miR-409-3p Regulates IFNG and p16 Signaling in the Human Blood of Aging-Related Hearing Loss
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Study Population
2.2. Isolation of Peripheral Blood Mononuclear Cells (PBMCs) from Human Blood and the mRNA Differentiation Expression Assay
2.3. Quantifying Gene Expression and Differentially Expressed Gene Analysis
2.4. Validation of Target Gene Expression (RT-qPCR)
2.5. Gene Ontology Enrichment Analysis for miRNA Related to IFNG
2.6. UTR Vector Construction and miRNA
2.7. Cell Culture and Transfection
2.8. Dual-Luciferase Reporter Assay
2.9. Western Blot Analysis
2.10. Statistical Analysis
3. Results
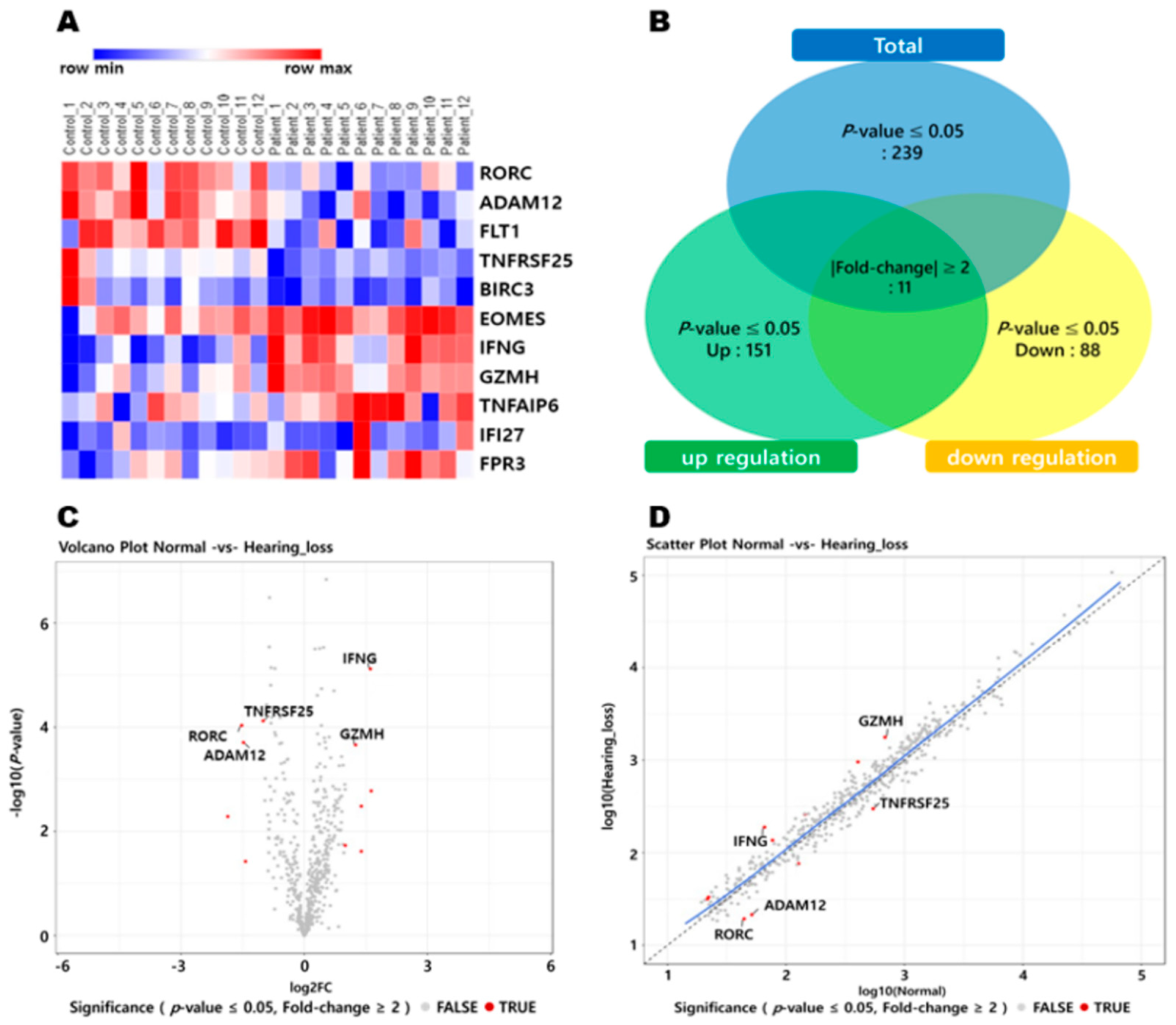
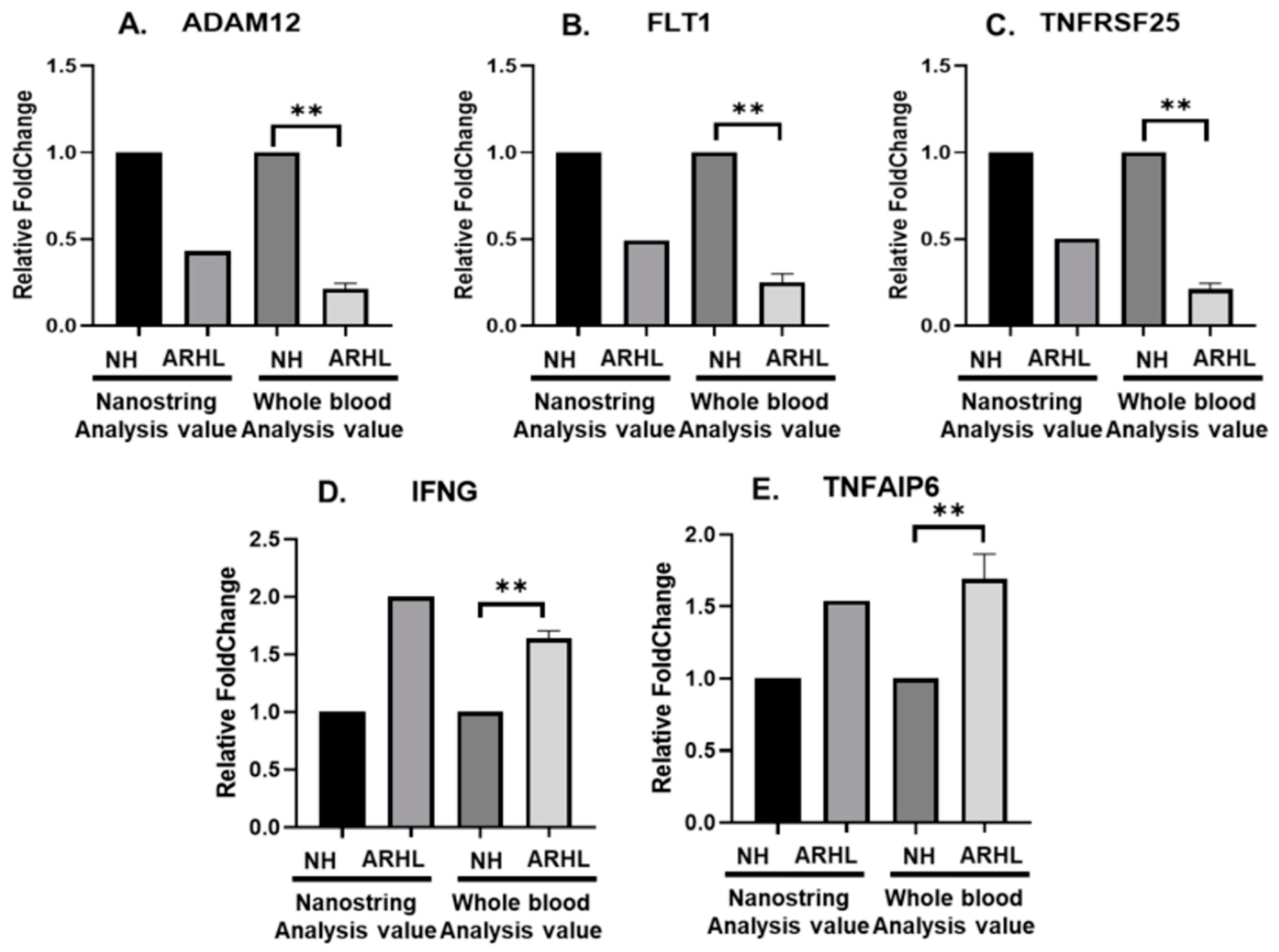
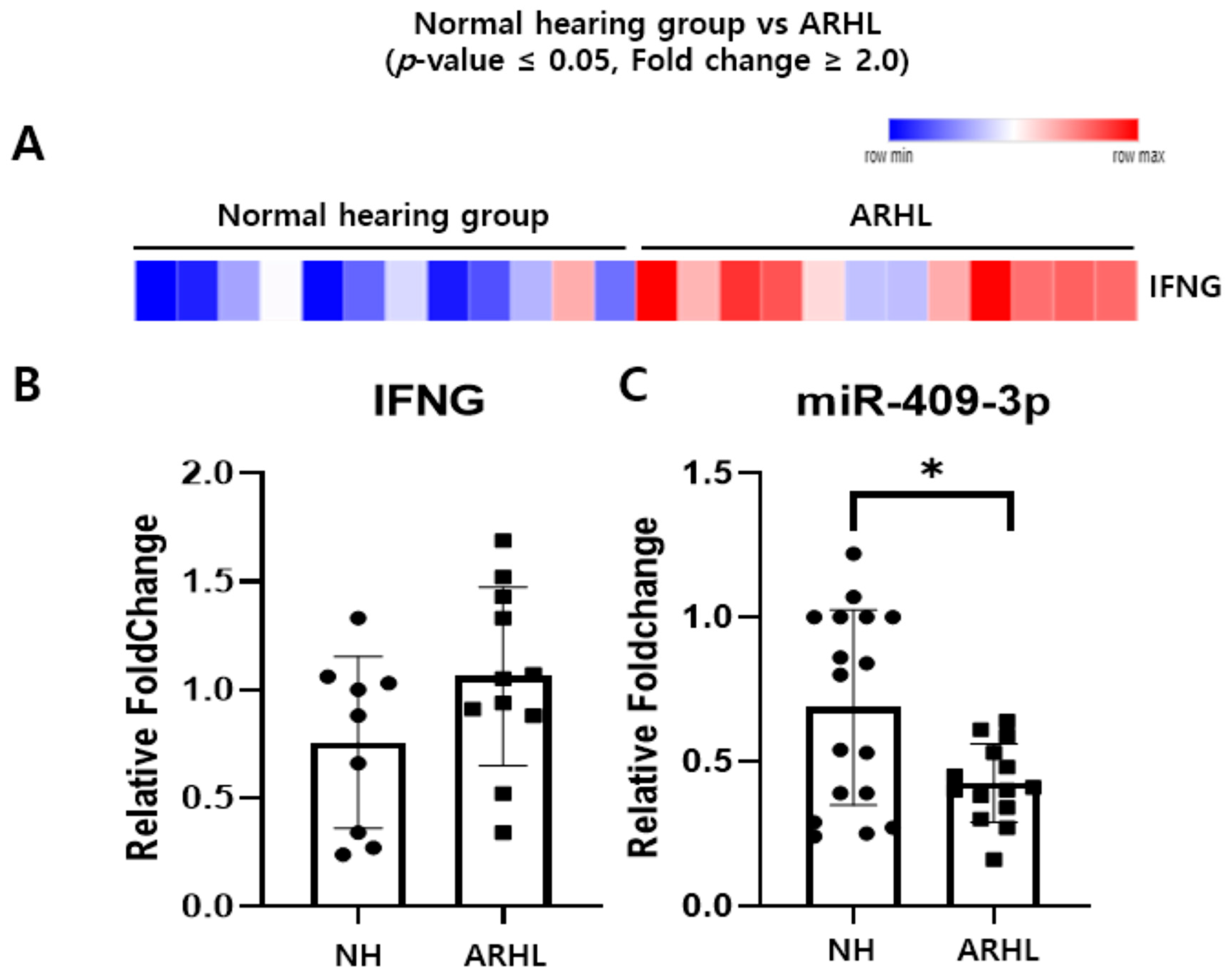
3.1. Inverse Correlation of IFNG and Putative Target miRNAs in the Whole Blood between NH and ARHL Groups
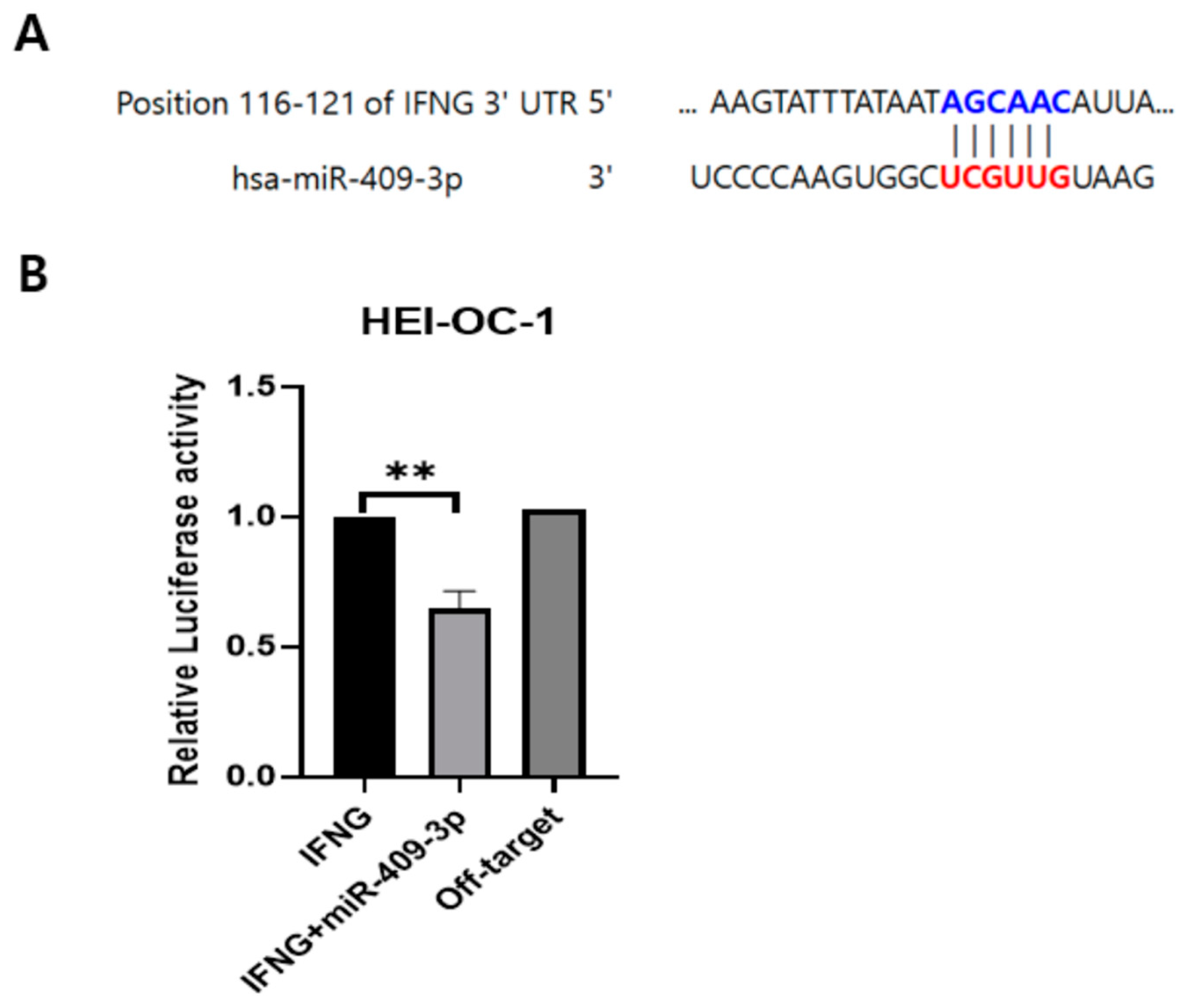
3.2. IFNG Is a Direct Target of miR-409-3p in HEI-OC-1 Cells
3.3. miR-409-3p Regulates IFNG and p16 Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Watson, N.; Ding, B.; Zhu, X.; Frisina, R.D. Chronic Inflammation—Inflammaging—in the Ageing Cochlea: A Novel Target for Future Presbycusis Therapy. Ageing Res. Rev. 2017, 40, 142–148. [Google Scholar] [CrossRef]
- Harris, J.P. Immunology of the Inner Ear: Response of the Inner Ear to Antigen Challenge. Otolaryngol. Head Neck Surg. 1983, 91, 18–32. [Google Scholar] [CrossRef] [PubMed]
- Kociszewska, D.; Chan, J.; Thorne, P.R.; Vlajkovic, S.M. The Link between Gut Dysbiosis Caused by a High-Fat Diet and Hearing Loss. Int. J. Mol. Sci. 2021, 22, 13177. [Google Scholar] [CrossRef] [PubMed]
- Kämpfe Nordström, C.; Danckwardt-Lillieström, N.; Laurell, G.; Liu, W.; Rask-Andersen, H. The Human Endolymphatic Sac and Inner Ear Immunity: Macrophage Interaction and Molecular Expression. Front. Immunol. 2018, 9, 3181. [Google Scholar] [CrossRef] [PubMed]
- Satoh, H.; Firestein, G.S.; Billings, P.B.; Harris, J.P.; Keithley, E.M. Pro-inflammatory Cytokine Expression in the Endolymphatic Sac during Inner Ear Inflammation. J. Assoc. Res. Otolaryngol. 2003, 4, 139–147. [Google Scholar] [CrossRef]
- Satoh, H.; Billings, P.; Firestein, G.S.; Harris, J.P.; Keithley, E.M. Transforming Growth Factor Beta Expression during an Inner Ear Immune Response. Ann. Otol. Rhinol. Laryngol. 2006, 115, 81–88. [Google Scholar] [CrossRef]
- Verschuur, C.A.; Dowell, A.; Syddall, H.E.; Ntani, G.; Simmonds, S.J.; Baylis, D.; Gale, C.R.; Walsh, B.; Cooper, C.; Lord, J.M.; et al. Markers of Inflammatory Status Are Associated with Hearing Threshold in Older People: Findings from the Hertfordshire Ageing Study. Age Ageing 2012, 41, 92–97. [Google Scholar] [CrossRef]
- Lowthian, J.A.; Britt, C.J.; Rance, G.; Lin, F.R.; Woods, R.L.; Wolfe, R.; Nelson, M.R.; Dillon, H.A.; Ward, S.; Reid, C.M.; et al. Slowing the Progression of Age-Related Hearing Loss: Rationale and Study Design of the ASPIRIN in HEARING, Retinal Vessels Imaging and Neurocognition in Older Generations (ASPREE-HEARING) Trial. Contemp. Clin. Trials 2016, 46, 60–66. [Google Scholar] [CrossRef]
- Trpchevska, N.; Freidin, M.B.; Broer, L.; Oosterloo, B.C.; Yao, S.; Zhou, Y.; Vona, B.; Bishop, C.; Bizaki-Vallaskangas, A.; Canlon, B.; et al. Genome-Wide Association Meta-Analysis Identifies 48 Risk Variants and Highlights the Role of the Stria Vascularis in Hearing Loss. Am. J. Hum. Genet. 2022, 109, 1077–1091. [Google Scholar] [CrossRef]
- Ivarsdottir, E.V.; Holm, H.; Benonisdottir, S.; Olafsdottir, T.; Sveinbjornsson, G.; Thorleifsson, G.; Eggertsson, H.P.; Halldorsson, G.H.; Hjorleifsson, K.E.; Melsted, P.; et al. The Genetic Architecture of Age-Related Hearing Impairment Revealed by Genome-Wide Association Analysis. Commun. Biol. 2021, 4, 706. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, H.; McGee, J.; Walsh, E.J.; Soukup, G.A.; He, D.Z. Identifying MicroRNAs Involved in Degeneration of the Organ of Corti during Age-Related Hearing Loss. PLoS ONE 2013, 8, e62786. [Google Scholar] [CrossRef] [PubMed]
- Frucht, C.S.; Santos-Sacchi, J.; Navaratnam, D.S. MicroRNA181a Plays a Key Role in Hair Cell Regeneration in the Avian Auditory Epithelium. Neurosci. Lett. 2011, 493, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Hermeking, H. The MiR-34 Family in Cancer and Apoptosis. Cell Death Differ. 2010, 17, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Herron, S.; Silveira, S.; Kleemann, K.; Gauthier, C.; Mallah, D.; Cheng, Y.; Margeta, M.A.; Pitts, K.M.; Barry, J.L.; et al. Identification of a Protective Microglial State Mediated by MiR-155 and Interferon-γ Signaling in a Mouse Model of Alzheimer’s Disease. Nat. Neurosci. 2023, 26, 1196–1207. [Google Scholar] [CrossRef] [PubMed]
- Amado, T.; Amorim, A.; Enguita, F.J.; Romero, P.V.; Inácio, D.; de Miranda, M.P.; Winter, S.J.; Simas, J.P.; Krueger, A.; Schmolka, N.; et al. MicroRNA-181a Regulates IFN-γ Expression in Effector CD8(+) T Cell Differentiation. J. Mol. Med. 2020, 98, 309–320. [Google Scholar] [CrossRef]
- Guo, W.; Wu, Z.; Chen, J.; Guo, S.; You, W.; Wang, S.; Ma, J.; Wang, H.; Wang, X.; Wang, H.; et al. Nanoparticle Delivery of MiR-21-3p Sensitizes Melanoma to Anti-PD-1 Immunotherapy by Promoting Ferroptosis. J. Immunother. Cancer 2022, 10, e004381. [Google Scholar] [CrossRef]
- NanoString Technologies®, Inc. nSolverTM Analysis Software. Available online: https://www.nanostring.com/products/analysis-software/nsolver (accessed on 27 December 2019).
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, Research0034. [Google Scholar] [CrossRef]
- NanoStringTechnologies®, Inc. nSolverTM Adavanced Analysis Software. Available online: https://www.nanostring.com/products/analysis-software/advanced-analysis (accessed on 27 December 2019).
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. G:Profiler-a Web Server for Functional Interpretation of Gene Lists (2016 Update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.D.; Lin, F.M.; Wu, W.Y.; Liang, C.; Huang, W.C.; Chan, W.L.; Tsai, W.T.; Chen, G.Z.; Lee, C.J.; Chiu, C.M.; et al. MiRTarBase: A Database Curates Experimentally Validated MicroRNA-Target Interactions. Nucleic Acids Res. 2011, 39, D163–D169. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Annotating High Confidence MicroRNAs Using Deep Sequencing Data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Ivashkiv, L.B. IFNγ: Signalling, Epigenetics and Roles in Immunity, Metabolism, Disease and Cancer Immunotherapy. Nat. Rev. Immunol. 2018, 18, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Hubackova, S.; Kucerova, A.; Michlits, G.; Kyjacova, L.; Reinis, M.; Korolov, O.; Bartek, J.; Hodny, Z. IFNγ Induces Oxidative Stress, DNA Damage and Tumor Cell Senescence via TGFβ/SMAD Signaling-Dependent Induction of Nox4 and Suppression of ANT2. Oncogene 2016, 35, 1236–1249. [Google Scholar] [CrossRef]
- Volpe, E.A.; Henriksson, J.T.; Wang, C.; Barbosa, F.L.; Zaheer, M.; Zhang, X.; Pflugfelder, S.C.; de Paiva, C.S. Interferon-Gamma Deficiency Protects against Aging-Related Goblet Cell Loss. Oncotarget 2016, 7, 64605–64614. [Google Scholar] [CrossRef]
- Lorenz, R.R.; Solares, C.A.; Williams, P.; Sikora, J.; Pelfrey, C.M.; Hughes, G.B.; Tuohy, V.K. Interferon-Gamma Production to Inner Ear Antigens by T Cells from Patients with Autoimmune Sensorineural Hearing Loss. J. Neuroimmunol. 2002, 130, 173–178. [Google Scholar] [CrossRef]
- Moon, S.K.; Woo, J.I.; Lim, D.J. Involvement of TNF-α and IFN-γ in Inflammation-Mediated Cochlear Injury. Ann. Otol. Rhinol. Laryngol. 2019, 128 (Suppl. S6), 8s–15s. [Google Scholar] [CrossRef]
- Miguel, V.; Cui, J.Y.; Daimiel, L.; Espinosa-Díez, C.; Fernández-Hernando, C.; Kavanagh, T.J.; Lamas, S. The Role of MicroRNAs in Environmental Risk Factors, Noise-Induced Hearing Loss, and Mental Stress. Antioxid. Redox. Signal. 2018, 28, 773–796. [Google Scholar] [CrossRef]
- Pang, J.; Xiong, H.; Yang, H.; Ou, Y.; Xu, Y.; Huang, Q.; Lai, L.; Chen, S.; Zhang, Z.; Cai, Y.; et al. Circulating MiR-34a Levels Correlate with Age-Related Hearing Loss in Mice and Humans. Exp. Gerontol. 2016, 76, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.N.; Wu, Y.J.; Lee, H.I.; Wang, H.H.; Hung, C.L.; Chang, C.Y.; Chou, Y.H.; Tien, T.Y.; Lee, C.W.; Lin, C.F.; et al. Hsa-MiR-409-3p Regulates Endothelial Progenitor Senescence via PP2A-P38 and Is a Potential Ageing Marker in Humans. J. Cell. Mol. Med. 2023, 27, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Toyama, K.; Spin, J.M.; Deng, A.C.; Abe, Y.; Tsao, P.S.; Mogi, M. Role of MicroRNAs in Acceleration of Vascular Endothelial Senescence. Biochem. Biophys. Rep. 2022, 30, 101281. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhao, H.; Xue, F.; Zhang, X.; Zhang, D.; Ge, J.; Yang, Y.; Xuan, M.; Fu, R.; Yang, R. Reduced Expression of MIR409-3p in Primary Immune Thrombocytopenia. Br. J. Haematol. 2013, 161, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Yin and Yang Interplay of IFN-Gamma in Inflammation and Autoimmune Disease. J. Clin. Investig. 2007, 117, 871–873. [Google Scholar] [CrossRef]
- Sharpless, N.E.; Sherr, C.J. Forging a Signature of in Vivo Senescence. Nat. Rev. Cancer 2015, 15, 397–408. [Google Scholar] [CrossRef]
- Chen, J.; Huang, X.; Halicka, D.; Brodsky, S.; Avram, A.; Eskander, J.; Bloomgarden, N.A.; Darzynkiewicz, Z.; Goligorsky, M.S. Contribution of P16INK4a and P21CIP1 Pathways to Induction of Premature Senescence of Human Endothelial Cells: Permissive Role of P53. Am. J. Physiol. Heart Circ. Physiol. 2006, 290, H1575–H1586. [Google Scholar] [CrossRef]
- Chen, J.; Li, S.; Li, C. Mechanisms of Melanocyte Death in Vitiligo. Med. Res. Rev. 2021, 41, 1138–1166. [Google Scholar] [CrossRef]
- Sviderskaya, E.V.; Gray-Schopfer, V.C.; Hill, S.P.; Smit, N.P.; Evans-Whipp, T.J.; Bond, J.; Hill, L.; Bataille, V.; Peters, G.; Kipling, D.; et al. P16/Cyclin-Dependent Kinase Inhibitor 2A Deficiency in Human Melanocyte Senescence, Apoptosis, and Immortalization: Possible Implications for Melanoma Progression. J. Natl. Cancer Inst. 2003, 95, 723–732. [Google Scholar] [CrossRef]
- Rayess, H.; Wang, M.B.; Srivatsan, E.S. Cellular Senescence and Tumor Suppressor Gene P16. Int. J. Cancer 2012, 130, 1715–1725. [Google Scholar] [CrossRef]
- Safwan-Zaiter, H.; Wagner, N.; Wagner, K.-D. P16INK4A—More Than a Senescence Marker. Life 2022, 12, 1332. [Google Scholar] [CrossRef] [PubMed]
- LaPak, K.M.; Burd, C.E. The Molecular Balancing Act of P16(INK4a) in Cancer and Aging. Mol. Cancer Res. 2014, 12, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Almontashiri, N.A.; Fan, M.; Cheng, B.L.; Chen, H.H.; Roberts, R.; Stewart, A.F. Interferon-γ Activates Expression of P15 and P16 Regardless of 9p21.3 Coronary Artery Disease Risk Genotype. J. Am. Coll. Cardiol. 2013, 61, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Seelig, H.P.; Ehrfeld, H.; Renz, M. Interferon-Gamma-Inducible Protein P16. A New Target of Antinuclear Antibodies in Patients with Systemic Lupus Erythematosus. Arthritis Rheum. 1994, 37, 1672–1683. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, V.; Mukherjea, D.; Dhukhwa, A.; Rybak, L.P. Oxidative Stress and Inflammation Caused by Cisplatin Ototoxicity. Antioxidants 2021, 10, 1919. [Google Scholar] [CrossRef]
- Prasad, K.N.; Bondy, S.C. MicroRNAs in Hearing Disorders: Their Regulation by Oxidative Stress, Inflammation and Antioxidants. Front. Cell. Neurosci. 2017, 11, 276. [Google Scholar] [CrossRef]
- Linton, P.J.; Dorshkind, K. Age-Related Changes in Lymphocyte Development and Function. Nat. Immunol. 2004, 5, 133–139. [Google Scholar] [CrossRef]
- Liu, W.; Danckwardt-Lillieström, N.; Schrott-Fischer, A.; Glueckert, R.; Rask-Andersen, H. Distribution of Immune Cells Including Macrophages in the Human Cochlea. Front. Neurol. 2021, 12, 781702. [Google Scholar] [CrossRef]
- Liu, W.; Rask-Andersen, H. Super-Resolution Immunohistochemistry Study on CD4 and CD8 Cells and the Relation to Macrophages in Human Cochlea. J. Otol. 2019, 14, 1–5. [Google Scholar] [CrossRef]
- Hirose, K.; Discolo, C.M.; Keasler, J.R.; Ransohoff, R. Mononuclear Phagocytes Migrate into the Murine Cochlea after Acoustic Trauma. J. Comp. Neurol. 2005, 489, 180–194. [Google Scholar] [CrossRef]
- Okano, T.; Nakagawa, T.; Kita, T.; Kada, S.; Yoshimoto, M.; Nakahata, T.; Ito, J. Bone Marrow-Derived Cells Expressing Iba1 Are Constitutively Present as Resident Tissue Macrophages in the Mouse Cochlea. J. Neurosci. Res. 2008, 86, 1758–1767. [Google Scholar] [CrossRef] [PubMed]

| Name | Forward Primer | Reverse Primer |
|---|---|---|
| ADAM12 | CAGGAAGGCTTGTTGTGCTT | TTGGGATCTCTTGAGCTGCA |
| FLT1 | GACTGACAGCAAACCCAAGG | TAGATGGGTGGGGTGGAGTA |
| TNFRSF25 | TTCTAGCACCTCCTGACAGC | ACAGGAGAATGGGGTCAAGG |
| IFNG | GGGGCTCAGTTTCCTCATCT | TAGAGACTTGCAGTGGGGTG |
| TNFAIP6 | TACTGGGAAGTTTGGCGCTA | GTTCCTCTCCCTTCTCCCAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, J.; Lee, J.; Kang, H.; Park, K.; Kim, Y.S.; Ha, J.; So, S.; Sung, S.; Yun, J.H.; Jang, J.H.; et al. miR-409-3p Regulates IFNG and p16 Signaling in the Human Blood of Aging-Related Hearing Loss. Cells 2024, 13, 1595. https://doi.org/10.3390/cells13181595
Jung J, Lee J, Kang H, Park K, Kim YS, Ha J, So S, Sung S, Yun JH, Jang JH, et al. miR-409-3p Regulates IFNG and p16 Signaling in the Human Blood of Aging-Related Hearing Loss. Cells. 2024; 13(18):1595. https://doi.org/10.3390/cells13181595
Chicago/Turabian StyleJung, Junseo, Jeongmin Lee, Hyunsook Kang, Kyeongjin Park, Young Sun Kim, Jungho Ha, Seongjun So, Siung Sung, Jeong Hyeon Yun, Jeong Hun Jang, and et al. 2024. "miR-409-3p Regulates IFNG and p16 Signaling in the Human Blood of Aging-Related Hearing Loss" Cells 13, no. 18: 1595. https://doi.org/10.3390/cells13181595






