Synthetic Promoters in Gene Therapy: Design Approaches, Features and Applications
Abstract
1. Introduction
2. Natural Promoters
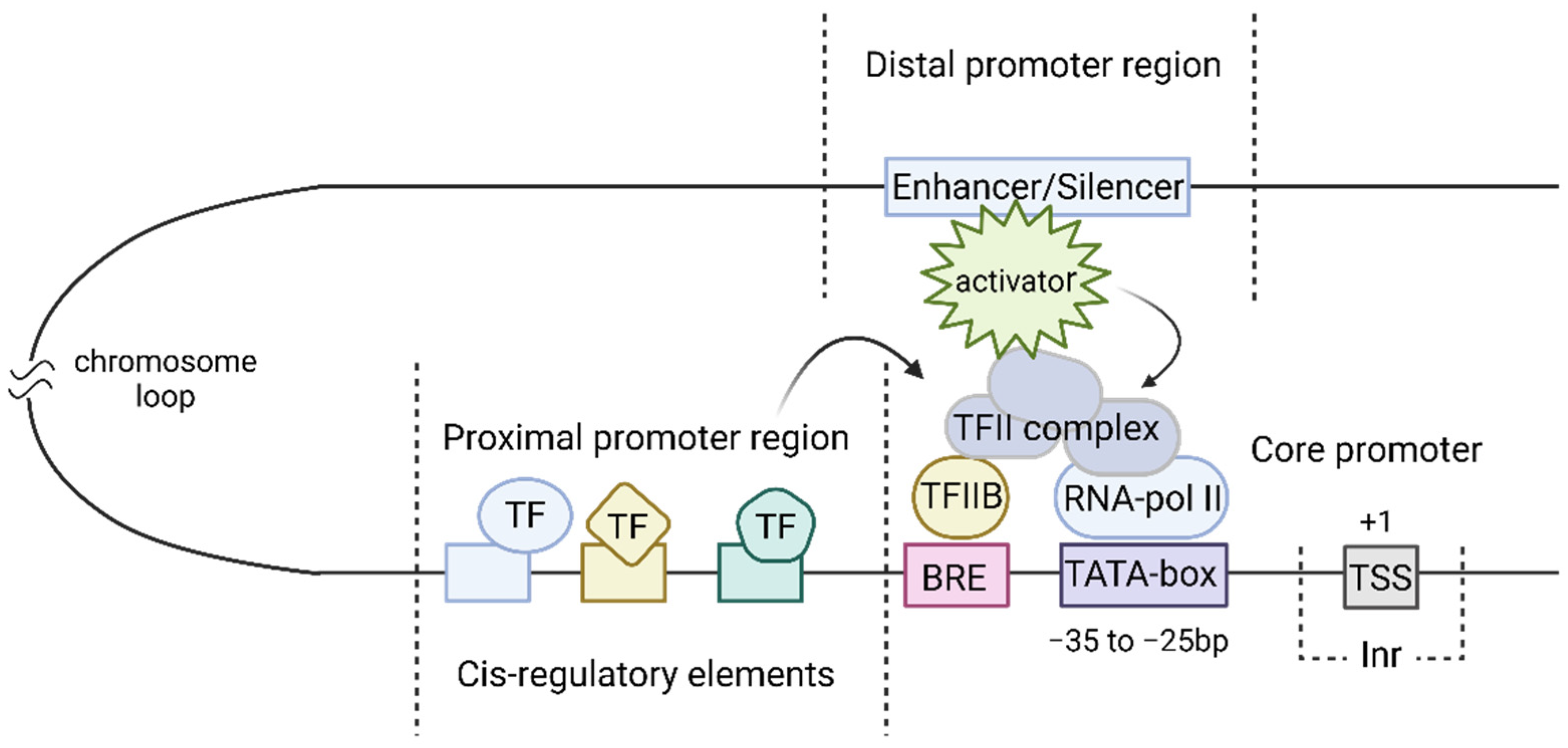
2.1. Structure of the Eukaryotic Natural Promoter
2.2. Modern Approaches for Recognizing CREs
3. Synthetic Promoters
3.1. Synthetic Promoters’ Manual Design
3.2. Computational Design of Synthetic Promoters
3.3. Synthetic Promoter Evaluation Methods
4. Synthetic Promoter Applications
4.1. Liver-Specific Promoters
4.2. Muscle-Specific Promoters
4.3. Eye-Specific Promoters
4.4. CNS-Specific Promoters
4.5. Tumor-Specific Promoters
5. Conclusions and Future Prospects
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baird, P.A.; Anderson, T.W.; Newcombe, H.B.; Lowry, R.B. Genetic Disorders in Children and Young Adults: A Population Study. Am. J. Hum. Genet. 1988, 42, 677–693. [Google Scholar] [PubMed]
- Spark Therapeutics Inc. LUXTURNA (Voretigene Neparvovec-Rzyl). Available online: https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/luxturna (accessed on 21 October 2024).
- Novartis Gene Therapies Inc. ZOLGENSMA (Onasemnogene Abeparvovec-Xioi). Available online: https://www.fda.gov/vaccines-blood-biologics/zolgensma (accessed on 21 October 2024).
- Sarepta Therapeutics Inc. ELEVIDYS (Delandistrogene Moxeparvovec-Rokl). Available online: https://www.fda.gov/vaccines-blood-biologics/tissue-tissue-products/elevidys (accessed on 21 October 2024).
- BioMarin Pharmaceutical Inc. ROCTAVIAN (Valoctocogene Roxaparvovec-Rvox). Available online: https://www.fda.gov/vaccines-blood-biologics/roctavian (accessed on 21 October 2024).
- CSL Behring LLC. HEMGENIX (Etranacogene Dezaparvovec-Drlb). Available online: https://www.fda.gov/vaccines-blood-biologics/vaccines/hemgenix (accessed on 21 October 2024).
- Wang, D.; Tai, P.W.L.; Gao, G. Adeno-Associated Virus Vector as a Platform for Gene Therapy Delivery. Nat. Rev. Drug Discov. 2019, 18, 358–378. [Google Scholar] [CrossRef] [PubMed]
- Chira, S.; Jackson, C.S.; Oprea, I.; Ozturk, F.; Pepper, M.S.; Diaconu, I.; Braicu, C.; Raduly, L.Z.; Calin, G.A.; Berindan-Neagoe, I. Progresses towards Safe and Efficient Gene Therapy Vectors. Oncotarget 2015, 6, 30675–30703. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Nasu, Y.; Kusumi, N.; Nagai, A.; Kumon, H.; Kashiwakura, Y. 238: Adeno-Associated Virus 2-Mediated Intratumoral Prostate Cancer Gene Therapy: Long-Term Maspin Expression Efficiently Suppresses Tumor Growth. J. Urol. 2005, 173, 65. [Google Scholar] [CrossRef]
- Ng, S.S.M.; Gao, Y.; Chau, D.H.W.; Li, G.H.Y.; Lai, L.H.; Huang, P.T.; Huang, C.F.; Huang, J.J.; Chen, Y.C.; Kung, H.F.; et al. A Novel Glioblastoma Cancer Gene Therapy Using AAV-Mediated Long-Term Expression of Human TERT C-Terminal Polypeptide. Cancer Gene Ther. 2007, 14, 561–572. [Google Scholar] [CrossRef]
- Hacker, U.T.; Bentler, M.; Kaniowska, D.; Morgan, M.; Büning, H. Towards Clinical Implementation of Adeno-Associated Virus (Aav) Vectors for Cancer Gene Therapy: Current Status and Future Perspectives. Cancers 2020, 12, 1889. [Google Scholar] [CrossRef]
- He, L.F.; Wang, Y.G.; Xiao, T.; Zhang, K.J.; Li, G.C.; Gu, J.F.; Chu, L.; Tang, W.H.; Tan, W.S.; Liu, X.Y. Suppression of Cancer Growth in Mice by Adeno-Associated Virus Vector-Mediated IFN-β Expression Driven by HTERT Promoter. Cancer Lett. 2009, 286, 196–205. [Google Scholar] [CrossRef]
- Münch, R.C.; Muth, A.; Muik, A.; Friedel, T.; Schmatz, J.; Dreier, B.; Trkola, A.; Plückthun, A.; Büning, H.; Buchholz, C.J. Off-Target-Free Gene Delivery by Affinity-Purified Receptor-Targeted Viral Vectors. Nat. Commun. 2015, 6, 6246. [Google Scholar] [CrossRef]
- Duan, D. Lethal Immunotoxicity in High-Dose Systemic AAV Therapy. Mol. Ther. 2023, 31, 3123–3126. [Google Scholar] [CrossRef]
- Korneyenkov, M.A.; Zamyatnin, A.A. Next Step in Gene Delivery: Modern Approaches and Further Perspectives of Aav Tropism Modification. Pharmaceutics 2021, 13, 750. [Google Scholar] [CrossRef]
- Lee, E.J.; Guenther, C.M.; Suh, J. Adeno-Associated Virus (AAV) Vectors: Rational Design Strategies for Capsid Engineering. Curr. Opin. Biomed. Eng. 2018, 7, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Ghauri, M.S.; Ou, L. AAV Engineering for Improving Tropism to the Central Nervous System. Biology 2023, 12, 186. [Google Scholar] [CrossRef] [PubMed]
- Paulk, N.K.; Pekrun, K.; Zhu, E.; Nygaard, S.; Li, B.; Xu, J.; Chu, K.; Leborgne, C.; Dane, A.P.; Haft, A.; et al. Bioengineered AAV Capsids with Combined High Human Liver Transduction In Vivo and Unique Humoral Seroreactivity. Mol. Ther. 2018, 26, 289–303. [Google Scholar] [CrossRef]
- Cabanes-Creus, M.; Hallwirth, C.V.; Westhaus, A.; Ng, B.H.; Liao, S.H.Y.; Zhu, E.; Gale, R.; Baltazar, G.; Drouyer, M.; Scott, S.; et al. Restoring the Natural Tropism of AAV2 Vectors for Human Liver. Sci. Transl. Med. 2020, 12, eaba3312. [Google Scholar] [CrossRef] [PubMed]
- Pavlou, M.; Schön, C.; Occelli, L.M.; Rossi, A.; Meumann, N.; Boyd, R.F.; Bartoe, J.T.; Siedlecki, J.; Gerhardt, M.J.; Babutzka, S.; et al. Novel AAV Capsids for Intravitreal Gene Therapy of Photoreceptor Disorders. EMBO Mol. Med. 2021, 13, e13392. [Google Scholar] [CrossRef]
- Yin, L.; Greenberg, K.; Hunter, J.J.; Dalkara, D.; Kolstad, K.D.; Masella, B.D.; Wolfe, R.; Visel, M.; Stone, D.; Libby, R.T.; et al. Intravitreal Injection of AAV2 Transduces Macaque Inner Retina. Investig. Ophthalmol. Vis. Sci. 2011, 52, 2775–2783. [Google Scholar] [CrossRef]
- Fraldi, A.; Hemsley, K.; Crawley, A.; Lombardi, A.; Lau, A.; Sutherland, L.; Auricchio, A.; Ballabio, A.; Hopwood, J.J. Functional Correction of CNS Lesions in an MPS-IIIA Mouse Model by Intracerebral AAV-Mediated Delivery of Sulfamidase and SUMF1 Genes. Hum. Mol. Genet. 2007, 16, 2693–2702. [Google Scholar] [CrossRef]
- Bosch, A.; Perret, E.; Desmaris, N.; Heard, J.M. Long-Term and Significant Correction of Brain Lesions in Adult Mucopolysaccharidosis Type VII Mice Using Recombinant AAV Vectors. Mol. Ther. 2000, 1, 63–70. [Google Scholar] [CrossRef]
- Zhao, L.; Gottesdiener, A.J.; Parmar, M.; Li, M.; Kaminsky, S.M.; Chiuchiolo, M.J.; Sondhi, D.; Sullivan, P.M.; Holtzman, D.M.; Crystal, R.G.; et al. Intracerebral Adeno-Associated Virus Gene Delivery of Apolipoprotein E2 Markedly Reduces Brain Amyloid Pathology in Alzheimer’s Disease Mouse Models. Neurobiol. Aging 2016, 44, 159–172. [Google Scholar] [CrossRef]
- Fraites, T.J.; Schleissing, M.R.; Shanely, R.A.; Walter, G.A.; Cloutier, D.A.; Zolotukhin, I.; Pauly, D.F.; Raben, N.; Plotz, P.H.; Powers, S.K.; et al. Correction of the Enzymatic and Functional Deficits in a Model of Pompe Disease Using Adeno-Associated Virus Vectors. Mol. Ther. 2002, 5, 571–578. [Google Scholar] [CrossRef]
- Massaro, G.; Geard, A.F.; Nelvagal, H.R.; Gore, K.; Clemo, N.K.; Waddington, S.N.; Rahim, A.A. Comparison of Different Promoters to Improve AAV Vector-Mediated Gene Therapy for Neuronopathic Gaucher Disease. Hum. Mol. Genet. 2024, 33, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Salabarria, S.M.; Nair, J.; Clement, N.; Smith, B.K.; Raben, N.; Fuller, D.D.; Byrne, B.J.; Corti, M. Advancements in AAV-Mediated Gene Therapy for Pompe Disease. J. Neuromuscul. Dis. 2020, 7, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.; Chen, T.; Sidransky, E.; Han, T.U. Advancements in Viral Gene Therapy for Gaucher Disease. Genes 2024, 15, 364. [Google Scholar] [CrossRef]
- Milenkovic, I.; Blumenreich, S.; Hochfelder, A.; Azulay, A.; Biton, I.E.; Zerbib, M.; Oren, R.; Tsoory, M.; Joseph, T.; Fleishman, S.J.; et al. Efficacy of an AAV Vector Encoding a Thermostable Form of Glucocerebrosidase in Alleviating Symptoms in a Gaucher Disease Mouse Model. Gene Ther. 2024, 31, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Au, H.K.E.; Isalan, M.; Mielcarek, M. Gene Therapy Advances: A Meta-Analysis of AAV Usage in Clinical Settings. Front. Med. 2022, 8, 809118. [Google Scholar] [CrossRef]
- Brooks, A.R.; Harkins, R.N.; Wang, P.; Qian, H.S.; Liu, P.; Rubanyi, G.M. Transcriptional Silencing Is Associated with Extensive Methylation of the CMV Promoter Following Adenoviral Gene Delivery to Muscle. J. Gene Med. 2004, 6, 395–404. [Google Scholar] [CrossRef]
- Shirley, J.L.; de Jong, Y.P.; Terhorst, C.; Herzog, R.W. Immune Responses to Viral Gene Therapy Vectors. Mol. Ther. 2020, 28, 709–722. [Google Scholar] [CrossRef]
- Xiong, W.; Wu, D.M.; Xue, Y.; Wang, S.K.; Chung, M.J.; Ji, X.; Rana, P.; Zhao, S.R.; Mai, S.; Cepko, C.L. AAV Cis-Regulatory Sequences Are Correlated with Ocular Toxicity. Proc. Natl. Acad. Sci. USA 2019, 116, 5785–5794. [Google Scholar] [CrossRef]
- Novartis Pharmaceuticals Corporation KYMRIAH (Tisagenlecleucel). Available online: https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/kymriah (accessed on 21 October 2024).
- Labbé, R.P.; Vessillier, S.; Rafiq, Q.A. Lentiviral Vectors for t Cell Engineering: Clinical Applications, Bioprocessing and Future Perspectives. Viruses 2021, 13, 1528. [Google Scholar] [CrossRef]
- Booth, C.; Gaspar, H.B.; Thrasher, A.J. Treating Immunodeficiency through HSC Gene Therapy. Trends Mol. Med. 2016, 22, 317–327. [Google Scholar] [CrossRef]
- Rintz, E.; Higuchi, T.; Kobayashi, H.; Galileo, D.S.; Wegrzyn, G.; Tomatsu, S. Promoter Considerations in the Design of Lentiviral Vectors for Use in Treating Lysosomal Storage Diseases. Mol. Ther. Methods Clin. Dev. 2022, 24, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Parr-Brownlie, L.C.; Bosch-Bouju, C.; Schoderboeck, L.; Sizemore, R.J.; Abraham, W.C.; Hughes, S.M. Lentiviral Vectors as Tools to Understand Central Nervous System Biology in Mammalian Model Organisms. Front. Mol. Neurosci. 2015, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Michels, K.R.; Sheih, A.; Hernandez, S.A.; Brandes, A.H.; Parrilla, D.; Irwin, B.; Perez, A.M.; Ting, H.A.; Nicolai, C.J.; Gervascio, T.; et al. Preclinical Proof of Concept for VivoVec, a Lentiviral-Based Platform for in Vivo CAR T-Cell Engineering. J. Immunother. Cancer 2023, 11, e006292. [Google Scholar] [CrossRef] [PubMed]
- Santilli, G.; Almarza, E.; Brendel, C.; Choi, U.; Beilin, C.; Blundell, M.P.; Haria, S.; Parsley, K.L.; Kinnon, C.; Malech, H.L.; et al. Biochemical Correction of X-CGD by a Novel Chimeric Promoter Regulating High Levels of Transgene Expression in Myeloid Cells. Mol. Ther. 2011, 19, 122–132. [Google Scholar] [CrossRef]
- Garcia-Perez, L.; van Eggermond, M.; van Roon, L.; Vloemans, S.A.; Cordes, M.; Schambach, A.; Rothe, M.; Berghuis, D.; Lagresle-Peyrou, C.; Cavazzana, M.; et al. Successful Preclinical Development of Gene Therapy for Recombinase-Activating Gene-1-Deficient SCID. Mol. Ther. Methods Clin. Dev. 2020, 17, 666–682. [Google Scholar] [CrossRef]
- Levin, M.C.; Lidberg, U.; Jirholt, P.; Adiels, M.; Wramstedt, A.; Gustafsson, K.; Greaves, D.R.; Li, S.; Fazio, S.; Linton, M.F.; et al. Evaluation of Macrophage-Specific Promoters Using Lentiviral Delivery in Mice. Gene Ther. 2012, 19, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Latorre-Rey, L.J.; Wintterle, S.; Dütting, S.; Kohlscheen, S.; Abel, T.; Schenk, F.; Wingert, S.; Rieger, M.A.; Nieswandt, B.; Heinz, N.; et al. Targeting Expression to Megakaryocytes and Platelets by Lineage-Specific Lentiviral Vectors. J. Thromb. Haemost. 2017, 15, 341–355. [Google Scholar] [CrossRef]
- Kerns, H.M.; Ryu, B.Y.; Stirling, B.V.; Sather, B.D.; Astrakhan, A.; Humblet-Baron, S.; Liggitt, D.; Rawlings, D.J. B Cell-Specific Lentiviral Gene Therapy Leads to Sustained B-Cell Functional Recovery in a Murine Model of X-Linked Agammaglobulinemia. Blood 2010, 115, 2146–2155. [Google Scholar] [CrossRef]
- Pucci, F.; Rickelt, S.; Newton, A.P.; Garris, C.; Nunes, E.; Evavold, C.; Pfirschke, C.; Engblom, C.; Mino-Kenudson, M.; Hynes, R.O.; et al. PF4 Promotes Platelet Production and Lung Cancer Growth. Cell Rep. 2016, 17, 1764–1772. [Google Scholar] [CrossRef]
- Montiel-Equihua, C.A.; Zhang, L.; Knight, S.; Saadeh, H.; Scholz, S.; Carmo, M.; Alonso-Ferrero, M.E.; Blundell, M.P.; Monkeviciute, A.; Schulz, R.; et al. The Β-Globin Locus Control Region in Combination with the EF1α Short Promoter Allows Enhanced Lentiviral Vector-Mediated Erythroid Gene Expression with Conserved Multilineage Activity. Mol. Ther. 2012, 20, 1400–1409. [Google Scholar] [CrossRef]
- Moreau-Gaudry, F.; Xia, P.; Jiang, G.; Perelman, N.P.; Bauer, G.; Ellis, J.; Surinya, K.H.; Mavilio, F.; Shen, C.K.; Malik, P. High-Level Erythroid-Specific Gene Expression in Primary Human and Murine Hematopoietic Cells with Self-Inactivating Lentiviral Vectors. Blood 2001, 98, 2664–2672. [Google Scholar] [CrossRef] [PubMed]
- Milone, M.C.; O’Doherty, U. Clinical Use of Lentiviral Vectors. Leukemia 2018, 32, 1529–1541. [Google Scholar] [CrossRef]
- Masiuk, K.E.; Laborada, J.; Roncarolo, M.G.; Hollis, R.P.; Kohn, D.B. Lentiviral Gene Therapy in HSCs Restores Lineage-Specific Foxp3 Expression and Suppresses Autoimmunity in a Mouse Model of IPEX Syndrome. Cell Stem Cell 2019, 24, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, C.T.; VanLith, C.J.; Hickey, R.D.; Du, Z.; Hillin, L.G.; Guthman, R.M.; Cao, W.J.; Haugo, B.; Lillegard, A.; Roy, D.; et al. In Vivo Lentiviral Vector Gene Therapy to Cure Hereditary Tyrosinemia Type 1 and Prevent Development of Precancerous and Cancerous Lesions. Nat. Commun. 2022, 13, 1–15. [Google Scholar] [CrossRef]
- Houghton, B.C.; Booth, C. Gene Therapy for Primary Immunodeficiency. HemaSphere 2021, 5, e509. [Google Scholar] [CrossRef] [PubMed]
- Fassler, J.S.; Gussin, G.N. Promoters and Basal Transcription Machinery in Eubacteria and Eukaryotes: Concepts, Definitions, and Analogies. Methods Enzymol. 1996, 273, 3–29. [Google Scholar] [CrossRef]
- Cooper, S.J.; Trinklein, N.D.; Anton, E.D.; Nguyen, L.; Myers, R.M. Comprehensive Analysis of Transcriptional Promoter Structure and Function in 1% of the Human Genome. Genome Res. 2006, 16, 1–10. [Google Scholar] [CrossRef]
- Carninci, P.; Sandelin, A.; Lenhard, B.; Katayama, S.; Shimokawa, K.; Ponjavic, J.; Semple, C.A.M.; Taylor, M.S.; Engström, P.G.; Frith, M.C.; et al. Genome-Wide Analysis of Mammalian Promoter Architecture and Evolution. Nat. Genet. 2006, 38, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Juven-Gershon, T.; Kadonaga, J.T. Regulation of Gene Expression via the Core Promoter and the Basal Transcriptional Machinery. Dev. Biol. 2010, 339, 225–229. [Google Scholar] [CrossRef]
- Emami, K.H.; Jain, A.; Smale, S.T. Mechanism of Synergy between TATA and Initiator: Synergistic Binding of TFIID Following a Putative TFIIA-Induced Isomerization. Genes Dev. 1997, 11, 3007–3019. [Google Scholar] [CrossRef]
- Zabidi, M.A.; Arnold, C.D.; Schernhuber, K.; Pagani, M.; Rath, M.; Frank, O.; Stark, A. Enhancer-Core-Promoter Specificity Separates Developmental and Housekeeping Gene Regulation. Nature 2015, 518, 556–559. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.E.F.; Kadonaga, J.T. Enhancer-Promoter Specificity Mediated by DPE or TATA Core Promoter Motifs. Genes Dev. 2001, 15, 2515–2519. [Google Scholar] [CrossRef] [PubMed]
- Gershenzon, N.I.; Trifonov, E.N.; Ioshikhes, I.P. The Features of Drosophila Core Promoters Revealed by Statistical Analysis. BMC Genomics 2006, 7, 161. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thomas, M.C.; Chiang, C.M. The General Transcription Machinery and General Cofactors. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 105–178. [Google Scholar] [CrossRef]
- Kadonaga, J.T. Perspectives on the RNA Polymerase II Core Promoter. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 40–51. [Google Scholar] [CrossRef]
- Spitz, F.; Furlong, E.E.M. Transcription Factors: From Enhancer Binding to Developmental Control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and Validation of Promoters and Cis-Acting Regulatory Elements. Plant Sci. 2014, 217, 109–119. [Google Scholar] [CrossRef]
- Powell, S.K.; Rivera-Soto, R.; Gray, S.J. Viral Expression Cassette Elements to Enhance Transgene Target Specificity and Expression in Gene Therapy. Discov. Med. 2015, 19, 49. [Google Scholar]
- Calo, E.; Wysocka, J. Modification of Enhancer Chromatin: What, How, and Why? Mol. Cell 2013, 49, 825–837. [Google Scholar] [CrossRef]
- Huminiecki, Ł.; Horbańczuk, J. Can We Predict Gene Expression by Understanding Proximal Promoter Architecture? Trends Biotechnol. 2017, 35, 530–546. [Google Scholar] [CrossRef]
- Haberle, V.; Stark, A. Eukaryotic Core Promoters and the Functional Basis of Transcription Initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Shlyueva, D.; Stampfel, G.; Stark, A. Transcriptional Enhancers: From Properties to Genome-Wide Predictions. Nat. Rev. Genet. 2014, 15, 272–286. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F. Gene Regulation at a Distance: From Remote Enhancers to 3D Regulatory Ensembles. Semin. Cell Dev. Biol. 2016, 57, 57–67. [Google Scholar] [CrossRef]
- Beagrie, R.A.; Scialdone, A.; Schueler, M.; Kraemer, D.C.A.; Chotalia, M.; Xie, S.Q.; Barbieri, M.; De Santiago, I.; Lavitas, L.M.; Branco, M.R.; et al. Complex Multi-Enhancer Contacts Captured by Genome Architecture Mapping. Nature 2017, 543, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.C.L.; Yang, Y. Identifying and Engineering Promoters for High Level and Sustainable Therapeutic Recombinant Protein Production in Cultured Mammalian Cells. Biotechnol. Lett. 2014, 36, 1569–1579. [Google Scholar] [CrossRef]
- Song, E.S.; Lee, Y.H.; So, M.K.; Kuk, M.U.; Park, J.H.; Yoon, J.H.; Lee, Y.J.; Kim, D.; So, B.; Byun, Y.; et al. Establishment of a New Promoter Trapping Vector Using 2A Peptide. Biotechnol. Bioprocess Eng. 2024, 29, 520–528. [Google Scholar] [CrossRef]
- Nakato, R.; Sakata, T. Methods for ChIP-Seq Analysis: A Practical Workflow and Advanced Applications. Methods 2021, 187, 44–53. [Google Scholar] [CrossRef]
- Zhou, P.; VanDusen, N.J.; Zhang, Y.; Cao, Y.; Sethi, I.; Hu, R.; Zhang, S.; Wang, G.; Ye, L.; Mazumdar, N.; et al. Dynamic Changes in P300 Enhancers and Enhancer-Promoter Contacts Control Mouse Cardiomyocyte Maturation. Dev. Cell 2023, 58, 898–914. [Google Scholar] [CrossRef]
- He, H.H.; Meyer, C.A.; Hu, S.S.; Chen, M.W.; Zang, C.; Liu, Y.; Rao, P.K.; Fei, T.; Xu, H.; Long, H.; et al. Refined DNase-Seq Protocol and Data Analysis Reveals Intrinsic Bias in Transcription Factor Footprint Identification. Nat. Methods 2014, 11, 73–78. [Google Scholar] [CrossRef]
- Giresi, P.G.; Kim, J.; McDaniell, R.M.; Iyer, V.R.; Lieb, J.D. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) Isolates Active Regulatory Elements from Human Chromatin. Genome Res. 2007, 17, 877–885. [Google Scholar] [CrossRef]
- Starks, R.R.; Biswas, A.; Jain, A.; Tuteja, G. Combined Analysis of Dissimilar Promoter Accessibility and Gene Expression Profiles Identifies Tissue-Specific Genes and Actively Repressed Networks. Epigenetics Chromatin 2019, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.R.; Blankvoort, S.; Lagartos, M.J.; Kentros, C. Enhancer-Driven Gene Expression (EDGE) Enables the Generation of Viral Vectors Specific to Neuronal Subtypes. iScience 2020, 23, 100888. [Google Scholar] [CrossRef] [PubMed]
- Graybuck, L.T.; Daigle, T.L.; Sedeño-Cortés, A.E.; Walker, M.; Kalmbach, B.; Lenz, G.H.; Morin, E.; Nguyen, T.N.; Garren, E.; Bendrick, J.L.; et al. Enhancer Viruses for Combinatorial Cell-Subclass-Specific Labeling. Neuron 2021, 109, 1449–1464. [Google Scholar] [CrossRef] [PubMed]
- Schlaeger, T.M.; Daheron, L.; Brickler, T.R.; Entwisle, S.; Chan, K.; Cianci, A.; DeVine, A.; Ettenger, A.; Fitzgerald, K.; Godfrey, M.; et al. A Comparison of Non-Integrating Reprogramming Methods. Nat. Biotechnol. 2015, 33, 58–63. [Google Scholar] [CrossRef]
- Zhang, M.; Jia, C.; Li, F.; Li, C.; Zhu, Y.; Akutsu, T.; Webb, G.I.; Zou, Q.; Coin, L.J.M.; Song, J. Critical Assessment of Computational Tools for Prokaryotic and Eukaryotic Promoter Prediction. Brief. Bioinform. 2022, 23, bbab551. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Yapp, E.K.Y.; Nagasundaram, N.; Yeh, H.Y. Classifying Promoters by Interpreting the Hidden Information of DNA Sequences via Deep Learning and Combination of Continuous FastText N-Grams. Front. Bioeng. Biotechnol. 2019, 7, 305. [Google Scholar] [CrossRef]
- Xiao, X.; Xu, Z.C.; Qiu, W.R.; Wang, P.; Ge, H.T.; Chou, K.C. IPSW(2L)-PseKNC: A Two-Layer Predictor for Identifying Promoters and Their Strength by Hybrid Features via Pseudo K-Tuple Nucleotide Composition. Genomics 2019, 111, 1785–1793. [Google Scholar] [CrossRef]
- Wang, Y.; Tai, S.; Zhang, S.; Sheng, N.; Xie, X. PromGER: Promoter Prediction Based on Graph Embedding and Ensemble Learning for Eukaryotic Sequence. Genes 2023, 14, 1441. [Google Scholar] [CrossRef]
- Huang, G.; Wu, L.; Ma, X.; Zhang, W.; Fan, J.; Yu, X.; Zeng, W.; Zhou, H. Evaluation of CatBoost Method for Prediction of Reference Evapotranspiration in Humid Regions. J. Hydrol. 2019, 574, 1029–1041. [Google Scholar] [CrossRef]
- Kari, H.; Bandi, S.M.S.; Kumar, A.; Yella, V.R. DeePromClass: Delineator for Eukaryotic Core Promoters Employing Deep Neural Networks. IEEE/ACM Trans. Comput. Biol. Bioinforma. 2023, 20, 802–807. [Google Scholar] [CrossRef]
- Meylan, P.; Dreos, R.; Ambrosini, G.; Groux, R.; Bucher, P. EPD in 2020: Enhanced Data Visualization and Extension to NcRNA Promoters. Nucleic Acids Res. 2020, 48, D65–D69. [Google Scholar] [CrossRef] [PubMed]
- Kolchanov, N.A.; Podkolodnaya, O.A.; Ananko, E.A.; Ignatieva, E.V.; Stepanenko, I.L.; Kel-Margoulis, O.V.; Kel, A.E.; Merkulova, T.I.; Goryachkovskaya, T.N.; Busygina, T.V.; et al. Transcription Regulatory Regions Database (TRRD): Its Status in 2000. Nucleic Acids Res. 2000, 28, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, R.; Sugano, S.; Suzuki, Y.; Nakai, K. DBTSS: DataBase of Transcriptional Start Sites Progress Report in 2012. Nucleic Acids Res. 2012, 40, D150–D154. [Google Scholar] [CrossRef]
- Forrest, A.R.R.; Kawaji, H.; Rehli, M.; Baillie, J.K.; De Hoon, M.J.L.; Haberle, V.; Lassmann, T.; Kulakovskiy, I.V.; Lizio, M.; Itoh, M.; et al. A Promoter-Level Mammalian Expression Atlas. Nature 2014, 507, 462–470. [Google Scholar] [CrossRef]
- Salgado, H.; Gama-Castro, S.; Lara, P.; Mejia-Almonte, C.; Alarcón-Carranza, G.; López-Almazo, A.G.; Betancourt-Figueroa, F.; Peña-Loredo, P.; Alquicira-Hernández, S.; Ledezma-Tejeida, D.; et al. RegulonDB V12.0:Ã Comprehensive Resource of Transcriptional Regulation in E. Coli K-12. Nucleic Acids Res. 2024, 52, D255–D264. [Google Scholar] [CrossRef] [PubMed]
- Sierro, N.; Makita, Y.; De Hoon, M.; Nakai, K. DBTBS: A Database of Transcriptional Regulation in Bacillus Subtilis Containing Upstream Intergenic Conservation Information. Nucleic Acids Res. 2008, 36, D93–D96. [Google Scholar] [CrossRef]
- Roberts, M.L.; Katsoupi, P.; Tseveleki, V.; Taoufik, E. Bioinformatically Informed Design of Synthetic Mammalian Promoters. Methods Mol. Biol. 2017, 1651, 93–112. [Google Scholar]
- Roberts, M.L. The Use of Functional Genomics in Synthetic Promoter Design. In Computational Biology and Applied Bioinformatics; IntechOpen: London, UK, 2011. [Google Scholar]
- Sarcar, S.; Tulalamba, W.; Rincon, M.Y.; Tipanee, J.; Pham, H.Q.; Evens, H.; Boon, D.; Samara-Kuko, E.; Keyaerts, M.; Loperfido, M.; et al. Next-Generation Muscle-Directed Gene Therapy by in Silico Vector Design. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Mogno, I.; Vallania, F.; Mitra, R.D.; Cohen, B.A. TATA Is a Modular Component of Synthetic Promoters. Genome Res. 2010, 20, 1391–1397. [Google Scholar] [CrossRef]
- Domenger, C.; Grimm, D. Next-Generation AAV Vectors-Do Not Judge a Virus (Only) by Its Cover. Hum. Mol. Genet. 2019, 28, R3–R14. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Jones, R.D.; Snavely, A.R.; Pfenning, A.R.; Kirchner, R.; Hemberg, M.; Gray, J.M. High-Throughput Functional Comparison of Promoter and Enhancer Activities. Genome Res. 2016, 26, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J. What Have We Learned about Synthetic Promoter Construction? Methods Mol. Biol. 2016, 1482, 1–13. [Google Scholar] [PubMed]
- Aysha, J.; Noman, M.; Wang, F.; Liu, W.; Zhou, Y.; Li, H.; Li, X. Synthetic Promoters: Designing the Cis Regulatory Modules for Controlled Gene Expression. Mol. Biotechnol. 2018, 60, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Cazier, A.P.; Blazeck, J. Advances in Promoter Engineering: Novel Applications and Predefined Transcriptional Control. Biotechnol. J. 2021, 16, 2100239. [Google Scholar] [CrossRef] [PubMed]
- Salva, M.Z.; Himeda, C.L.; Tai, P.W.L.; Nishiuchi, E.; Gregorevic, P.; Allen, J.M.; Finn, E.E.; Nguyen, Q.G.; Blankinship, M.J.; Meuse, L.; et al. Design of Tissue-Specific Regulatory Cassettes for High-Level RAAV-Mediated Expression in Skeletal and Cardiac Muscle. Mol. Ther. 2007, 15, 320–329. [Google Scholar] [CrossRef]
- Lee, Y.; Messing, A.; Su, M.; Brenner, M. GFAP Promoter Elements Required for Region-Specific and Astrocyte-Specific Expression. Glia 2008, 56, 481–493. [Google Scholar] [CrossRef]
- Yasmeen, E.; Wang, J.; Riaz, M.; Zhang, L.; Zuo, K. Designing Artificial Synthetic Promoters for Accurate, Smart, and Versatile Gene Expression in Plants. Plant Commun. 2023, 4, 100558. [Google Scholar] [CrossRef]
- Miao, C.H.; Ohashi, K.; Patijn, G.A.; Meuse, L.; Ye, X.; Thompson, A.R.; Kay, M.A. Inclusion of the Hepatic Locus Control Region, an Intron, and Untranslated Region Increases and Stabilizes Hepatic Factor IX Gene Expression in Vivo but Not in Vitro. Mol. Ther. 2000, 1, 522–532. [Google Scholar] [CrossRef]
- Davidoff, A.M.; Gray, J.T.; Ng, C.Y.C.; Zhang, Y.; Zhou, J.; Spence, Y.; Bakar, Y.; Nathwani, A.C. Comparison of the Ability of Adeno-Associated Viral Vectors Pseudotyped with Serotype 2, 5, and 8 Capsid Proteins to Mediate Efficient Transduction of the Liver in Murine and Nonhuman Primate Models. Mol. Ther. 2005, 11, 875–888. [Google Scholar] [CrossRef]
- Nathwani, A.C.; Gray, J.T.; Ng, C.Y.C.; Zhou, J.; Spence, Y.; Waddington, S.N.; Tuddenham, E.G.D.; Kemball-Cook, G.; McIntosh, J.; Boon-Spijker, M.; et al. Self-Complementary Adeno-Associated Virus Vectors Containing a Novel Liver-Specific Human Factor IX Expression Cassette Enable Highly Efficient Transduction of Murine and Nonhuman Primate Liver. Blood 2006, 107, 2653–2661. [Google Scholar] [CrossRef]
- McIntosh, J.; Lenting, P.J.; Rosales, C.; Lee, D.; Rabbanian, S.; Raj, D.; Patel, N.; Tuddenham, E.G.D.; Christophe, O.D.; McVey, J.H.; et al. Therapeutic Levels of FVIII Following a Single Peripheral Vein Administration of RAAV Vector Encoding a Novel Human Factor VIII Variant. Blood 2013, 121, 3335–3344. [Google Scholar] [CrossRef] [PubMed]
- Ede, C.; Chen, X.; Lin, M.Y.; Chen, Y.Y. Quantitative Analyses of Core Promoters Enable Precise Engineering of Regulated Gene Expression in Mammalian Cells. ACS Synth. Biol. 2016, 5, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Brown, H.C.; Zakas, P.M.; George, S.N.; Parker, E.T.; Spencer, H.T.; Doering, C.B. Target-Cell-Directed Bioengineering Approaches for Gene Therapy of Hemophilia A. Mol. Ther. Methods Clin. Dev. 2018, 9, 57–69. [Google Scholar] [CrossRef]
- Müller, O.J.; Leuchs, B.; Pleger, S.T.; Grimm, D.; Franz, W.M.; Katus, H.A.; Kleinschmidt, J.A. Improved Cardiac Gene Transfer by Transcriptional and Transductional Targeting of Adeno-Associated Viral Vectors. Cardiovasc. Res. 2006, 70, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Yew, N.S.; Wysokenski, D.M.; Wang, K.X.; Ziegler, R.J.; Marshall, J.; Mcneilly, D.; Cherry, M.; Osburn, W.; Cheng, S.H. Optimization of Plasmid Vectors for High-Level Expression in Lung Epithelial Cells. Hum. Gene Ther. 1997, 8, 575–584. [Google Scholar] [CrossRef]
- Liu, Y.L.; Mingozzi, F.; Rodriguéz-Colôn, S.M.; Joseph, S.; Dobrzynski, E.; Suzuki, T.; High, K.A.; Herzog, R.W. Therapeutic Levels of Factor IX Expression Using a Muscle-Specific Promoter and Adeno-Associated Virus Serotype 1 Vector. Hum. Gene Ther. 2004, 15, 783–792. [Google Scholar] [CrossRef]
- Hitoshi, N.; Ken-ichi, Y.; Jun-ichi, M. Efficient Selection for High-Expression Transfectants with a Novel Eukaryotic Vector. Gene 1991, 108, 193–199. [Google Scholar] [CrossRef]
- Li, X.; Eastman, E.M.; Schwartz, R.J.; Draghia-Akli, R. Synthetic Muscle Promoters: Activities Exceeding Naturally Occurring Regulatory Sequences. Nat. Biotechnol. 1999, 17, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.R.; Nissim, L.; Stupp, D.; Pery, E.; Binder-Nissim, A.; Weisinger, K.; Enghuus, C.; Palacios, S.R.; Humphrey, M.; Zhang, Z.; et al. A High-Throughput Screening and Computation Platform for Identifying Synthetic Promoters with Enhanced Cell-State Specificity (SPECS). Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Shibata, T.; Giaccia, A.J.; Brown, J.M. Development of a Hypoxia-Responsive Vector for Tumor-Specific Gene Therapy. Gene Ther. 2000, 7, 493–498. [Google Scholar] [CrossRef]
- Xia, J.B.; Wu, H.Y.; Lai, B.L.; Zheng, L.; Zhou, D.C.; Chang, Z.S.; Mao, C.Z.; Liu, G.H.; Park, K.S.; Zhao, H.; et al. Gene Delivery of Hypoxia-Inducible VEGF Targeting Collagen Effectively Improves Cardiac Function after Myocardial Infarction. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Lemken, M.L.; Wybranietz, W.A.; Schmidt, U.; Graepler, F.; Armeanu, S.; Bitzer, M.; Lauer, U.M. Expression Liver-Directed Genes by Employing Synthetic Transcriptional Control Units. World J. Gastroenterol. 2005, 11, 5295–5302. [Google Scholar] [CrossRef]
- Maturana, C.J. Engineered Compact Pan-Neuronal Promoter from Alphaherpesvirus LAP2 Enhances Target Gene Expression in the Mouse Brain and Reduces Tropism in the Liver. Gene Ther. 2024, 31, 335–344. [Google Scholar] [CrossRef]
- Korecki, A.J.; Cueva-Vargas, J.L.; Fornes, O.; Agostinone, J.; Farkas, R.A.; Hickmott, J.W.; Lam, S.L.; Mathelier, A.; Zhou, M.; Wasserman, W.W.; et al. Human MiniPromoters for Ocular-RAAV Expression in ON Bipolar, Cone, Corneal, Endothelial, Müller Glial, and PAX6 Cells. Gene Ther. 2021, 28, 351–372. [Google Scholar] [CrossRef]
- Simpson, E.M.; Korecki, A.J.; Fornes, O.; McGill, T.J.; Cueva-Vargas, J.L.; Agostinone, J.; Farkas, R.A.; Hickmott, J.W.; Lam, S.L.; Mathelier, A.; et al. New MiniPromoter Ple345 (NEFL) Drives Strong and Specific Expression in Retinal Ganglion Cells of Mouse and Primate Retina. Hum. Gene Ther. 2019, 30, 257–272. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Wei, L.; Li, S.; Liu, L.; Wang, X. Synthetic Promoter Design in Escherichia Coli Based on a Deep Generative Network. Nucleic Acids Res. 2020, 48, 6403–6412. [Google Scholar] [CrossRef]
- Qiao, H.; Zhang, S.; Xue, T.; Wang, J.; Wang, B. IPro-GAN: A Novel Model Based on Generative Adversarial Learning for Identifying Promoters and Their Strength. Comput. Methods Programs Biomed. 2022, 215, 106625. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, H.; Xu, H.; Wei, L.; Liu, L.; Hu, Z.; Wang, X. Deep Flanking Sequence Engineering for Efficient Promoter Design Using DeepSEED. Nat. Commun. 2023, 14, 1–14. [Google Scholar] [CrossRef]
- Seo, E.; Choi, Y.N.; Shin, Y.R.; Kim, D.; Lee, J.W. Design of Synthetic Promoters for Cyanobacteria with Generative Deep-Learning Model. Nucleic Acids Res. 2023, 51, 7071–7082. [Google Scholar] [CrossRef]
- Gosai, S.J.; Castro, R.I.; Fuentes, N.; Butts, J.C.; Mouri, K.; Alasoadura, M.; Kales, S.; Nguyen, T.T.L.; Noche, R.R.; Rao, A.S.; et al. Machine-Guided Design of Cell-Type-Targeting Cis-Regulatory Elements. Nature 2024, 634, 1211–1220. [Google Scholar] [CrossRef]
- Linder, J.; Seelig, G. Fast Activation Maximization for Molecular Sequence Design. BMC Bioinformatics 2021, 22, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Du, Q.; Wang, Y.; Xu, H.; Wei, Z.; Wang, X. GPro: Generative AI-Empowered Toolkit for Promoter Design. Bioinformatics 2024, 40, btae123. [Google Scholar] [CrossRef]
- Belokopytova, P.S.; Nuriddinov, M.A.; Mozheiko, E.A.; Fishman, D.; Fishman, V. Quantitative Prediction of Enhancer–Promoter Interactions. Genome Res. 2020, 30, 72–84. [Google Scholar] [CrossRef]
- Zheng, L.; Liu, L.; Zhu, W.; Ding, Y.; Wu, F. Predicting Enhancer-Promoter Interaction Based on Epigenomic Signals. Front. Genet. 2023, 14, 1133775. [Google Scholar] [CrossRef]
- Noman, M.Z.; Desantis, G.; Janji, B.; Hasmim, M.; Karray, S.; Dessen, P.; Bronte, V.; Chouaib, S. PD-L1 Is a Novel Direct Target of HIF-1α, and Its Blockade under Hypoxia Enhanced: MDSC-Mediated T Cell Activation. J. Exp. Med. 2014, 211, 781–790. [Google Scholar] [CrossRef]
- de Wet, J.R.; Wood, K.V.; DeLuca, M.; Helinski, D.R.; Subramani, S. Firefly Luciferase Gene: Structure and Expression in Mammalian Cells. Mol. Cell. Biol. 1987, 7, 725–737. [Google Scholar] [CrossRef]
- Cayrou, C.; Bayliss, C.D. Assessment of FHbp Expression Level by Reverse Transcriptase Quantitative PCR and Promoter Sequence Analysis. Methods Mol. Biol. 2019, 1969, 237–249. [Google Scholar]
- Rizzuto, R.; Brini, M.; Pizzo, P.; Murgia, M.; Pozzan, T. Chimeric Green Fluorescent Protein as a Tool for Visualizing Subcellular Organelles in Living Cells. Curr. Biol. 1995, 5, 635–642. [Google Scholar] [CrossRef]
- Kang, J.; Huang, L.; Zheng, W.; Luo, J.; Zhang, X.; Song, Y.; Liu, A. Promoter CAG Is More Efficient than Hepatocyte-Targeting TBG for Transgene Expression via RAAV8 in Liver Tissues. Mol. Med. Rep. 2022, 25, 1–9. [Google Scholar] [CrossRef]
- Mogno, I.; Kwasnieski, J.C.; Cohen, B.A. Massively Parallel Synthetic Promoter Assays Reveal the in Vivo Effects of Binding Site Variants. Genome Res. 2013, 23, 1908–1915. [Google Scholar] [CrossRef]
- Shen, S.Q.; Myers, C.A.; Hughes, A.E.O.; Byrne, L.C.; Flannery, J.G.; Corbo, J.C. Massively Parallel Cis-Regulatory Analysis in the Mammalian Central Nervous System. Genome Res. 2016, 26, 238–255. [Google Scholar] [CrossRef]
- Kuzmin, D.A.; Shutova, M.V.; Johnston, N.R.; Smith, O.P.; Fedorin, V.V.; Kukushkin, Y.S.; van der Loo, J.C.M.; Johnstone, E.C. The Clinical Landscape for AAV Gene Therapies. Nat. Rev. Drug Discov. 2021, 20, 173–174. [Google Scholar] [CrossRef]
- Burdett, T.; Nuseibeh, S. Changing Trends in the Development of AAV-Based Gene Therapies: A Meta-Analysis of Past and Present Therapies. Gene Ther. 2023, 30, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Rheaume, B.A.; Jereen, A.; Bolisetty, M.; Sajid, M.S.; Yang, Y.; Renna, K.; Sun, L.; Robson, P.; Trakhtenberg, E.F. Single Cell Transcriptome Profiling of Retinal Ganglion Cells Identifies Cellular Subtypes. Nat. Commun. 2018, 9, 2759. [Google Scholar] [CrossRef]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-Wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Darmanis, S.; Sloan, S.A.; Zhang, Y.; Enge, M.; Caneda, C.; Shuer, L.M.; Gephart, M.G.H.; Barres, B.A.; Quake, S.R. A Survey of Human Brain Transcriptome Diversity at the Single Cell Level. Proc. Natl. Acad. Sci. USA 2015, 112, 7285–7290. [Google Scholar] [CrossRef]
- Raj, B.; Wagner, D.E.; McKenna, A.; Pandey, S.; Klein, A.M.; Shendure, J.; Gagnon, J.A.; Schier, A.F. Simultaneous Single-Cell Profiling of Lineages and Cell Types in the Vertebrate Brain. Nat. Biotechnol. 2018, 36, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Visel, A.; Taher, L.; Girgis, H.; May, D.; Golonzhka, O.; Hoch, R.V.; McKinsey, G.L.; Pattabiraman, K.; Silberberg, S.N.; Blow, M.J.; et al. A High-Resolution Enhancer Atlas of the Developing Telencephalon. Cell 2013, 152, 895–908. [Google Scholar] [CrossRef]
- Silberberg, S.N.; Taher, L.; Lindtner, S.; Sandberg, M.; Nord, A.S.; Vogt, D.; Mckinsey, G.L.; Hoch, R.; Pattabiraman, K.; Zhang, D.; et al. Subpallial Enhancer Transgenic Lines: A Data and Tool Resource to Study Transcriptional Regulation of GABAergic Cell Fate. Neuron 2016, 92, 59–74. [Google Scholar] [CrossRef]
- Jüttner, J.; Szabo, A.; Gross-Scherf, B.; Morikawa, R.K.; Rompani, S.B.; Hantz, P.; Szikra, T.; Esposti, F.; Cowan, C.S.; Bharioke, A.; et al. Targeting Neuronal and Glial Cell Types with Synthetic Promoter AAVs in Mice, Non-Human Primates and Humans. Nat. Neurosci. 2019, 22, 1345–1356. [Google Scholar] [CrossRef]
- Markenscoff-Papadimitriou, E.; Whalen, S.; Przytycki, P.; Thomas, R.; Binyameen, F.; Nowakowski, T.J.; Kriegstein, A.R.; Sanders, S.J.; State, M.W.; Pollard, K.S.; et al. A Chromatin Accessibility Atlas of the Developing Human Telencephalon. Cell 2020, 182, 754–769. [Google Scholar] [CrossRef] [PubMed]
- Mich, J.K.; Graybuck, L.T.; Hess, E.E.; Mahoney, J.T.; Kojima, Y.; Ding, Y.; Somasundaram, S.; Miller, J.A.; Kalmbach, B.E.; Radaelli, C.; et al. Functional Enhancer Elements Drive Subclass-Selective Expression from Mouse to Primate Neocortex. Cell Rep. 2021, 34, 108754. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, E.M. Getting a Foot IN the Door: GABAergic INterneuron-Specific Enhancers. Epilepsy Curr. 2021, 21, 114–116. [Google Scholar] [CrossRef]
- Vormstein-Schneider, D.; Lin, J.D.; Pelkey, K.A.; Chittajallu, R.; Guo, B.; Arias-Garcia, M.A.; Allaway, K.; Sakopoulos, S.; Schneider, G.; Stevenson, O.; et al. Viral Manipulation of Functionally Distinct Interneurons in Mice, Non-Human Primates and Humans. Nat. Neurosci. 2020, 23, 1629–1636. [Google Scholar] [CrossRef]
- Schiedner, G.; Morral, N.; Parks, R.J.; Wu, Y.; Koopmans, S.C.; Langston, C.; Graham, F.L.; Beaudet, A.L.; Kochanek, S. Genomic DNA Transfer with a High-Capacity Adenovirus Vector Results in Improved in Vivo Gene Expression and Decreased Toxicity. Nat. Genet. 1998, 18, 180–183. [Google Scholar] [CrossRef]
- FDA. Available online: https://www.fda.gov/ (accessed on 21 October 2024).
- Hauser, M.A.; Robinson, A.; Hartigan-O’Connor, D.; Williams-Gregory, D.A.; Buskin, J.N.; Apone, S.; Kirk, C.J.; Hardy, S.; Hauschka, S.D.; Chamberlain, J.S. Analysis of Muscle Creatine Kinase Regulatory Elements in Recombinant Adenoviral Vectors. Mol. Ther. 2000, 2, 16–25. [Google Scholar] [CrossRef]
- Martari, M.; Sagazio, A.; Mohamadi, A.; Nguyen, Q.; Hauschka, S.D.; Kim, E.; Salvatori, R. Partial Rescue of Growth Failure in Growth Hormone (GH)-Deficient Mice by a Single Injection of a Double-Stranded Adeno-Associated Viral Vector Expressing the GH Gene Driven by a Muscle-Specific Regulatory Cassette. Hum. Gene Ther. 2009, 20, 759–766. [Google Scholar] [CrossRef]
- Wang, B.; Li, J.; Fu, F.H.; Chen, C.; Zhu, X.; Zhou, L.; Jiang, X.; Xiao, X. Construction and Analysis of Compact Muscle-Specific Promoters for AAV Vectors. Gene Ther. 2008, 15, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Skopenkova, V.V.; Egorova, T.V.; Bardina, M.V. Muscle-Specific Promoters for Gene Therapy. Acta Naturae 2021, 13, 47–58. [Google Scholar] [CrossRef]
- Piekarowicz, K.; Bertrand, A.T.; Azibani, F.; Beuvin, M.; Julien, L.; Machowska, M.; Bonne, G.; Rzepecki, R. A Muscle Hybrid Promoter as a Novel Tool for Gene Therapy. Mol. Ther. Methods Clin. Dev. 2019, 15, 157–169. [Google Scholar] [CrossRef]
- Georgiadis, A.; Duran, Y.; Ribeiro, J.; Abelleira-Hervas, L.; Robbie, S.J.; Sünkel-Laing, B.; Fourali, S.; Gonzalez-Cordero, A.; Cristante, E.; Michaelides, M.; et al. Development of an Optimized AAV2/5 Gene Therapy Vector for Leber Congenital Amaurosis Owing to Defects in RPE65. Gene Ther. 2016, 23, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.J.; Budzynski, E.; Sonnentag, P.; Nork, T.M.; Sheibani, N.; Gurel, Z.; Boye, S.L.; Peterson, J.J.; Boye, S.E.; Hauswirth, W.W.; et al. Cone-Specific Promoters for Gene Therapy of Achromatopsia and Other Retinal Diseases. Hum. Gene Ther. 2016, 27, 72–82. [Google Scholar] [CrossRef]
- Khabou, H.; Garita-Hernandez, M.; Chaffiol, A.; Reichman, S.; Jaillard, C.; Brazhnikova, E.; Bertin, S.; Forster, V.; Desrosiers, M.; Winckler, C.; et al. Noninvasive Gene Delivery to Foveal Cones for Vision Restoration. JCI Insight 2018, 3, e96029. [Google Scholar] [CrossRef]
- Kralik, J.; van Wyk, M.; Stocker, N.; Kleinlogel, S. Bipolar Cell Targeted Optogenetic Gene Therapy Restores Parallel Retinal Signaling and High-Level Vision in the Degenerated Retina. Commun. Biol. 2022, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Bi, A.; Cui, J.; Ma, Y.P.; Olshevskaya, E.; Pu, M.; Dizhoor, A.M.; Pan, Z.H. Ectopic Expression of a Microbial-Type Rhodopsin Restores Visual Responses in Mice with Photoreceptor Degeneration. Neuron 2006, 50, 23–33. [Google Scholar] [CrossRef]
- Lagali, P.S.; Balya, D.; Awatramani, G.B.; Münch, T.A.; Kim, D.S.; Busskamp, V.; Cepko, C.L.; Roska, B. Light-Activated Channels Targeted to ON Bipolar Cells Restore Visual Function in Retinal Degeneration. Nat. Neurosci. 2008, 11, 667–675. [Google Scholar] [CrossRef]
- Busskamp, V.; Picaud, S.; Sahel, J.A.; Roska, B. Optogenetic Therapy for Retinitis Pigmentosa. Gene Ther. 2012, 19, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Hulliger, E.C.; Hostettler, S.M.; Kleinlogel, S. Empowering Retinal Gene Therapy with a Specific Promoter for Human Rod and Cone ON-Bipolar Cells. Mol. Ther. Methods Clin. Dev. 2020, 17, 505–519. [Google Scholar] [CrossRef]
- Nieuwenhuis, B.; Haenzi, B.; Hilton, S.; Carnicer-Lombarte, A.; Hobo, B.; Verhaagen, J.; Fawcett, J.W. Optimization of Adeno-Associated Viral Vector-Mediated Transduction of the Corticospinal Tract: Comparison of Four Promoters. Gene Ther. 2021, 28, 56–74. [Google Scholar] [CrossRef]
- Matsuzaki, Y.; Oue, M.; Hirai, H. Generation of a Neurodegenerative Disease Mouse Model Using Lentiviral Vectors Carrying an Enhanced Synapsin I Promoter. J. Neurosci. Methods 2014, 223, 133–143. [Google Scholar] [CrossRef]
- Rubin, A.N.; Malik, R.; Cho, K.K.A.; Lim, K.J.; Lindtner, S.; Schwartz, S.E.R.; Vogt, D.; Sohal, V.S.; Rubenstein, J.L.R. Regulatory Elements Inserted into Aavs Confer Preferential Activity in Cortical Interneurons. eNeuro 2020, 7, ENEURO.0211-20.2020. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yue, D.; Lei, L.; Wang, H.; Lu, J.; Zhou, Y.; Liu, S.; Ding, T.; Guo, M.; Xu, L. Promoter-Operating Targeted Expression of Gene Therapy in Cancer: Current Stage and Prospect. Mol. Ther. Nucleic Acids 2018, 11, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Montaño-Samaniego, M.; Bravo-Estupiñan, D.M.; Méndez-Guerrero, O.; Alarcón-Hernández, E.; Ibáñez-Hernández, M. Strategies for Targeting Gene Therapy in Cancer Cells With Tumor-Specific Promoters. Front. Oncol. 2020, 10, 605380. [Google Scholar] [CrossRef]
- Liu, S.H.; Yu, J.; Sanchez, R.; Liu, X.; Heidt, D.; Willey, J.; Nemunaitis, J.; Brunicardi, F.C. A Novel Synthetic Human Insulin Super Promoter for Targeting PDX-1-Expressing Pancreatic Cancer. Cancer Lett. 2018, 418, 75–83. [Google Scholar] [CrossRef]
- Reddy, A.J.; Geng, X.; Herschl, M.H.; Kolli, S.; Kumar, A.; Hsu, P.D.; Levine, S.; Ioannidis, N.M. Designing Cell-Type-Specific Promoter Sequences Using Conservative Model-Based Optimization. bioRxiv 2024. [Google Scholar] [CrossRef]
- Le Guiner, C.; Stieger, K.; Toromanoff, A.; Guilbaud, M.; Mendes-Madeira, A.; Devaux, M.; Guigand, L.; Cherel, Y.; Moullier, P.; Rolling, F.; et al. Transgene Regulation Using the Tetracycline-Inducible TetR-KRAB System after AAV-Mediated Gene Transfer in Rodents and Nonhuman Primates. PLoS ONE 2014, 9, e102538. [Google Scholar] [CrossRef] [PubMed]
- Sohn, J.; Takahashi, M.; Okamoto, S.; Ishida, Y.; Furuta, T.; Hioki, H. A Single Vector Platform for High-Level Gene Transduction of Central Neurons: Adeno-Associated Virus Vector Equipped with the Tet-off System. PLoS ONE 2017, 12, e0169611. [Google Scholar] [CrossRef]
- O’Callaghan, J.; Crosbie, D.E.; Cassidy, P.S.; Sherwood, J.M.; Flügel-Koch, C.; Lütjen-Drecoll, E.; Humphries, M.M.; Reina-Torres, E.; Wallace, D.; Kiang, A.S.; et al. Therapeutic Potential of AAV-Mediated MMP-3 Secretion from Corneal Endothelium in Treating Glaucoma. Hum. Mol. Genet. 2017, 26, 1230–1246. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, B.; García-Díaz, P.; Foight, G.W. Synthetic Transcription Factor Engineering for Cell and Gene Therapy. Trends Biotechnol. 2024, 42, 449–463. [Google Scholar] [CrossRef]
- Galvan, S.; Madderson, O.; Xue, S.; Teixeira, A.P.; Fussenegger, M. Regulation of Transgene Expression by the Natural Sweetener Xylose. Adv. Sci. 2022, 9, e2203193. [Google Scholar] [CrossRef]
- uniQure. Glybera (Alipogene Tiparvovec). Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/glybera (accessed on 21 October 2024).
- Pfizer Inc. BEQVEZ (Fidanacogene Elaparvovec-Dzkt). Available online: https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/beqvez (accessed on 21 October 2024).

| Gene of Origin | Promoter | Disease | Specificity | Design | Therapy | Trial | Status |
|---|---|---|---|---|---|---|---|
| Liver-specific | |||||||
| TTR | TTR | Hemophilia B | hepatocytes | murine TTR promoter/enhancer | SHP648 | NCT04394286, phase 1/2 | terminated |
| Hemophilia B | TTR promoter/enhancer | AskBio009, Bax335 | NCT01687608, phase 1/2 | active, not recruiting | |||
| Wilson’s Disease | murine TTR enhancer/human TTR promoter + SV40 intron | UX701 | NCT04884815, phase 1/2 | active, not recruiting | |||
| Hemophilia A | truncated TTR promoter/enhancer | SPK-8011 | NCT06297486, phase 3 | recruiting | |||
| Hemophilia A | TTR promoter/enhancer | Bax 888, AAV8-BDD-FVIIIopt | NCT03370172, phase 1/2 | active, not recruiting | |||
| ApoE, hAAT | LP1 | Hemophilia B | hepatocytes | truncated HCR/hAAT hybrid promoter (448 bp) | AMT-061 | NCT03569891, phase 3 | active, not recruiting |
| LP1 | Hemophilia B | truncated HCR/hAAT hybrid promoter (448 bp) | scAAV 2/8-LP1-hFIXco | NCT00979238, phase 1 | active, not recruiting | ||
| HLP | Hemophilia A | truncated HCR/hAAT hybrid promoter (252 bp) | BMN 270-301 | NCT03370913, phase 3 | active, not recruiting | ||
| HLP | Severe Crigler Najjar Syndrome | truncated HCR/hAAT hybrid promoter (252 bp) | GNT0003 | NCT03466463, not applicable | recruiting | ||
| ApoE/ hAAT | Fabry disease | enhancer and HCR from human ApoE gene + hAAT promoter + modified chimeric intron (HBB-IgG) | ST-920 | NCT05039866, phase 1/2 follow-up | enrolling by invitation | ||
| hAAT | hAAT | Wilson’s Disease | hepatocytes | hAAT promoter | VTX-801 | NCT04537377, phase 1/2 | recruiting |
| AAT, albumin | EalbAAT | Acute Intermittent Porphyria | hepatocytes | albumin enhancer + AAT promoter | rAAV2/5-PBGD | NCT02082860, phase 1 | completed |
| Albumin | Albumin | MPS I | hepatocytes | albumin promoter | SB-318 | NCT02702115, phase 1/2 | terminated |
| MPS II | SB-913 | NCT03041324, phase 1/2 | terminated | ||||
| Hemophilia B | SB-FIX | NCT02695160, phase 1 | terminated | ||||
| TBG | TBG | MPS VI | hepatocytes | TBG promoter | AAV2/8.TBG.hARSB | NCT03173521, phase 1/2 | not recruiting |
| TBG | Late-onset OTC Deficiency | TBG promoter | DTX301, scAAV8OTC | NCT02991144, phase 1/2 | completed | ||
| LSP | Pompe Disease | α1-microglobulin/bikunin enhancer x2 + TBG promoter | ACTUS-101, AAV2/8LSPhGAA | NCT03533673, phase 1/2 | not recruiting | ||
| Combination | HCB | Hemophilia A | hepatocytes | combination of TFBS + minimal promoter SynO | ASC618 | NCT04676048, phase 1/2 | recruiting |
| Muscle-specific | |||||||
| MCK | CK8 | DMD | skeletal muscles | optimized MCK-enhancer + optimized CK6 (enhancer 2RS5 + proximal MCK promoter) | SGT-001 | NCT03368742, phase 1/2 | active, not recruiting |
| SGT-003 | NCT06138639, phase 1/2 | recruiting | |||||
| MHCK7 | Dysferlinopathy | α-MHC-enhancer + optimized CK cassette (enhancer 2RS5 + proximal MCK promoter) | rAAVrh.74.MHCK7.DYSF.DV | NCT02710500, phase 1 | completed | ||
| LGMD2E | SRP-9003 | NCT06246513, phase 3 | recruiting | ||||
| DMD | SRP-9001 | NCT05881408, phase 3 | recruiting | ||||
| LGMD2B/R2 | SRP-6004 | NCT05906251, phase 1 | active, not recruiting | ||||
| tMCK | DMD | enhancer 2RS5 x3 + proximal MCK promoter | PF-06939926 | NCT04281485, phase 3 | active, not recruiting | ||
| LGMD2D | SRP-9004 | NCT01976091, phase 1/2 NCT00494195, phase 1 | completed completed | ||||
| CMT | scAAV1.tMCK.NTF3 | NCT03520751, phase 1/2 | suspended (vector has not been produced) | ||||
| eMCK | Pompe Disease | MCK promoter/enhancer combination | AT845 | NCT04174105, phase 1/2 | completed | ||
| Des | Des | LGMD2C | skeletal muscles | desmin promoter | AAV1-gamma-sarcoglycan vector injection | NCT01344798, phase 1 | completed |
| X-Linked Myotubular Myopathy | truncated desmin enhancer/promoter (1.05 kb) | Resamirigene bilparvovec, AT132 | NCT03199469, phase 2/3 | not recruiting | |||
| Pompe Disease | desmin promoter | rAAV9-DES-hGAA | NCT02240407, phase 1 | completed | |||
| hCK | hCK | DMD | skeletal and cardiac muscle | hCK promoter | fordadistrogene movaparvovec | NCT05429372, phase 2 | active, not recruiting |
| Combination | Spc5-12 | DMD | skeletal and cardiac muscle | combination of TFBS (SRE, MEF-1, MEF-2, TEF-1) + chicken skeletal a-actin promoter | RGX-202 | NCT05693142, phase 1/2 | recruiting |
| OPMD | BB-301 | NCT06185673, phase 1/2 | recruiting | ||||
| Eye-specific | |||||||
| hRS1 | hRS1 | XLRS | PRs | native human RS1 promoter | AAV8-scRS/IRBPhRS, RS1 AAV Vector | NCT02317887, phase 1/2 | active, not recruiting |
| hRPE65 | hRPE65 | LCA | RPE | truncated hRPE65 (1.4 kb) | tgAAG76, rAAV 2/2.hRPE65p.hRPE65 | NCT00643747, phase 1/2 | completed |
| NA65p | LCA10 | optimized hRPE65 promoter (757 bp) | AAV2/5-OPTIRPE65 | NCT02946879, phase 1/2 | completed | ||
| hGRK1/GRK1 | hGRK1 | LCA10 | PRs(rods and cones) | hGRK1 promoter | EDIT-101, AAV5.SaCas9 AGN-151587 | NCT03872479, phase 1/2 | active, not recruiting |
| GRK1 | X-linked RP | GRK1 promoter | AGTC-501, rAAV2tYF-GRK1-RPGR | NCT06275620, phase 2 | enrolling by invitation | ||
| hRK | hRK | X-linked RP | PRs (rods and cones) | hPK promoter | Cotoretigene toliparvovec, BIIB112, AAV8-RPGR | NCT03116113, phase 2/3 | completed |
| X-linked RP | truncated hPK promoter | Botaretigene Sparoparvovec, AAV5-hRKp.RPGR | NCT03252847, phase 1/2 | completed | |||
| Autosomal Recessive RP | truncated hPK promoter | AAV2/5-hPDE6B | NCT03252847, phase 1/2 | recruiting | |||
| XLRS | hPK promoter | ATSN-201, rAAV.SPR-hGRK1-hRS1syn | NCT05878860, phase 1/2 | recruiting | |||
| human red opsin | PR1.7 | Achromatopsia | PRs (cones) | LCR enhancer fragment + red opsin promoter | (rAAV2tYF-PR1.7-hCNGB3), AGTC-401 | NCT02599922, phase 1/2 | active, not recruiting |
| (rAAV2tYF-PR1.7-hCNGA3), AGTC-402 | NCT02935517, phase 1/2 | active, not recruiting | |||||
| Neuron-specific | |||||||
| hSyn1 | hSyn1 | MTLE | neurons | hSyn1 promoter | AMT-260, AAV9-hSyn1-miGRIK | NCT06063850, phase 1/2 | recruiting |
| FTD | AVB-101 | NCT06064890, phase 1/2 | recruiting | ||||
| mPGK | mPGK | Sanfilippo Syndrome B | cortical neurons and oligodendrocytes | mPGK promoter | rAAV2/5-hNAGLU | NCT03300453, phase 1/2 | completed |
| MeCP | P546 | Batten Disease | neurons | truncated MeCP2 promoter (546 bp) | AT-GTX-502, scAAV9.P546.CLN3 | NCT03770572, phase 1/2 | active, not recruiting |
| MeP426 | Rett Syndrome | MeCP2 core promoter + regulatory elements (RE) | TSHA-102 | NCT05606614, phase 1/2 | phase 1/2 recruiting | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Artemyev, V.; Gubaeva, A.; Paremskaia, A.I.; Dzhioeva, A.A.; Deviatkin, A.; Feoktistova, S.G.; Mityaeva, O.; Volchkov, P.Y. Synthetic Promoters in Gene Therapy: Design Approaches, Features and Applications. Cells 2024, 13, 1963. https://doi.org/10.3390/cells13231963
Artemyev V, Gubaeva A, Paremskaia AI, Dzhioeva AA, Deviatkin A, Feoktistova SG, Mityaeva O, Volchkov PY. Synthetic Promoters in Gene Therapy: Design Approaches, Features and Applications. Cells. 2024; 13(23):1963. https://doi.org/10.3390/cells13231963
Chicago/Turabian StyleArtemyev, Valentin, Anna Gubaeva, Anastasiia Iu. Paremskaia, Amina A. Dzhioeva, Andrei Deviatkin, Sofya G. Feoktistova, Olga Mityaeva, and Pavel Yu. Volchkov. 2024. "Synthetic Promoters in Gene Therapy: Design Approaches, Features and Applications" Cells 13, no. 23: 1963. https://doi.org/10.3390/cells13231963
APA StyleArtemyev, V., Gubaeva, A., Paremskaia, A. I., Dzhioeva, A. A., Deviatkin, A., Feoktistova, S. G., Mityaeva, O., & Volchkov, P. Y. (2024). Synthetic Promoters in Gene Therapy: Design Approaches, Features and Applications. Cells, 13(23), 1963. https://doi.org/10.3390/cells13231963


.png)



