improves Functional Recovery in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Mouse Spinal Cord Contusion Model
2.2. Flow Cytometry
2.3. Total RNA Extraction and Real-Time PCR (qPCR)
2.4. Sample Processing and Western Blot Analysis
2.5. NanoString Analysis
2.6. Bulk RNA Sequencing and Transcriptomic Analysis
2.7. Tissue Processing and Histologic, Immunohistochemistry Analysis
2.8. Neurological Behavioral Tests
2.9. Statistical Analysis
3. Results
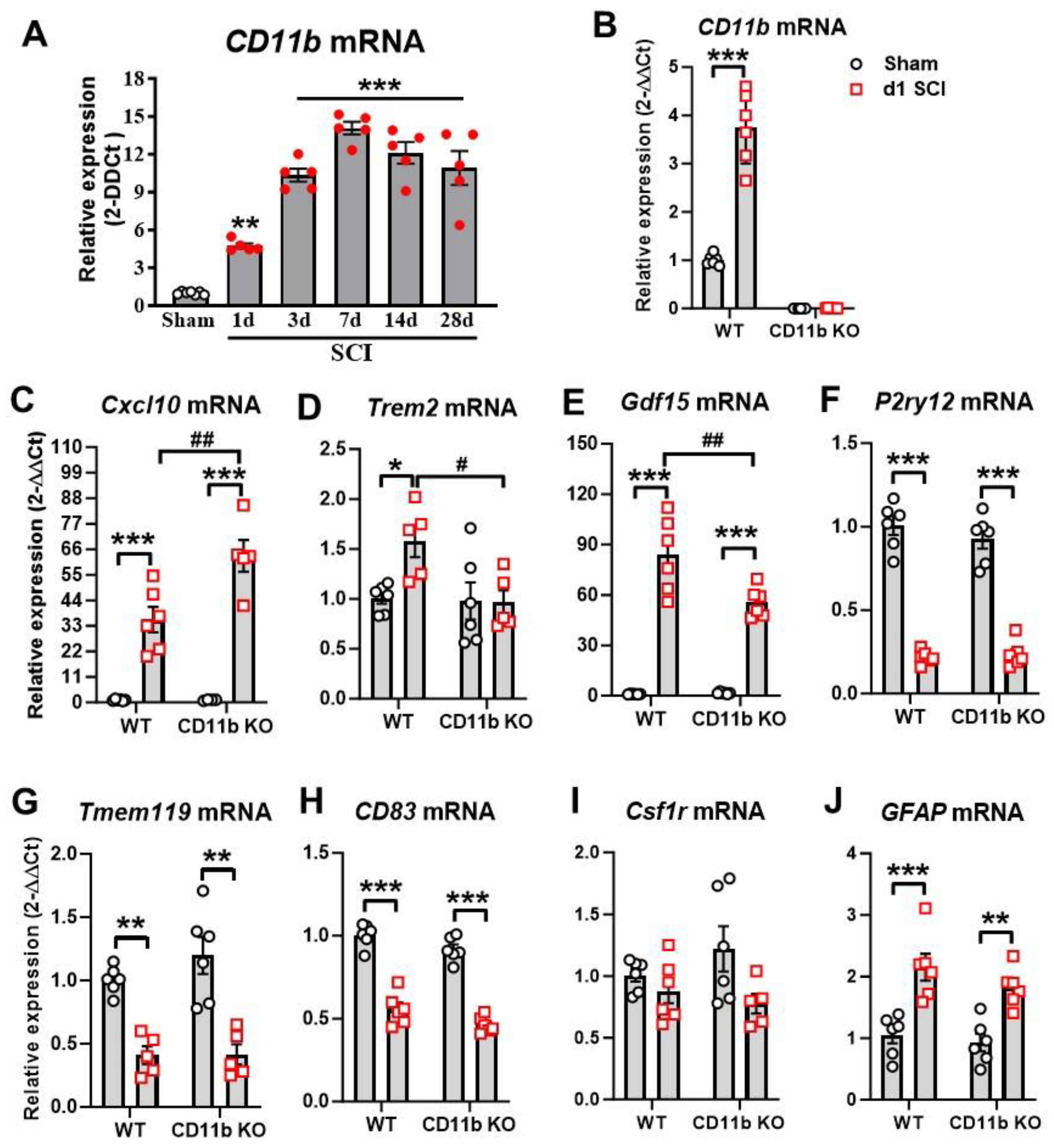
3.1. Ablation of CD11b Gene Limits Acute Proinflammatory Response to SCI
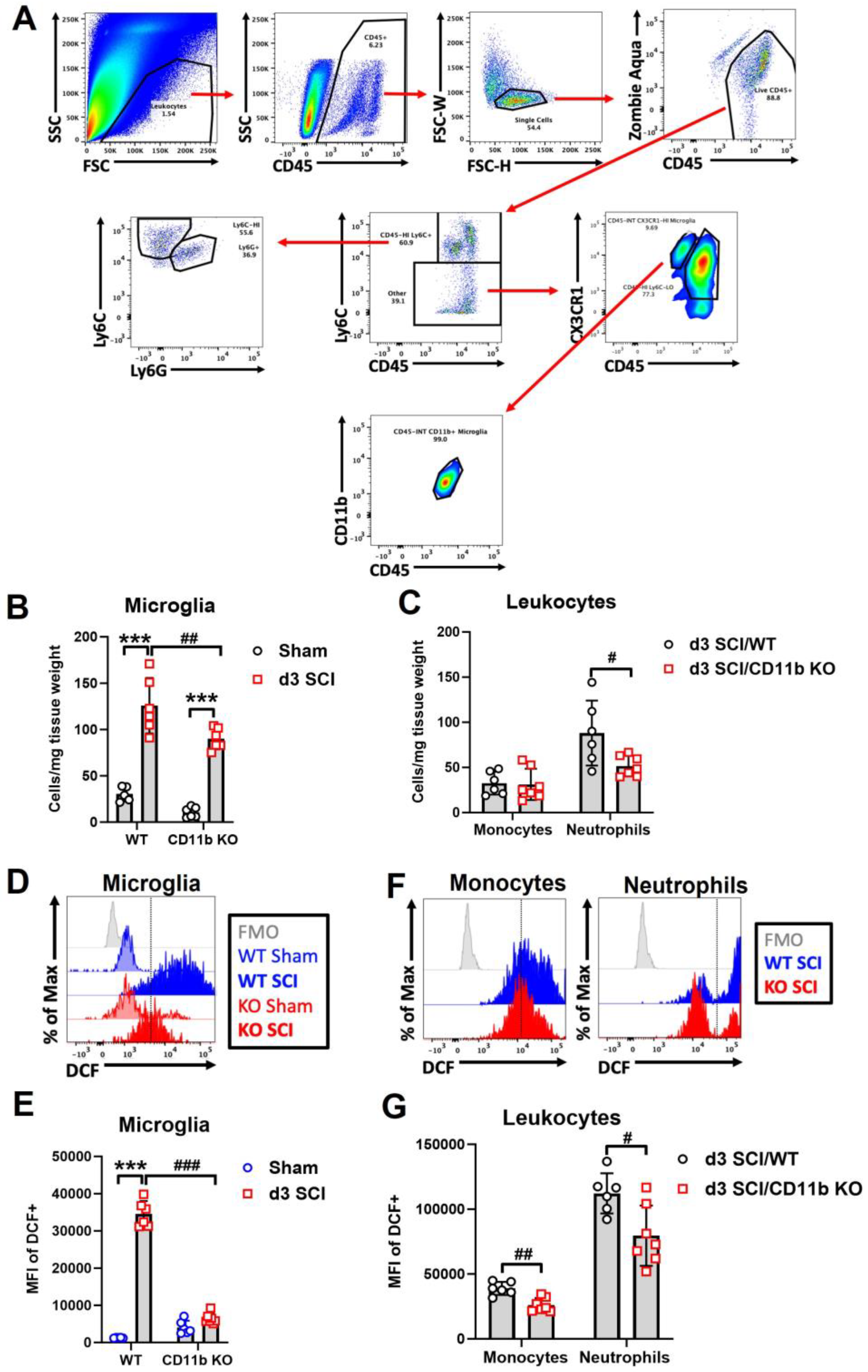
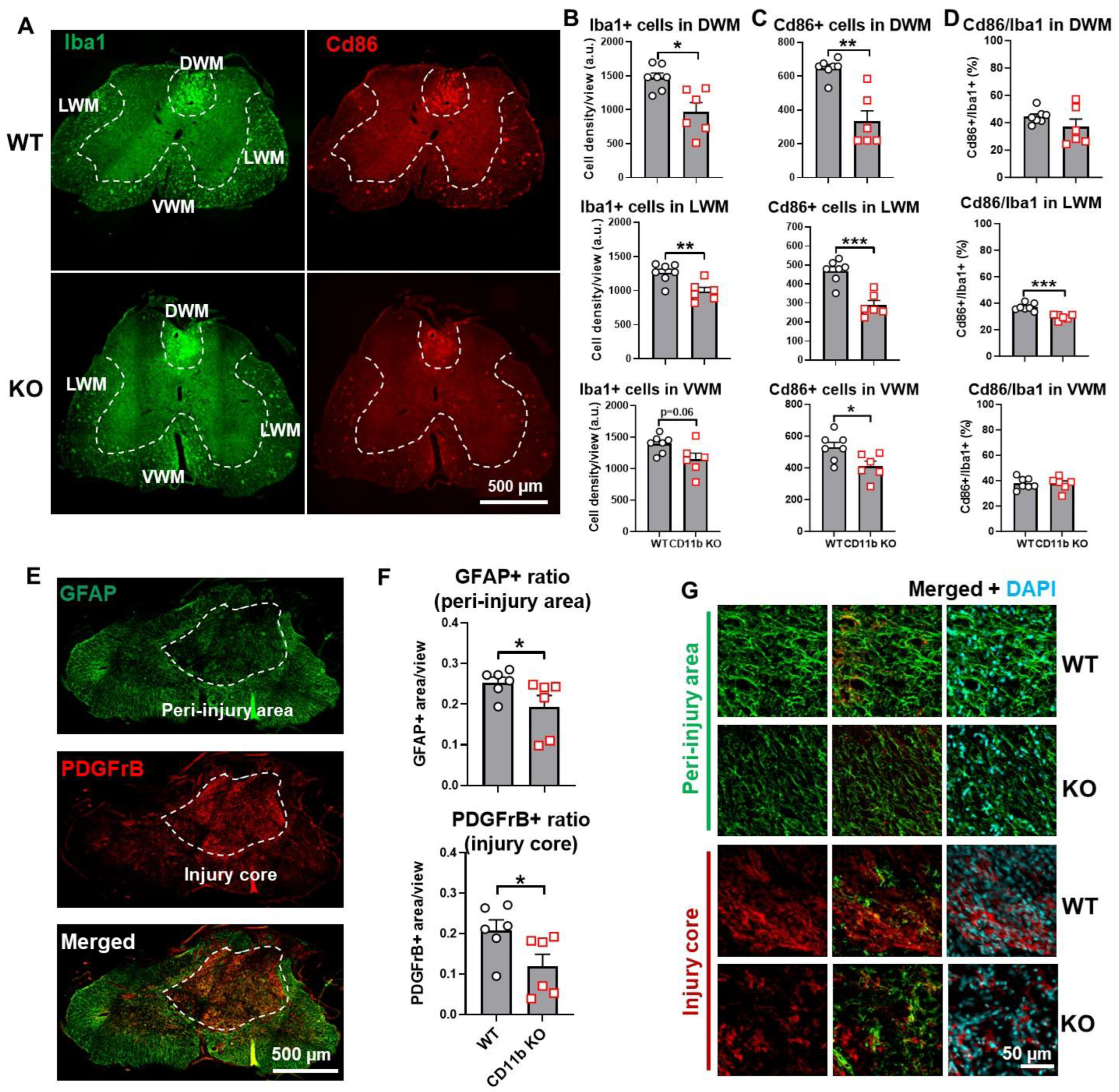
3.2. Depletion of CD11b Reduces the Number of Microglia and Infiltration of Neutrophils in the Injured Spinal Cord
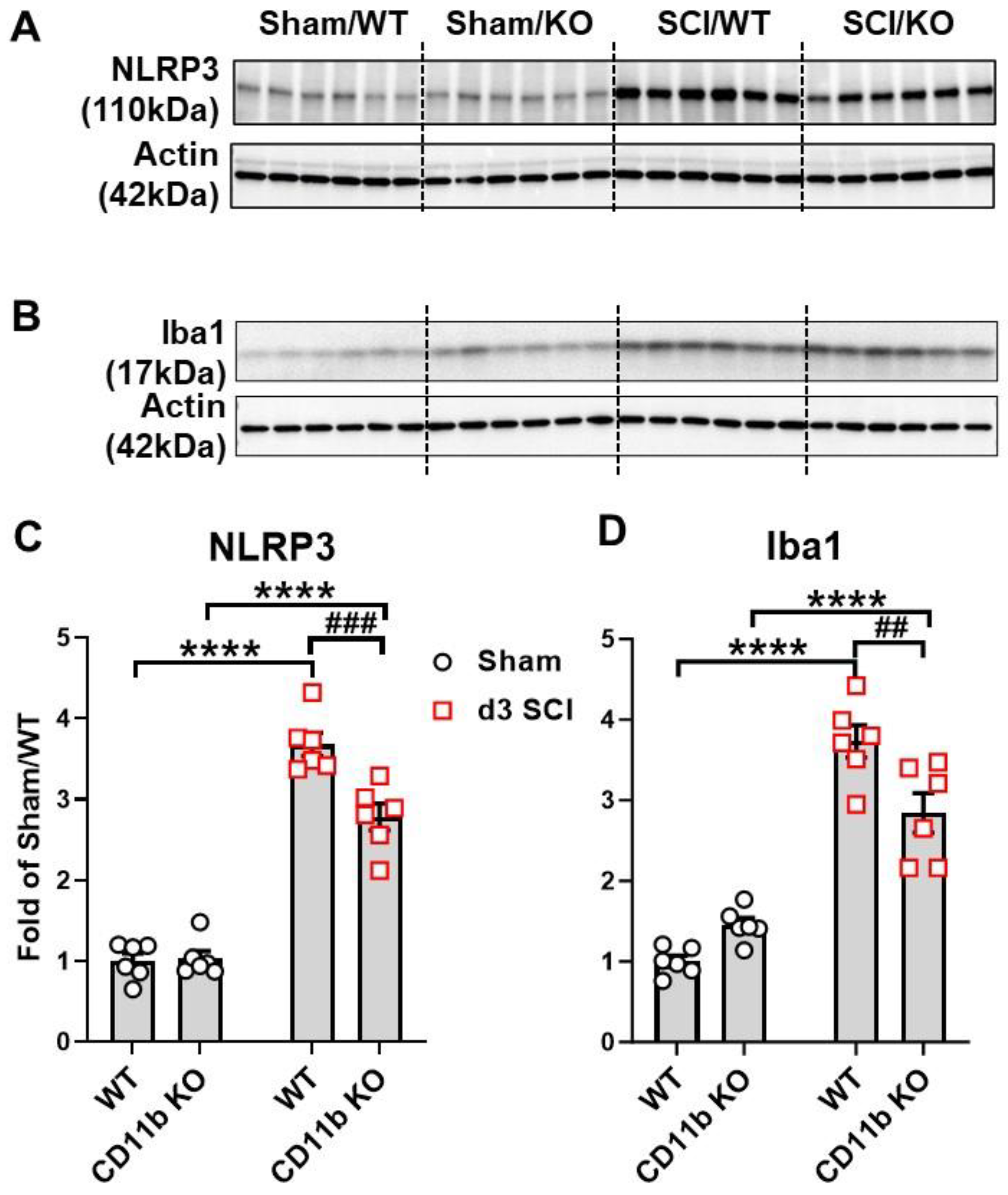
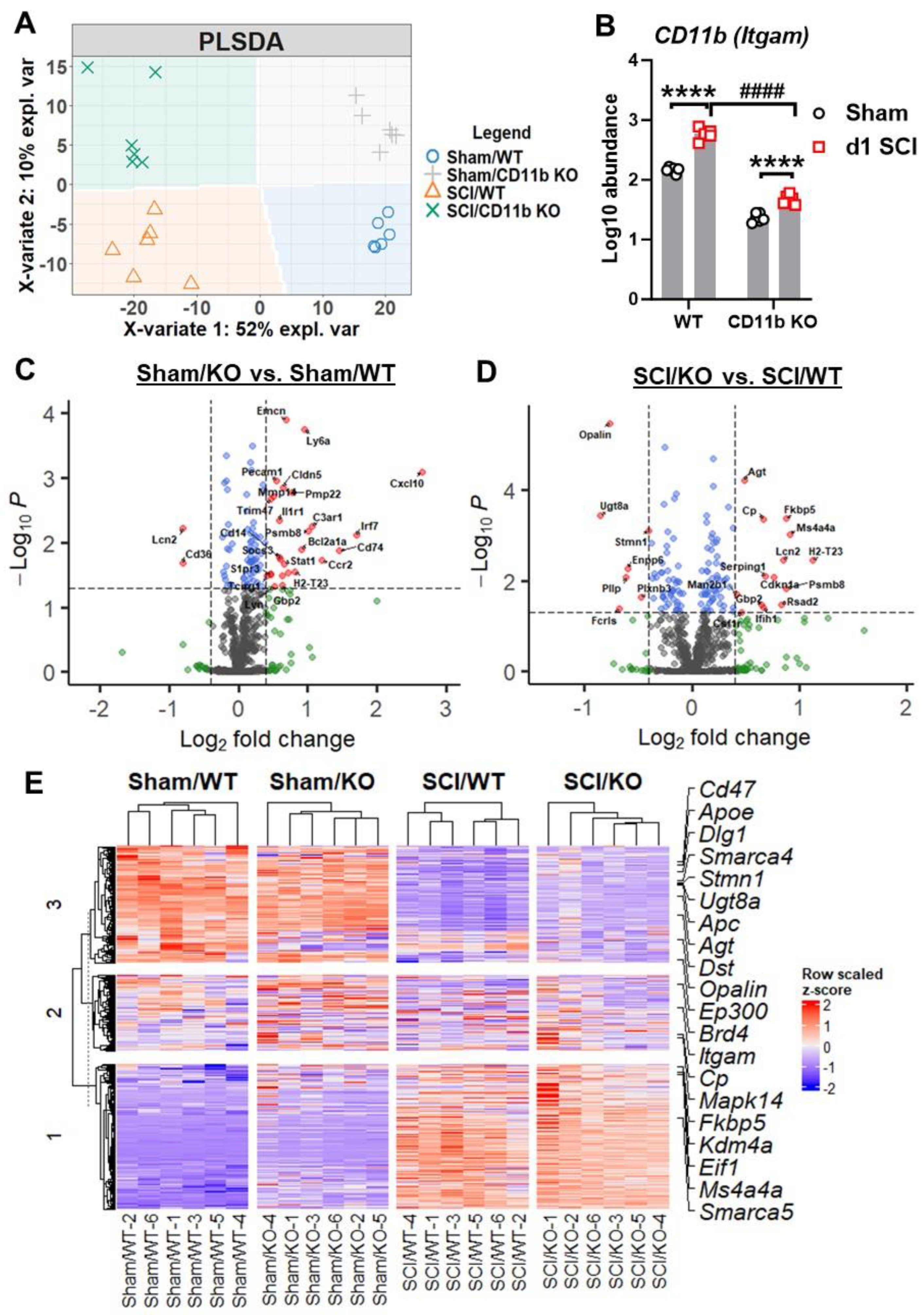
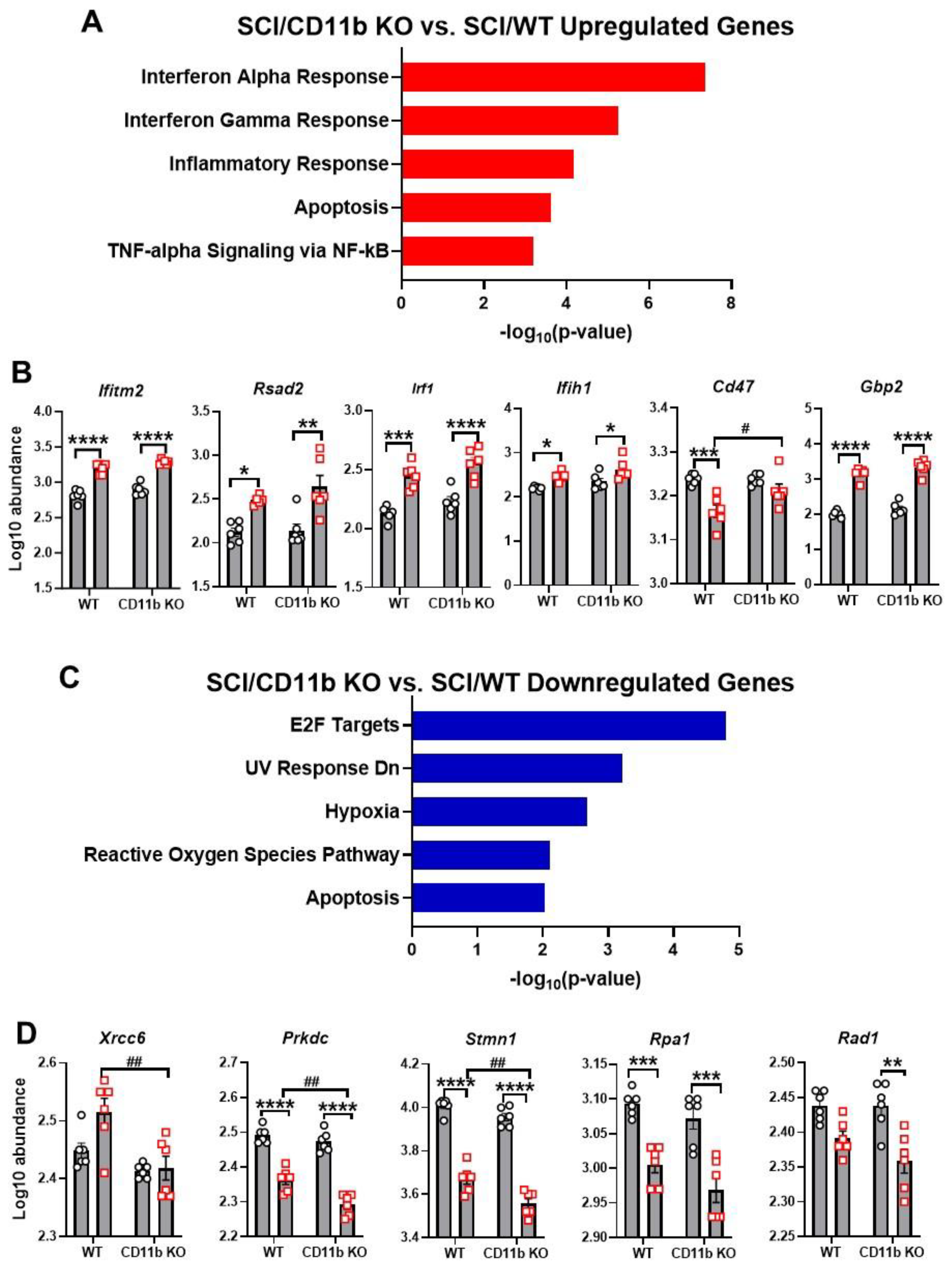
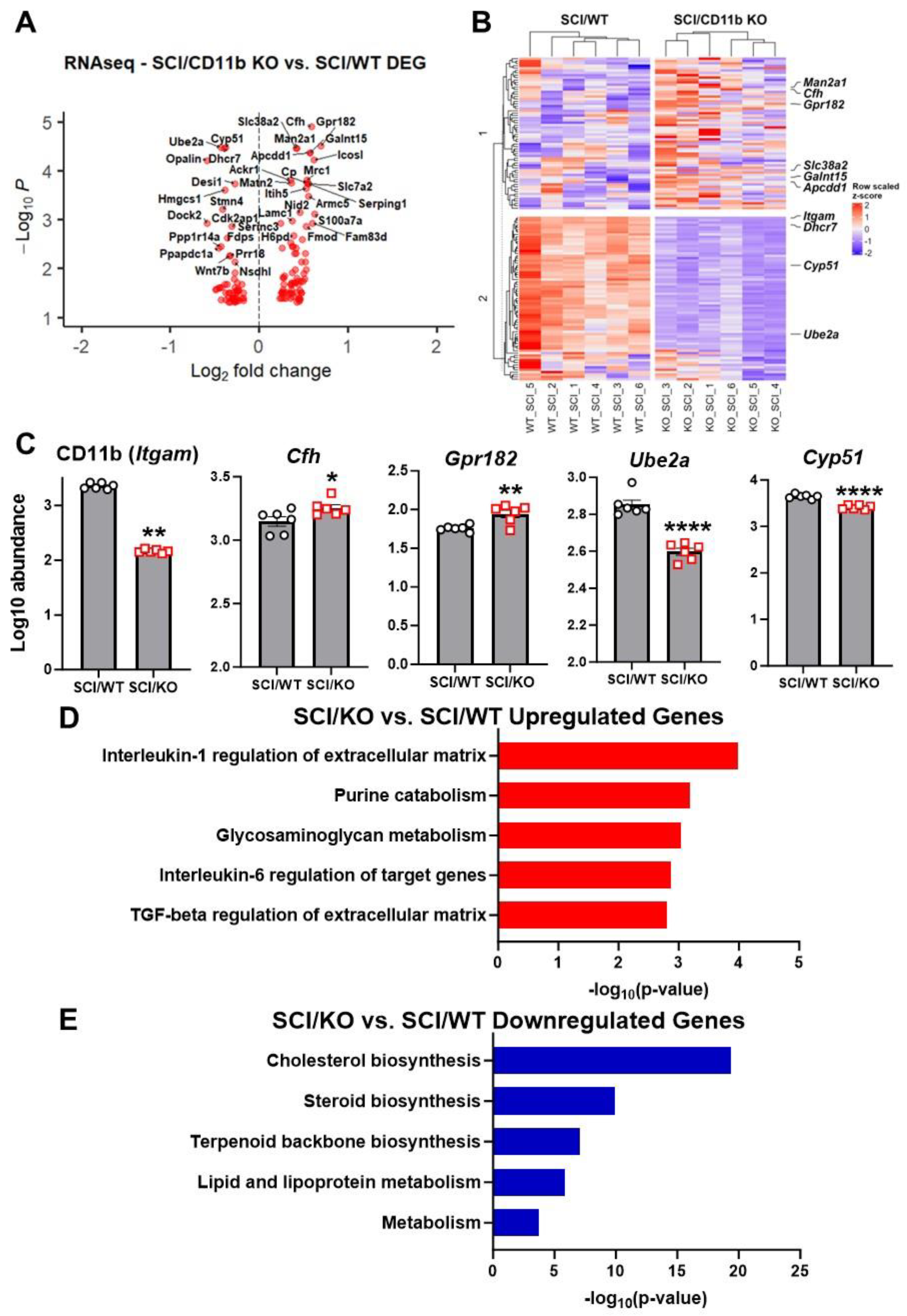
3.3. CD11b KO Mice Show Robust Changes in Neuroinflammation-Related Genes after SCI
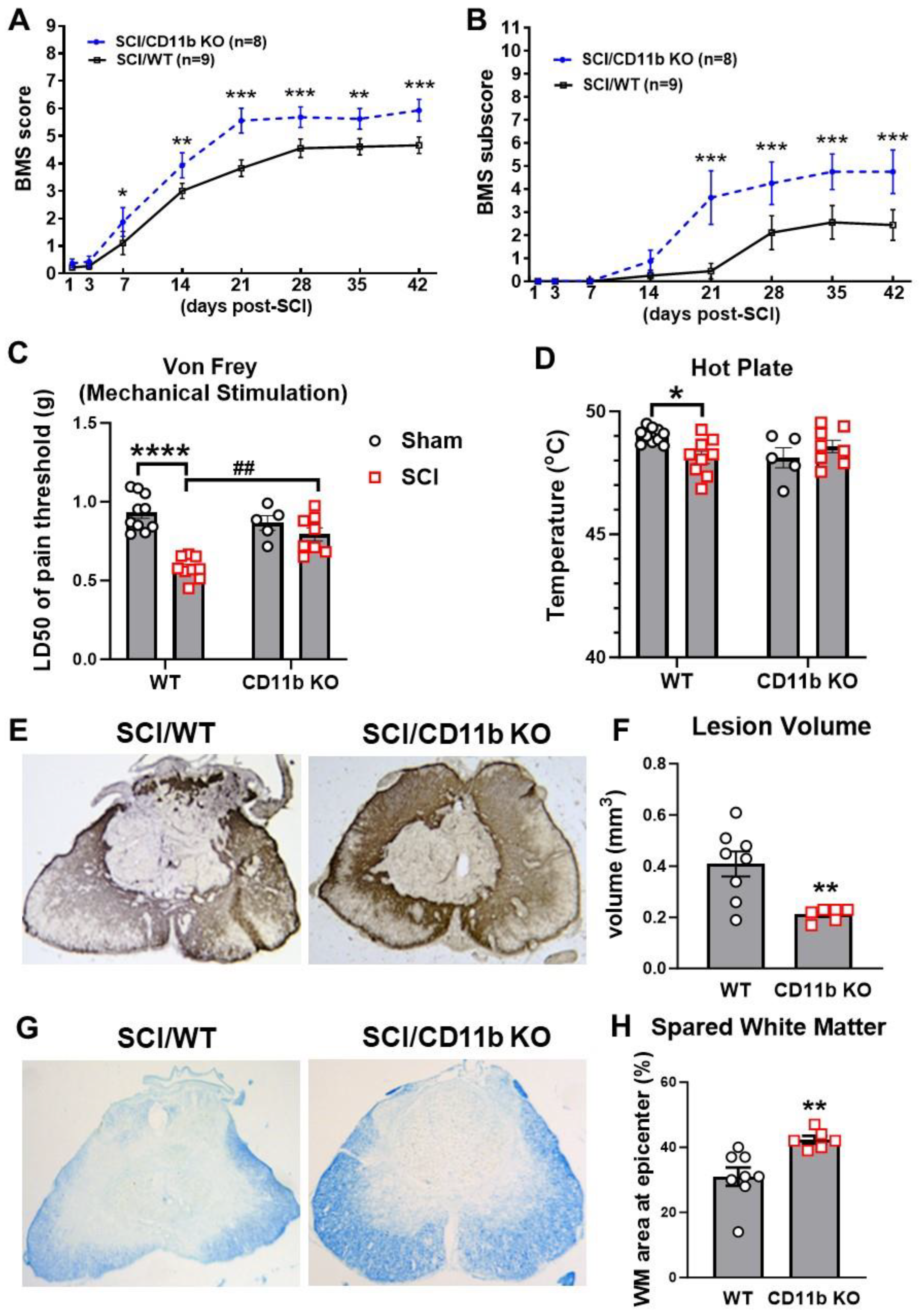
3.4. Ablation of CD11b Improves Functional Recovery and Reduces Tissue Damage after SCI
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hausmann, O.N. Post-traumatic inflammation following spinal cord injury. Spinal Cord 2003, 41, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Tator, C.H.; Fehlings, M.G. Review of the secondary injury theory of acute spinal cord trauma with emphasis on vascular mechanisms. J. Neurosurg. 1991, 75, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Houle, J.D.; Cote, M.P. Axon regeneration and exercise-dependent plasticity after spinal cord injury. Ann. N. Y. Acad. Sci. 2013, 1279, 154–163. [Google Scholar] [CrossRef]
- Lee, C.Y.; Chooi, W.H.; Ng, S.Y.; Chew, S.Y. Modulating neuroinflammation through molecular, cellular and biomaterial-based approaches to treat spinal cord injury. Bioeng. Transl. Med. 2023, 8, e10389. [Google Scholar] [CrossRef] [PubMed]
- Cha, B.H.; Shin, S.R.; Leijten, J.; Li, Y.C.; Singh, S.; Liu, J.C.; Annabi, N.; Abdi, R.; Dokmeci, M.R.; Vrana, N.E.; et al. Integrin-Mediated Interactions Control Macrophage Polarization in 3D Hydrogels. Adv. Healthc. Mater. 2017, 6, 1700289. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.; Flores-Borja, F.; Nassiri, S.; Miranda, E.; Lawler, K.; Grigoriadis, A.; Monypenny, J.; Gillet, C.; Owen, J.; Gordon, P.; et al. Integrin-Mediated Macrophage Adhesion Promotes Lymphovascular Dissemination in Breast Cancer. Cell Rep. 2019, 27, 1967–1978.e4. [Google Scholar] [CrossRef]
- Dash, S.P.; Chakraborty, P.; Sarangi, P.P. Inflammatory Monocytes and Subsets of Macrophages with Distinct Surface Phenotype Correlate with Specific Integrin Expression Profile during Murine Sepsis. J. Immunol. 2021, 207, 2841–2855. [Google Scholar] [CrossRef]
- Cai, X.F.; Lin, S.; Geng, Z.; Luo, L.L.; Liu, Y.J.; Zhang, Z.; Liu, W.Y.; Chen, X.; Li, X.; Yan, J.; et al. Integrin CD11b Deficiency Aggravates Retinal Microglial Activation and RGCs Degeneration After Acute Optic Nerve Injury. Neurochem. Res. 2020, 45, 1072–1085. [Google Scholar] [CrossRef]
- Bao, F.; Chen, Y.; Dekaban, G.A.; Weaver, L.C. An anti-CD11d integrin antibody reduces cyclooxygenase-2 expression and protein and DNA oxidation after spinal cord injury in rats. J. Neurochem. 2004, 90, 1194–1204. [Google Scholar] [CrossRef]
- Gris, D.; Marsh, D.R.; Oatway, M.A.; Chen, Y.; Hamilton, E.F.; Dekaban, G.A.; Weaver, L.C. Transient blockade of the CD11d/CD18 integrin reduces secondary damage after spinal cord injury, improving sensory, autonomic, and motor function. J. Neurosci. 2004, 24, 4043–4051. [Google Scholar] [CrossRef]
- Bao, F.; Omana, V.; Brown, A.; Weaver, L.C. The systemic inflammatory response after spinal cord injury in the rat is decreased by alpha4beta1 integrin blockade. J. Neurotrauma 2012, 29, 1626–1637. [Google Scholar] [CrossRef] [PubMed]
- Bao, F.; Brown, A.; Dekaban, G.A.; Omana, V.; Weaver, L.C. CD11d integrin blockade reduces the systemic inflammatory response syndrome after spinal cord injury. Exp. Neurol. 2011, 231, 272–283. [Google Scholar] [CrossRef] [PubMed]
- North, H.A.; Pan, L.; McGuire, T.L.; Brooker, S.; Kessler, J.A. beta1-Integrin alters ependymal stem cell BMP receptor localization and attenuates astrogliosis after spinal cord injury. J. Neurosci. 2015, 35, 3725–3733. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Wang, Y.; Wang, J.; Xu, Y.; Liu, H.; Guo, J.; Zhu, L. Perlecan Improves Blood Spinal Cord Barrier Repair Through the Integrin beta1/ROCK/MLC Pathway After Spinal Cord Injury. Mol. Neurobiol. 2023, 60, 51–67. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.L.; Li, N.; Xu, M.; Wang, L.; Sun, J.; Li, L.; Wang, Y.F.; Zhang, B.L.; Suo, H.Y.; Wang, T.H.; et al. Implantation of human urine stem cells promotes neural repair in spinal cord injury rats associated cadeharin-1 and integrin subunit beta 1 expression. J. Gene Med. 2023, 26, e3615. [Google Scholar] [CrossRef] [PubMed]
- Arnaout, M.A. Structure and function of the leukocyte adhesion molecules CD11/CD18. Blood 1990, 75, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Ley, K.; Laudanna, C.; Cybulsky, M.I.; Nourshargh, S. Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nat. Rev. Immunol. 2007, 7, 678–689. [Google Scholar] [CrossRef]
- Isaksson, J.; Farooque, M.; Holtz, A.; Hillered, L.; Olsson, Y. Expression of ICAM-1 and CD11b after experimental spinal cord injury in rats. J. Neurotrauma 1999, 16, 165–173. [Google Scholar] [CrossRef]
- Zhou, J.; Lin, S.; Hu, Q.; Li, X.; Chen, X.; Luo, L.; Ye, S.; Liu, W.; Ye, J. Microglial CD11b Knockout Contributes to Axonal Debris Clearance and Axonal Degradation Attenuation via IGF-1 After Acute Optic Nerve Injury. Investig. Ophthalmol. Vis. Sci. 2023, 64, 7. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Bai, J.; Yu, W.J.; Lin, Q.Y.; Li, H.H. CD11b mediates hypertensive cardiac remodeling by regulating macrophage infiltration and polarization. J. Adv. Res. 2023, 55, 17–31. [Google Scholar] [CrossRef]
- Zhang, S.; Wan, D.; Zhu, M.; Wang, G.; Zhang, X.; Huang, N.; Zhang, J.; Zhang, C.; Shang, Q.; Zhang, C.; et al. CD11b + CD43 hi Ly6C lo splenocyte-derived macrophages exacerbate liver fibrosis via spleen-liver axis. Hepatology 2023, 77, 1612–1629. [Google Scholar] [CrossRef] [PubMed]
- Zec, K.; Thiebes, S.; Bottek, J.; Siemes, D.; Spangenberg, P.; Trieu, D.V.; Kirstein, N.; Subramaniam, N.; Christ, R.; Klein, D.; et al. Comparative transcriptomic and proteomic signature of lung alveolar macrophages reveals the integrin CD11b as a regulatory hub during pneumococcal pneumonia infection. Front. Immunol. 2023, 14, 1227191. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Du, S.; Bimler, L.H.; Mauk, K.E.; Lortal, L.; Kichik, N.; Griffiths, J.S.; Osicka, R.; Song, L.; Polsky, K.; et al. Toll-like receptor 4 and CD11b expressed on microglia coordinate eradication of Candida albicans cerebral mycosis. Cell Rep. 2023, 42, 113240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, Z.G.; Zhang, R.L.; Lu, M.; Krams, M.; Chopp, M. Effects of a selective CD11b/CD18 antagonist and recombinant human tissue plasminogen activator treatment alone and in combination in a rat embolic model of stroke. Stroke 2003, 34, 1790–1795. [Google Scholar] [CrossRef]
- Soriano, S.G.; Coxon, A.; Wang, Y.F.; Frosch, M.P.; Lipton, S.A.; Hickey, P.R.; Mayadas, T.N. Mice deficient in Mac-1 (CD11b/CD18) are less susceptible to cerebral ischemia/reperfusion injury. Stroke 1999, 30, 134–139. [Google Scholar] [CrossRef]
- Garcia, J.H.; Liu, K.F.; Bree, M.P. Effects of CD11b/18 monoclonal antibody on rats with permanent middle cerebral artery occlusion. Am. J. Pathol. 1996, 148, 241–248. [Google Scholar] [PubMed]
- Su, E.J.; Cao, C.; Fredriksson, L.; Nilsson, I.; Stefanitsch, C.; Stevenson, T.K.; Zhao, J.; Ragsdale, M.; Sun, Y.Y.; Yepes, M.; et al. Microglial-mediated PDGF-CC activation increases cerebrovascular permeability during ischemic stroke. Acta Neuropathol. 2017, 134, 585–604. [Google Scholar] [CrossRef]
- Cao, C.; Gao, Y.; Li, Y.; Antalis, T.M.; Castellino, F.J.; Zhang, L. The efficacy of activated protein C in murine endotoxemia is dependent on integrin CD11b. J. Clin. Investig. 2010, 120, 1971–1980. [Google Scholar] [CrossRef]
- Ehirchiou, D.; Xiong, Y.; Xu, G.; Chen, W.; Shi, Y.; Zhang, L. CD11b facilitates the development of peripheral tolerance by suppressing Th17 differentiation. J. Exp. Med. 2007, 204, 1519–1524. [Google Scholar] [CrossRef]
- Han, C.; Jin, J.; Xu, S.; Liu, H.; Li, N.; Cao, X. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat. Immunol. 2010, 11, 734–742. [Google Scholar] [CrossRef]
- Nath, S.K.; Han, S.; Kim-Howard, X.; Kelly, J.A.; Viswanathan, P.; Gilkeson, G.S.; Chen, W.; Zhu, C.; McEver, R.P.; Kimberly, R.P.; et al. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat. Genet. 2008, 40, 152–154. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fang, S.C.; Zhou, L.; Mo, X.M.; Guo, H.D.; Deng, Y.B.; Yu, H.H.; Gong, W.Y. Complement Receptor 3 Pathway and NMDA Receptor 2B Subunit Involve Neuropathic Pain Associated with Spinal Cord Injury. J. Pain Res. 2022, 15, 1813–1823. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, X.; Chen, T.; Li, T.; Cao, K.; Lu, A.; Chen, Y.; Sun, D.; Luo, J.; Fan, J.; et al. Myelin activates FAK/Akt/NF-kappaB pathways and provokes CR3-dependent inflammatory response in murine system. PLoS ONE 2010, 5, e9380. [Google Scholar] [CrossRef]
- Stewart, A.N.; MacLean, S.M.; Stromberg, A.J.; Whelan, J.P.; Bailey, W.M.; Gensel, J.C.; Wilson, M.E. Considerations for Studying Sex as a Biological Variable in Spinal Cord Injury. Front. Neurol. 2020, 11, 802. [Google Scholar] [CrossRef]
- Hashimoto, M.; Karnup, S.; Daugherty, S.L.; Cho, K.J.; Banno, E.; Shimizu, N.; Fujita, K.; Hirayama, A.; Uemura, H.; de Groat, W.C.; et al. Sex differences in lower urinary tract function in mice with or without spinal cord injury. Neurourol. Urodyn. 2024, 43, 267–275. [Google Scholar] [CrossRef]
- Zhao, Z.; Sabirzhanov, B.; Wu, J.; Faden, A.I.; Stoica, B.A. Voluntary Exercise Preconditioning Activates Multiple Antiapoptotic Mechanisms and Improves Neurological Recovery after Experimental Traumatic Brain Injury. J. Neurotrauma 2015, 32, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Ritzel, R.M.; Li, Y.; Jiao, Y.; Lei, Z.; Doran, S.J.; He, J.; Shahror, R.A.; Henry, R.J.; Khan, R.; Tan, C.; et al. Brain injury accelerates the onset of a reversible age-related microglial phenotype associated with inflammatory neurodegeneration. Sci. Adv. 2023, 9, eadd1101. [Google Scholar] [CrossRef]
- Ritzel, R.M.; Li, Y.; Lei, Z.; Carter, J.; He, J.; Choi, H.M.C.; Khan, N.; Li, H.; Allen, S.; Lipinski, M.M.; et al. Functional and transcriptional profiling of microglial activation during the chronic phase of TBI identifies an age-related driver of poor outcome in old mice. Geroscience 2022, 44, 1407–1440. [Google Scholar] [CrossRef]
- Ritzel, R.M.; Doran, S.J.; Glaser, E.P.; Meadows, V.E.; Faden, A.I.; Stoica, B.A.; Loane, D.J. Old age increases microglial senescence, exacerbates secondary neuroinflammation, and worsens neurological outcomes after acute traumatic brain injury in mice. Neurobiol. Aging 2019, 77, 194–206. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Le Cao, K.A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer Science + Business Media: New York, NY, USA; London, UK, 2009; p. viii. 212p. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Res 2018, 7, 1338. [Google Scholar] [CrossRef]
- Kopylova, E.; Noe, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res 2015, 4, 1521. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Li, Y.; Lei, Z.; Ritzel, R.M.; He, J.; Li, H.; Choi, H.M.C.; Lipinski, M.M.; Wu, J. Impairment of autophagy after spinal cord injury potentiates neuroinflammation and motor function deficit in mice. Theranostics 2022, 12, 5364–5388. [Google Scholar] [CrossRef]
- Li, Y.; Ritzel, R.M.; He, J.; Cao, T.; Sabirzhanov, B.; Li, H.; Liu, S.; Wu, L.J.; Wu, J. The voltage-gated proton channel Hv1 plays a detrimental role in contusion spinal cord injury via extracellular acidosis-mediated neuroinflammation. Brain Behav. Immun. 2021, 91, 267–283. [Google Scholar] [CrossRef]
- Sabirzhanov, B.; Li, Y.; Coll-Miro, M.; Matyas, J.J.; He, J.; Kumar, A.; Ward, N.; Yu, J.; Faden, A.I.; Wu, J. Inhibition of NOX2 signaling limits pain-related behavior and improves motor function in male mice after spinal cord injury: Participation of IL-10/miR-155 pathways. Brain Behav. Immun. 2019, 80, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Basso, D.M.; Fisher, L.C.; Anderson, A.J.; Jakeman, L.B.; McTigue, D.M.; Popovich, P.G. Basso Mouse Scale for locomotion detects differences in recovery after spinal cord injury in five common mouse strains. J. Neurotrauma 2006, 23, 635–659. [Google Scholar] [CrossRef] [PubMed]
- Matyas, J.J.; O’Driscoll, C.M.; Yu, L.; Coll-Miro, M.; Daugherty, S.; Renn, C.L.; Faden, A.I.; Dorsey, S.G.; Wu, J. Truncated TrkB.T1-Mediated Astrocyte Dysfunction Contributes to Impaired Motor Function and Neuropathic Pain after Spinal Cord Injury. J. Neurosci. 2017, 37, 3956–3971. [Google Scholar] [CrossRef] [PubMed]
- Ritzel, R.M.; He, J.; Li, Y.; Cao, T.; Khan, N.; Shim, B.; Sabirzhanov, B.; Aubrecht, T.; Stoica, B.A.; Faden, A.I.; et al. Proton extrusion during oxidative burst in microglia exacerbates pathological acidosis following traumatic brain injury. Glia 2021, 69, 746–764. [Google Scholar] [CrossRef] [PubMed]
- Chaplan, S.R.; Bach, F.W.; Pogrel, J.W.; Chung, J.M.; Yaksh, T.L. Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods 1994, 53, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Bonin, R.P.; Bories, C.; De Koninck, Y. A simplified up-down method (SUDO) for measuring mechanical nociception in rodents using von Frey filaments. Mol. Pain 2014, 10, 26. [Google Scholar] [CrossRef]
- Dixon, W.J. Efficient analysis of experimental observations. Annu. Rev. Pharmacol. Toxicol. 1980, 20, 441–462. [Google Scholar] [CrossRef]
- Cohen, J. A power primer. Psychol. Bull. 1992, 112, 155–159. [Google Scholar] [CrossRef]
- Clarner, T.; Janssen, K.; Nellessen, L.; Stangel, M.; Skripuletz, T.; Krauspe, B.; Hess, F.M.; Denecke, B.; Beutner, C.; Linnartz-Gerlach, B.; et al. CXCL10 triggers early microglial activation in the cuprizone model. J. Immunol. 2015, 194, 3400–3413. [Google Scholar] [CrossRef]
- Mercurio, D.; Fumagalli, S.; Schafer, M.K.; Pedragosa, J.; Ngassam, L.D.C.; Wilhelmi, V.; Winterberg, S.; Planas, A.M.; Weihe, E.; De Simoni, M.G. Protein Expression of the Microglial Marker Tmem119 Decreases in Association with Morphological Changes and Location in a Mouse Model of Traumatic Brain Injury. Front. Cell Neurosci. 2022, 16, 820127. [Google Scholar] [CrossRef]
- Walker, D.G.; Tang, T.M.; Mendsaikhan, A.; Tooyama, I.; Serrano, G.E.; Sue, L.I.; Beach, T.G.; Lue, L.F. Patterns of Expression of Purinergic Receptor P2RY12, a Putative Marker for Non-Activated Microglia, in Aged and Alzheimer’s Disease Brains. Int. J. Mol. Sci. 2020, 21, 678. [Google Scholar] [CrossRef] [PubMed]
- Sunnemark, D.; Eltayeb, S.; Wallstrom, E.; Appelsved, L.; Malmberg, A.; Lassmann, H.; Ericsson-Dahlstrand, A.; Piehl, F.; Olsson, T. Differential expression of the chemokine receptors CX3CR1 and CCR1 by microglia and macrophages in myelin-oligodendrocyte-glycoprotein-induced experimental autoimmune encephalomyelitis. Brain Pathol. 2003, 13, 617–629. [Google Scholar] [CrossRef]
- de Faria, O., Jr.; Dhaunchak, A.S.; Kamen, Y.; Roth, A.D.; Kuhlmann, T.; Colman, D.R.; Kennedy, T.E. TMEM10 Promotes Oligodendrocyte Differentiation and is Expressed by Oligodendrocytes in Human Remyelinating Multiple Sclerosis Plaques. Sci. Rep. 2019, 9, 3606. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, G.C.; Smothers, C.G.; Gandhi, S.A. Newly-identified blood biomarkers of neurological damage are correlated with infarct volume in patients with acute ischemic stroke. J. Clin. Neurosci. 2021, 94, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Koshinaga, M.; Whittemore, S.R. The temporal and spatial activation of microglia in fiber tracts undergoing anterograde and retrograde degeneration following spinal cord lesion. J. Neurotrauma 1995, 12, 209–222. [Google Scholar] [CrossRef]
- Ahuja, C.S.; Wilson, J.R.; Nori, S.; Kotter, M.R.N.; Druschel, C.; Curt, A.; Fehlings, M.G. Traumatic spinal cord injury. Nat. Rev. Dis. Primers 2017, 3, 17018. [Google Scholar] [CrossRef]
- Neirinckx, V.; Coste, C.; Franzen, R.; Gothot, A.; Rogister, B.; Wislet, S. Neutrophil contribution to spinal cord injury and repair. J. Neuroinflammation 2014, 11, 150. [Google Scholar] [CrossRef]
- Bellver-Landete, V.; Bretheau, F.; Mailhot, B.; Vallieres, N.; Lessard, M.; Janelle, M.E.; Vernoux, N.; Tremblay, M.E.; Fuehrmann, T.; Shoichet, M.S.; et al. Microglia are an essential component of the neuroprotective scar that forms after spinal cord injury. Nat. Commun. 2019, 10, 518. [Google Scholar] [CrossRef]
- Fu, H.; Zhao, Y.; Hu, D.; Wang, S.; Yu, T.; Zhang, L. Depletion of microglia exacerbates injury and impairs function recovery after spinal cord injury in mice. Cell Death Dis. 2020, 11, 528. [Google Scholar] [CrossRef]
- Li, Y.; He, X.; Kawaguchi, R.; Zhang, Y.; Wang, Q.; Monavarfeshani, A.; Yang, Z.; Chen, B.; Shi, Z.; Meng, H.; et al. Microglia-organized scar-free spinal cord repair in neonatal mice. Nature 2020, 587, 613–618. [Google Scholar] [CrossRef]
- Kobayashi, M.; Konishi, H.; Sayo, A.; Takai, T.; Kiyama, H. TREM2/DAP12 Signal Elicits Proinflammatory Response in Microglia and Exacerbates Neuropathic Pain. J. Neurosci. 2016, 36, 11138–11150. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ulland, T.K.; Ulrich, J.D.; Song, W.; Tzaferis, J.A.; Hole, J.T.; Yuan, P.; Mahan, T.E.; Shi, Y.; Gilfillan, S.; et al. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 2016, 213, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Reifschneider, A.; Robinson, S.; van Lengerich, B.; Gnorich, J.; Logan, T.; Heindl, S.; Vogt, M.A.; Weidinger, E.; Riedl, L.; Wind, K.; et al. Loss of TREM2 rescues hyperactivation of microglia, but not lysosomal deficits and neurotoxicity in models of progranulin deficiency. EMBO J. 2022, 41, e109108. [Google Scholar] [CrossRef]
- Wiklund, F.E.; Bennet, A.M.; Magnusson, P.K.; Eriksson, U.K.; Lindmark, F.; Wu, L.; Yaghoutyfam, N.; Marquis, C.P.; Stattin, P.; Pedersen, N.L.; et al. Macrophage inhibitory cytokine-1 (MIC-1/GDF15): A new marker of all-cause mortality. Aging Cell 2010, 9, 1057–1064. [Google Scholar] [CrossRef]
- Breniere, C.; Meloux, A.; Pedard, M.; Marie, C.; Thouant, P.; Vergely, C.; Bejot, Y. Growth Differentiation Factor-15 (GDF-15) Is Associated with Mortality in Ischemic Stroke Patients Treated with Acute Revascularization Therapy. Front. Neurol. 2019, 10, 611. [Google Scholar] [CrossRef]
- Tan, Q.; Hu, C.; Chen, Z.; Jin, T.; Li, L.; Zhu, P.; Ma, Y.; Lin, Z.; Chen, W.; Shi, N.; et al. Growth differentiation factor 15 is an early predictor for persistent organ failure and mortality in acute pancreatitis. Pancreatology 2022, 22, 200–209. [Google Scholar] [CrossRef]
- Patsalos, A.; Halasz, L.; Medina-Serpas, M.A.; Berger, W.K.; Daniel, B.; Tzerpos, P.; Kiss, M.; Nagy, G.; Fischer, C.; Simandi, Z.; et al. A growth factor-expressing macrophage subpopulation orchestrates regenerative inflammation via GDF-15. J. Exp. Med. 2022, 219, e20210420. [Google Scholar] [CrossRef] [PubMed]
- Hassanpour Golakani, M.; Mohammad, M.G.; Li, H.; Gamble, J.; Breit, S.N.; Ruitenberg, M.J.; Brown, D.A. MIC-1/GDF15 Overexpression Is Associated with Increased Functional Recovery in Traumatic Spinal Cord Injury. J. Neurotrauma 2019, 36, 3410–3421. [Google Scholar] [CrossRef]
- Jutila, D.B.; Kurk, S.; Jutila, M.A. Differences in the expression of Ly-6C on neutrophils and monocytes following PI-PLC hydrolysis and cellular activation. Immunol. Lett. 1994, 41, 49–57. [Google Scholar] [CrossRef]
- Saiwai, H.; Kumamaru, H.; Ohkawa, Y.; Kubota, K.; Kobayakawa, K.; Yamada, H.; Yokomizo, T.; Iwamoto, Y.; Okada, S. Ly6C+ Ly6G- Myeloid-derived suppressor cells play a critical role in the resolution of acute inflammation and the subsequent tissue repair process after spinal cord injury. J. Neurochem. 2013, 125, 74–88. [Google Scholar] [CrossRef]
- Zhao, X.F.; Alam, M.M.; Liao, Y.; Huang, T.; Mathur, R.; Zhu, X.; Huang, Y. Targeting Microglia Using Cx3cr1-Cre Lines: Revisiting the Specificity. eNeuro 2019, 6, ENEURO.0114-19.2019. [Google Scholar] [CrossRef] [PubMed]
- Wieghofer, P.; Prinz, M. Genetic manipulation of microglia during brain development and disease. Biochim. Biophys. Acta 2016, 1862, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, X.M.; Yuan, J.B.; Li, L.L.; Wang, C.; Lin, X.M.; Miao, X.; Shi, Z.C. MiR-665 inhibits inflammatory response in microglia following spinal cord injury by targeting TREM2. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 65–70. [Google Scholar] [CrossRef]
- Gao, H.; Di, J.; Clausen, B.H.; Wang, N.; Zhu, X.; Zhao, T.; Chang, Y.; Pang, M.; Yang, Y.; He, R.; et al. Distinct myeloid population phenotypes dependent on TREM2 expression levels shape the pathology of traumatic versus demyelinating CNS disorders. Cell Rep. 2023, 42, 112773. [Google Scholar] [CrossRef]
- Guo, Y.; Xue, Z.; Yang, B.; Liu, L.; Zhang, P.; Shi, J.; Fu, X.; Xue, Y.; Hao, Y.; Ji, G. Effects of intrathecally administered interferon alpha on chronic constriction injury model rats’ mechanical pain threshold and G protein expression in the spinal cord. Folia Neuropathol. 2023, 61, 97–104. [Google Scholar] [CrossRef]
- Lehrman, E.K.; Wilton, D.K.; Litvina, E.Y.; Welsh, C.A.; Chang, S.T.; Frouin, A.; Walker, A.J.; Heller, M.D.; Umemori, H.; Chen, C.; et al. CD47 Protects Synapses from Excess Microglia-Mediated Pruning during Development. Neuron 2018, 100, 120–134.e6. [Google Scholar] [CrossRef]
- Wu, J.; Sabirzhanov, B.; Stoica, B.A.; Lipinski, M.M.; Zhao, Z.; Zhao, S.; Ward, N.; Yang, D.; Faden, A.I. Ablation of the transcription factors E2F1-2 limits neuroinflammation and associated neurological deficits after contusive spinal cord injury. Cell Cycle 2015, 14, 3698–3712. [Google Scholar] [CrossRef]
- Bullard, D.C.; Hu, X.; Schoeb, T.R.; Axtell, R.C.; Raman, C.; Barnum, S.R. Critical requirement of CD11b (Mac-1) on T cells and accessory cells for development of experimental autoimmune encephalomyelitis. J. Immunol. 2005, 175, 6327–6333. [Google Scholar] [CrossRef] [PubMed]
- Rotshenker, S. Microglia and macrophage activation and the regulation of complement-receptor-3 (CR3/MAC-1)-mediated myelin phagocytosis in injury and disease. J. Mol. Neurosci. 2003, 21, 65–72. [Google Scholar] [CrossRef]
- Tak, T.; Rygiel, T.P.; Karnam, G.; Bastian, O.W.; Boon, L.; Viveen, M.; Coenjaerts, F.E.; Meyaard, L.; Koenderman, L.; Pillay, J. Neutrophil-mediated Suppression of Influenza-induced Pathology Requires CD11b/CD18 (MAC-1). Am. J. Respir. Cell Mol. Biol. 2018, 58, 492–499. [Google Scholar] [CrossRef]
- Hyun, Y.M.; Choe, Y.H.; Park, S.A.; Kim, M. LFA-1 (CD11a/CD18) and Mac-1 (CD11b/CD18) distinctly regulate neutrophil extravasation through hotspots I and II. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kantor, A.B.; Akassoglou, K.; Stavenhagen, J.B. Fibrin-Targeting Immunotherapy for Dementia. J. Prev. Alzheimers Dis. 2023, 10, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Yan, L.; Zhang, D.; Zhao, Y. Passive immunotherapy for Alzheimer’s disease. Ageing Res. Rev. 2024, 94, 102192. [Google Scholar] [CrossRef] [PubMed]
- Delrieu, J.; Andrieu, S.; Vellas, B. Dementia research in 2023: The year of anti-amyloid immunotherapy. Lancet Neurol. 2024, 23, 13–15. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Tan, E.K. Immunotherapy trials in Parkinson’s disease: Challenges. J. Transl. Med. 2023, 21, 178. [Google Scholar] [CrossRef]
- Palermo, G.; Ceravolo, R. Clues to Reshaping Anti-alpha-Synuclein Immunotherapy in Early Parkinson’s Disease Patients. Mov. Disord. 2023, 38, 196–197. [Google Scholar] [CrossRef] [PubMed]
- Carreno-Campos, C.; Govea-Alonso, D.O.; Villarreal, M.L.; Caltempa, A.O.; Gonzalez-Ortega, O.; Rosales-Mendoza, S. Production and characterization of a chimeric protein targeting synuclein epitopes for immunotherapy against synucleinopathies. Biotechnol. Prog. 2023, 39, e3390. [Google Scholar] [CrossRef]
- Matsoukas, J.M.; Ligielli, I.; Chasapis, C.T.; Kelaidonis, K.; Apostolopoulos, V.; Mavromoustakos, T. Novel Approaches in the Immunotherapy of Multiple Sclerosis: Cyclization of Myelin Epitope Peptides and Conjugation with Mannan. Brain Sci. 2021, 11, 1583. [Google Scholar] [CrossRef]
- Alrashoudi, R.H. Unleashing the power of anti-CD20 immunotherapy: Mitigating multiple sclerosis risk in Epstein-Barr virus latent infections. Adv. Clin. Exp. Med. 2023, 33, 869–880. [Google Scholar] [CrossRef]
- Lehnert, T.; Rover, C.; Kopke, S.; Rio, J.; Chard, D.; Fittipaldo, A.V.; Friede, T.; Heesen, C.; Rahn, A.C. Immunotherapy for people with clinically isolated syndrome or relapsing-remitting multiple sclerosis: Treatment response by demographic, clinical, and biomarker subgroups (PROMISE)-a systematic review protocol. Syst. Rev. 2022, 11, 134. [Google Scholar] [CrossRef]
- Alipour, M.; Tebianian, M.; Tofigh, N.; Taheri, R.S.; Mousavi, S.A.; Naseri, A.; Ahmadi, A.; Munawar, N.; Shahpasand, K. Active immunotherapy against pathogenic Cis pT231-tau suppresses neurodegeneration in traumatic brain injury mouse models. Neuropeptides 2022, 96, 102285. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.; Anto-Michel, N.; Blankenbach, H.; Wiedemann, A.; Buscher, K.; Hohmann, J.D.; Lim, B.; Bauml, M.; Marki, A.; Mauler, M.; et al. A ligand-specific blockade of the integrin Mac-1 selectively targets pathologic inflammation while maintaining protective host-defense. Nat. Commun. 2018, 9, 525. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Lei, Z.; Ritzel, R.M.; He, J.; Liu, S.; Zhang, L.; Wu, J. Ablation of the Integrin CD11b Mac-1 Limits Deleterious Responses to Traumatic Spinal Cord Injury and Improves Functional Recovery in Mice. Cells 2024, 13, 1584. https://doi.org/10.3390/cells13181584
Li Y, Lei Z, Ritzel RM, He J, Liu S, Zhang L, Wu J. Ablation of the Integrin CD11b Mac-1 Limits Deleterious Responses to Traumatic Spinal Cord Injury and Improves Functional Recovery in Mice. Cells. 2024; 13(18):1584. https://doi.org/10.3390/cells13181584
Chicago/Turabian StyleLi, Yun, Zhuofan Lei, Rodney M. Ritzel, Junyun He, Simon Liu, Li Zhang, and Junfang Wu. 2024. "Ablation of the Integrin CD11b Mac-1 Limits Deleterious Responses to Traumatic Spinal Cord Injury and Improves Functional Recovery in Mice" Cells 13, no. 18: 1584. https://doi.org/10.3390/cells13181584
APA StyleLi, Y., Lei, Z., Ritzel, R. M., He, J., Liu, S., Zhang, L., & Wu, J. (2024). Ablation of the Integrin CD11b Mac-1 Limits Deleterious Responses to Traumatic Spinal Cord Injury and Improves Functional Recovery in Mice. Cells, 13(18), 1584. https://doi.org/10.3390/cells13181584






