Inflammatory Response of THP1 and U937 Cells: The RNAseq Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. THP1 and U937 Cells
2.2. Total RNA Isolation, Sequencing, and Data Analysis
2.3. PCA, Volcano Plot, Venn Diagram, Heat Map, Protein–Protein Interactions, and Gene Set Enrichment Analysis
3. Results
3.1. Principal Component Analysis of Gene Expression Changes by Activated THP1 and U937
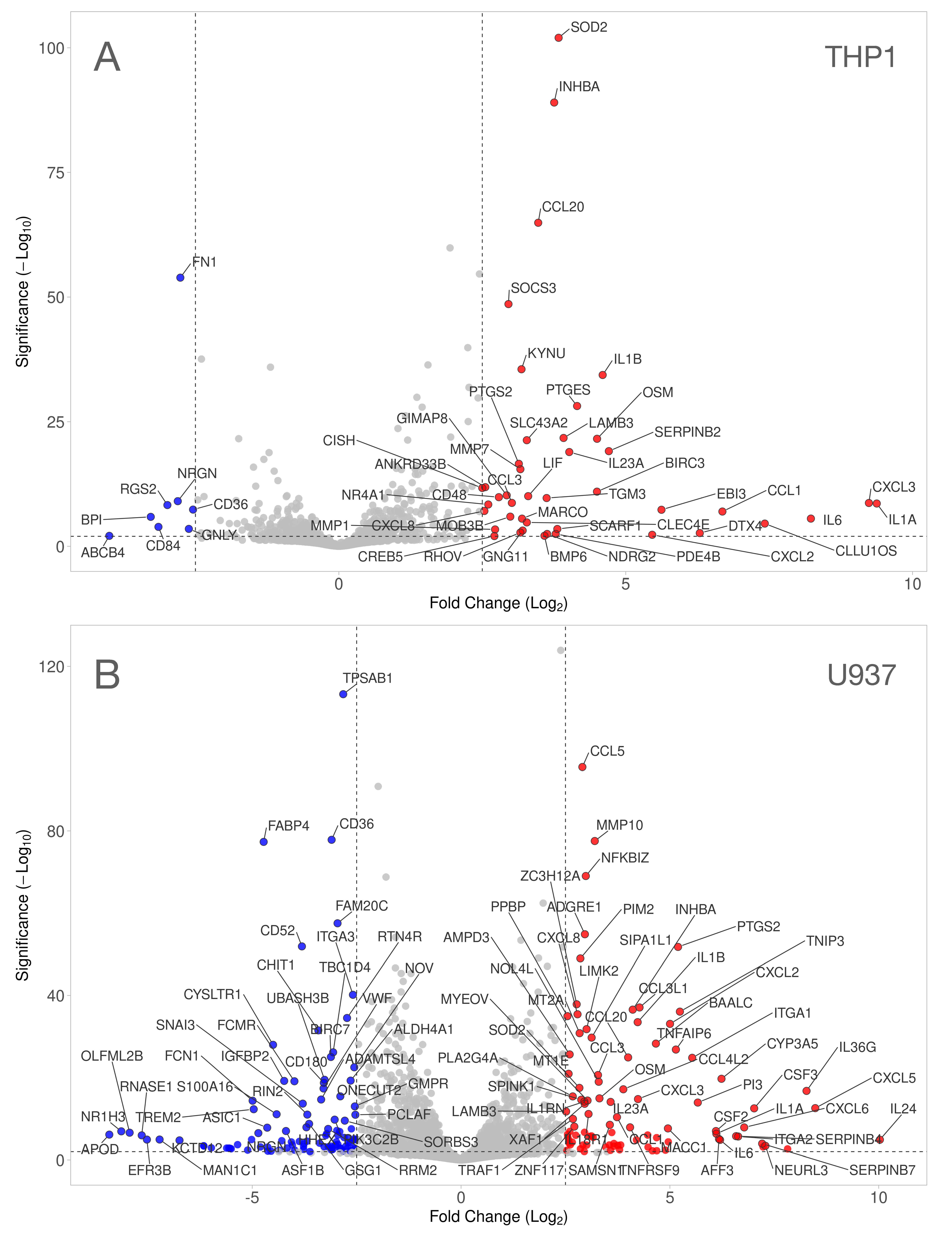
3.2. Analysis of Gene Expression Changes by LPS and IFNG Exposed THP1 and U937 Cells
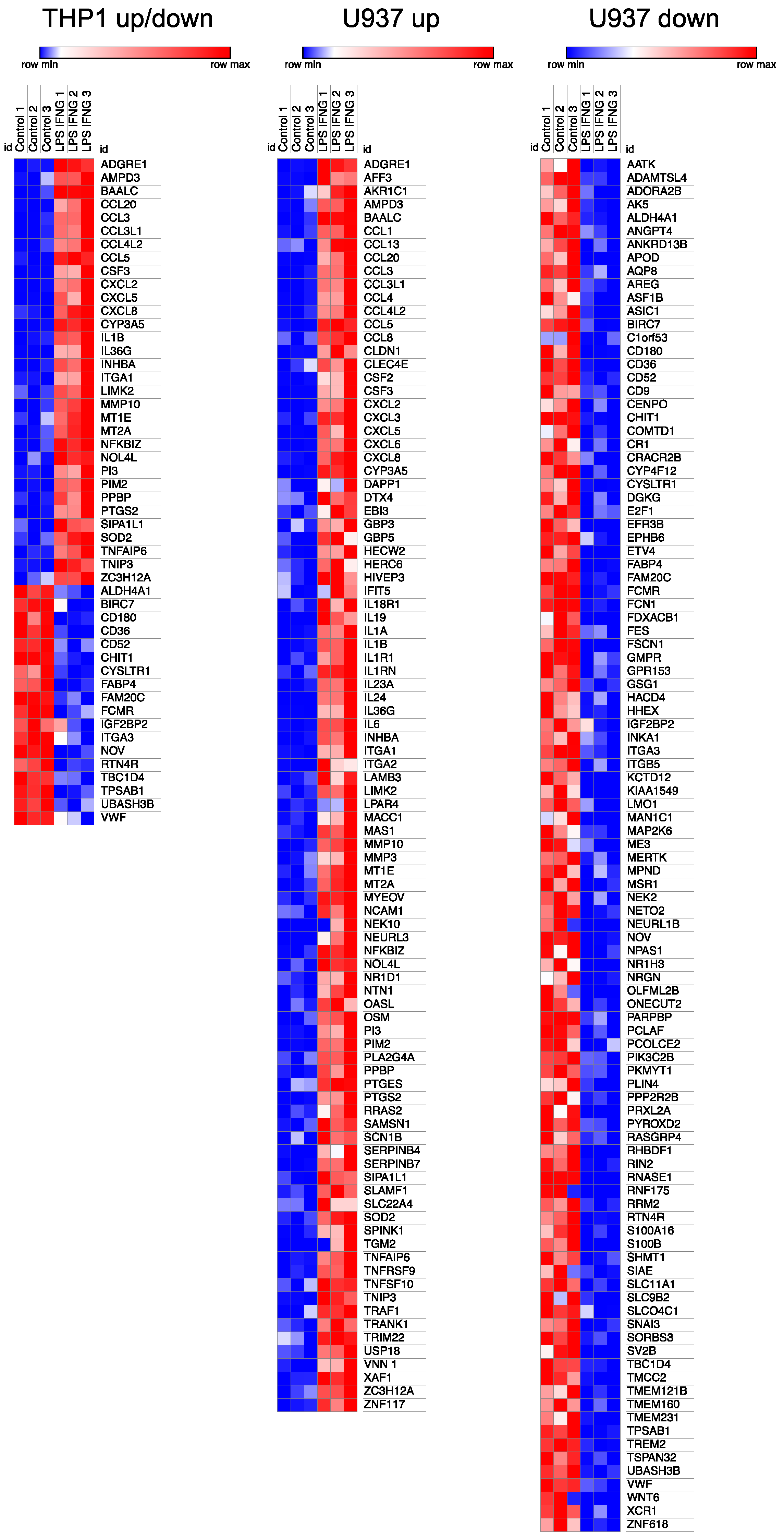
3.3. Heat Map of Gene Expression Changes by LPS and IFNG Exposed THP1 and U937 Cells
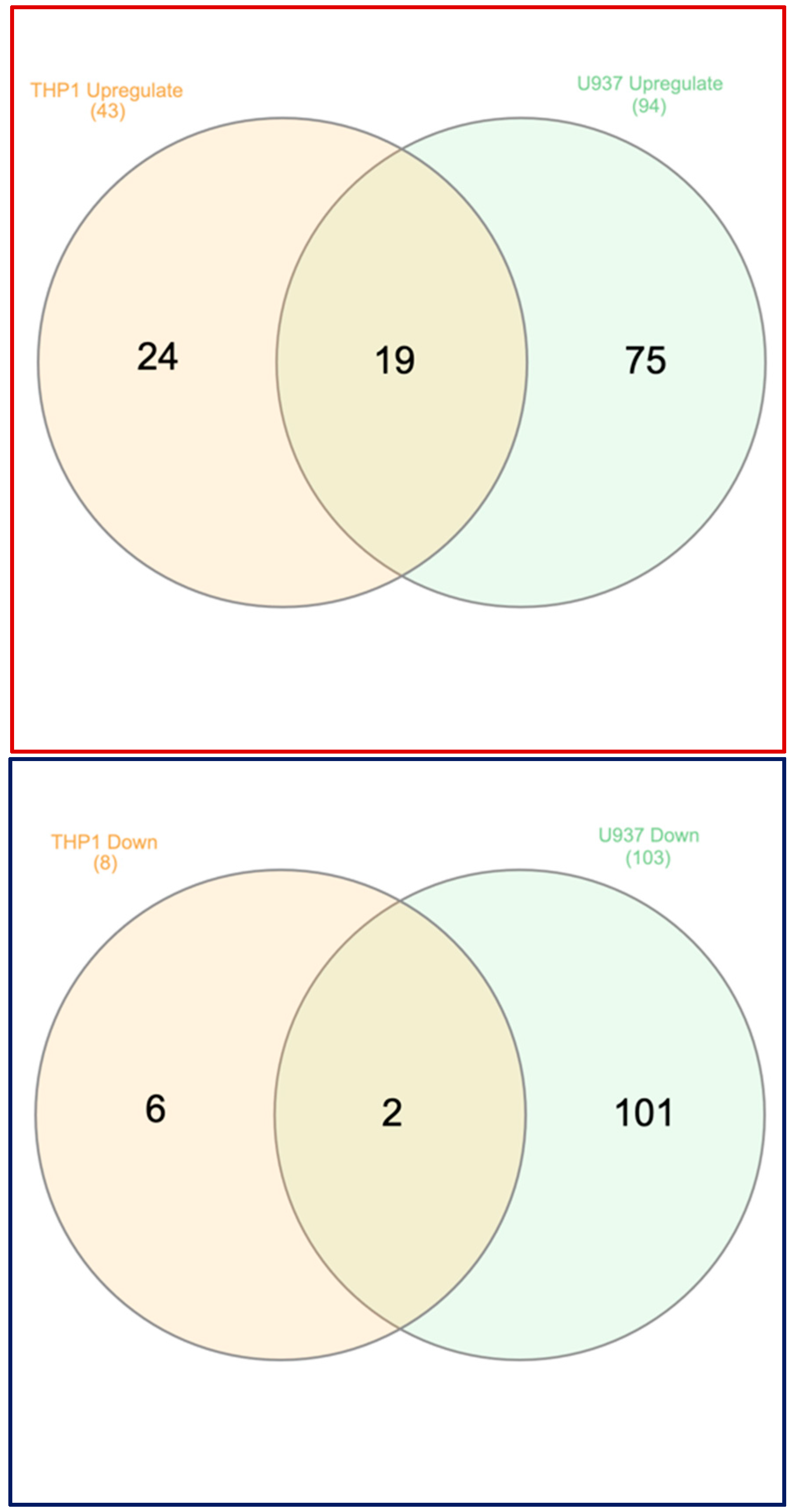
3.4. Analyzing Similarities and Differences Between LPS and IFNG Exposed THP1 and U937 Cells
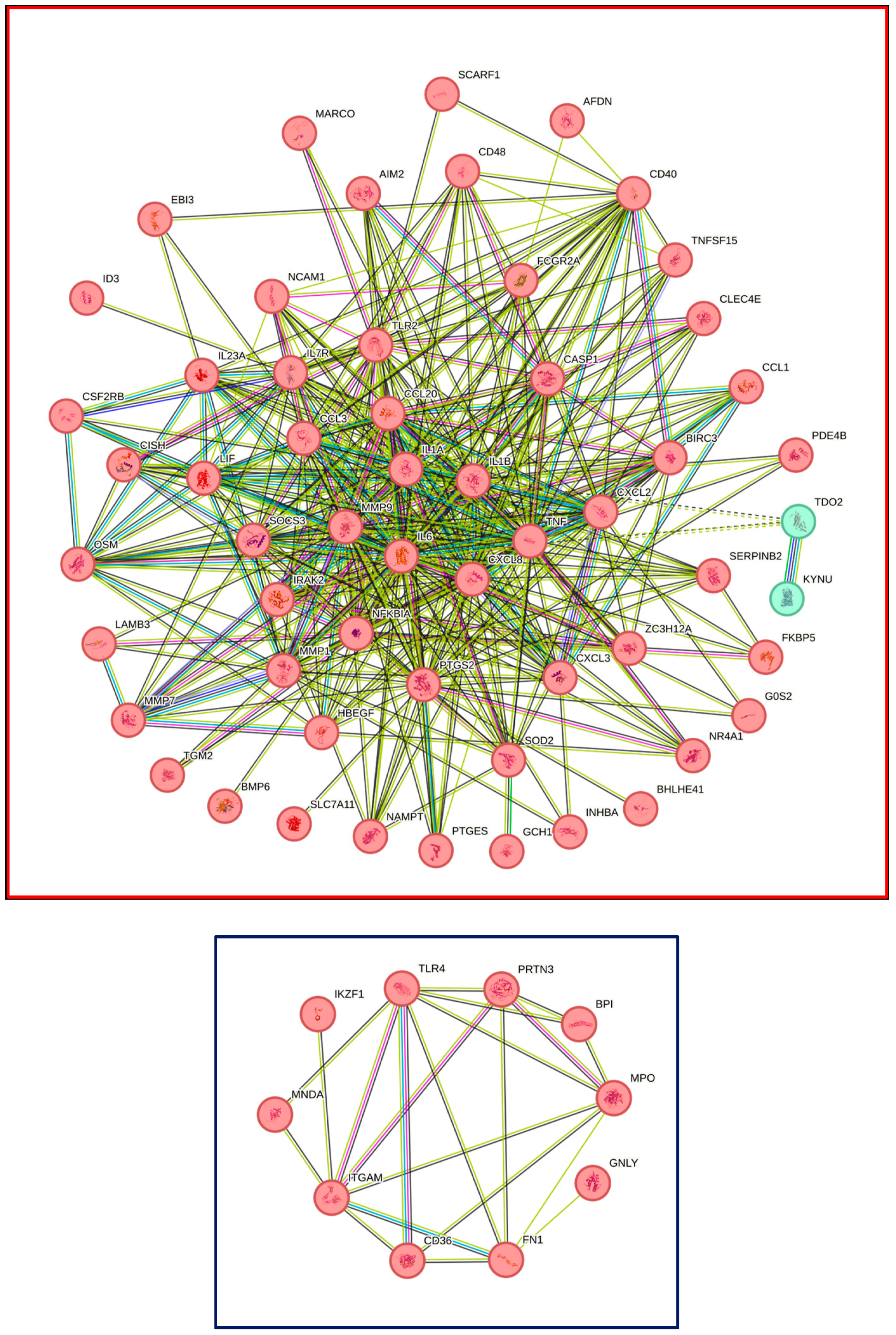
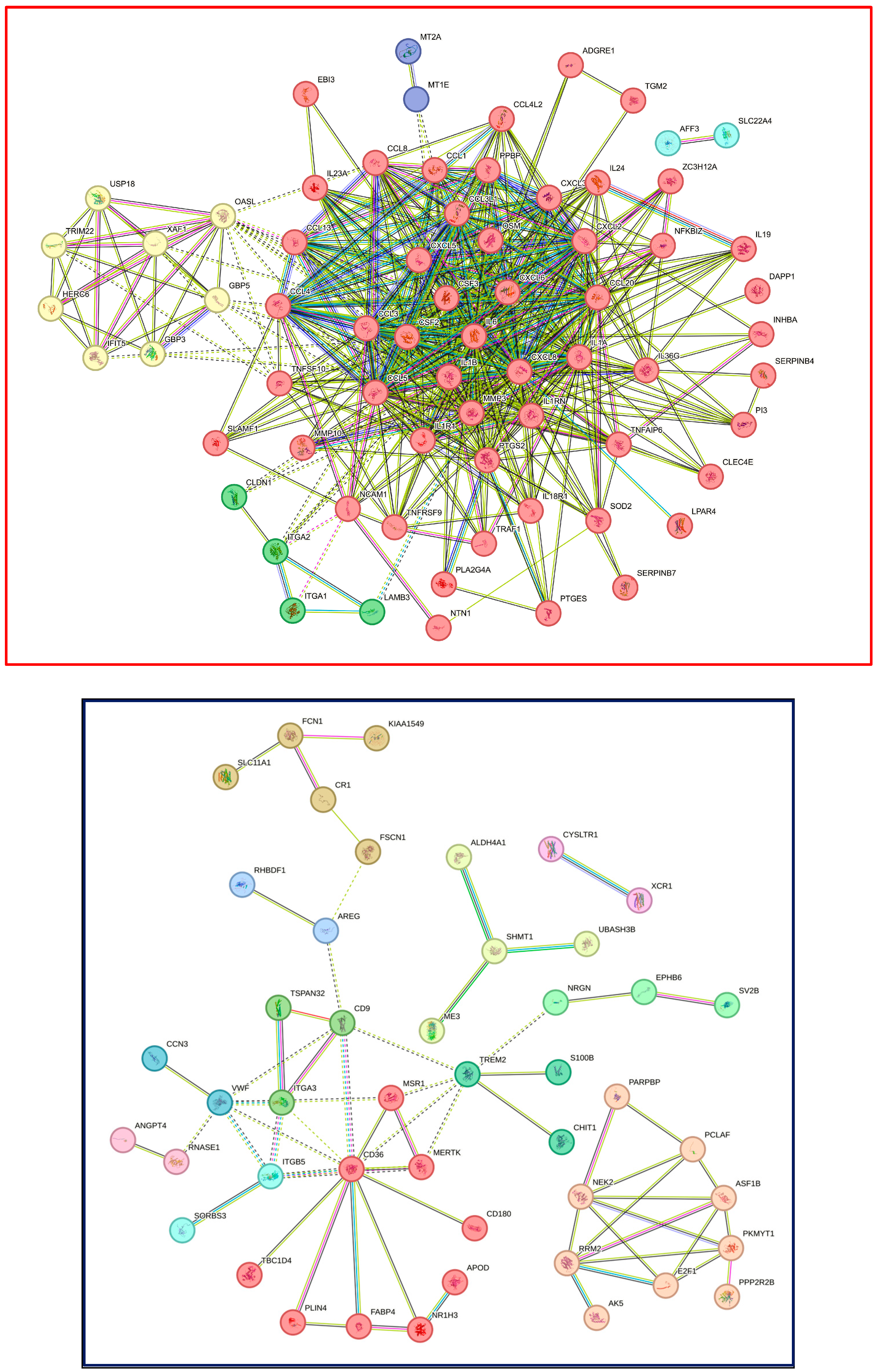
3.5. STRING Analysis of Expression Changes in THP1 and U937 Cells Exposed to LPS and IFNG
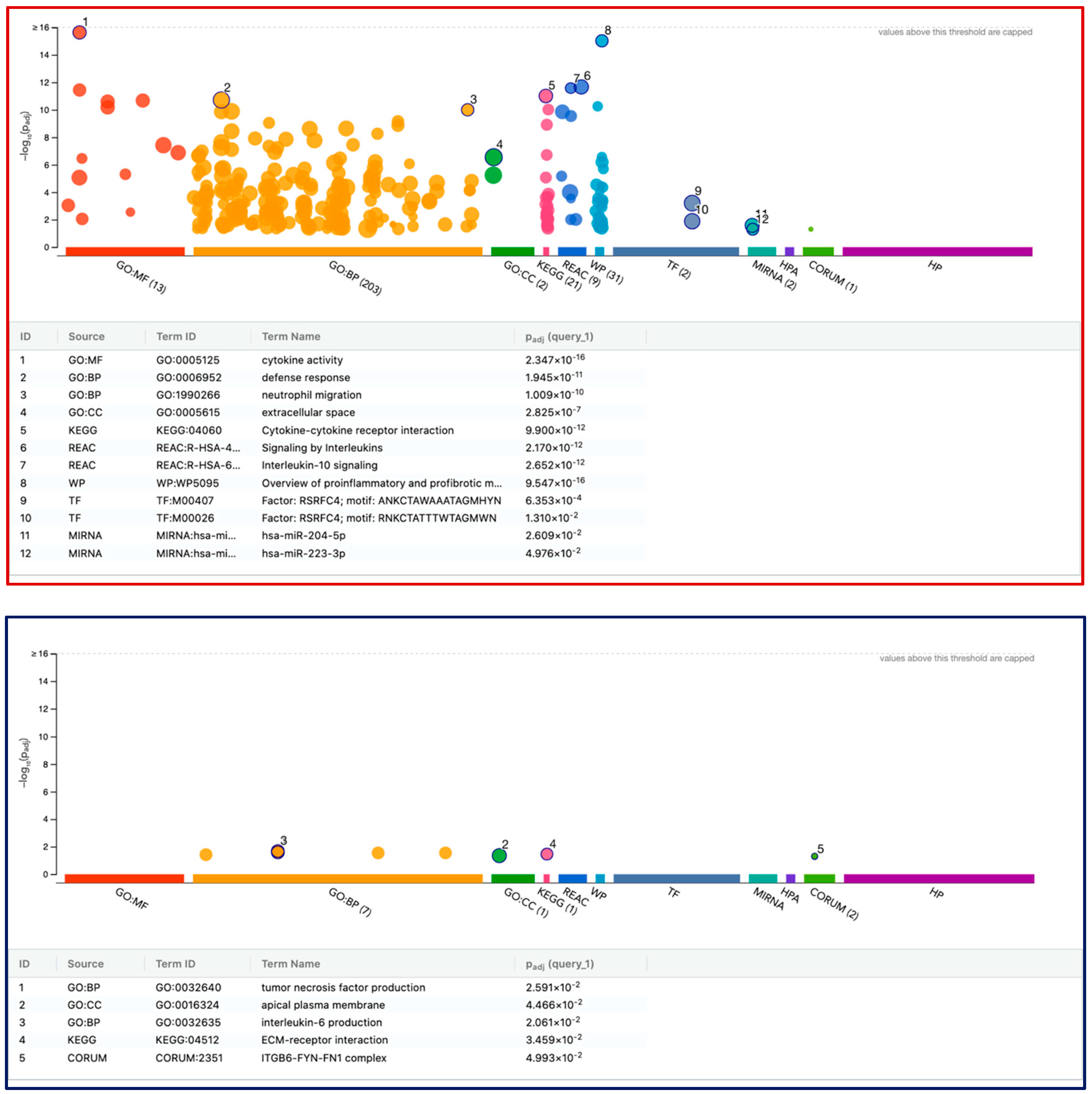
3.6. G:Profiler Analysis of Gene Expression Changes in THP1 and U937 Cells by LPS and IFNG
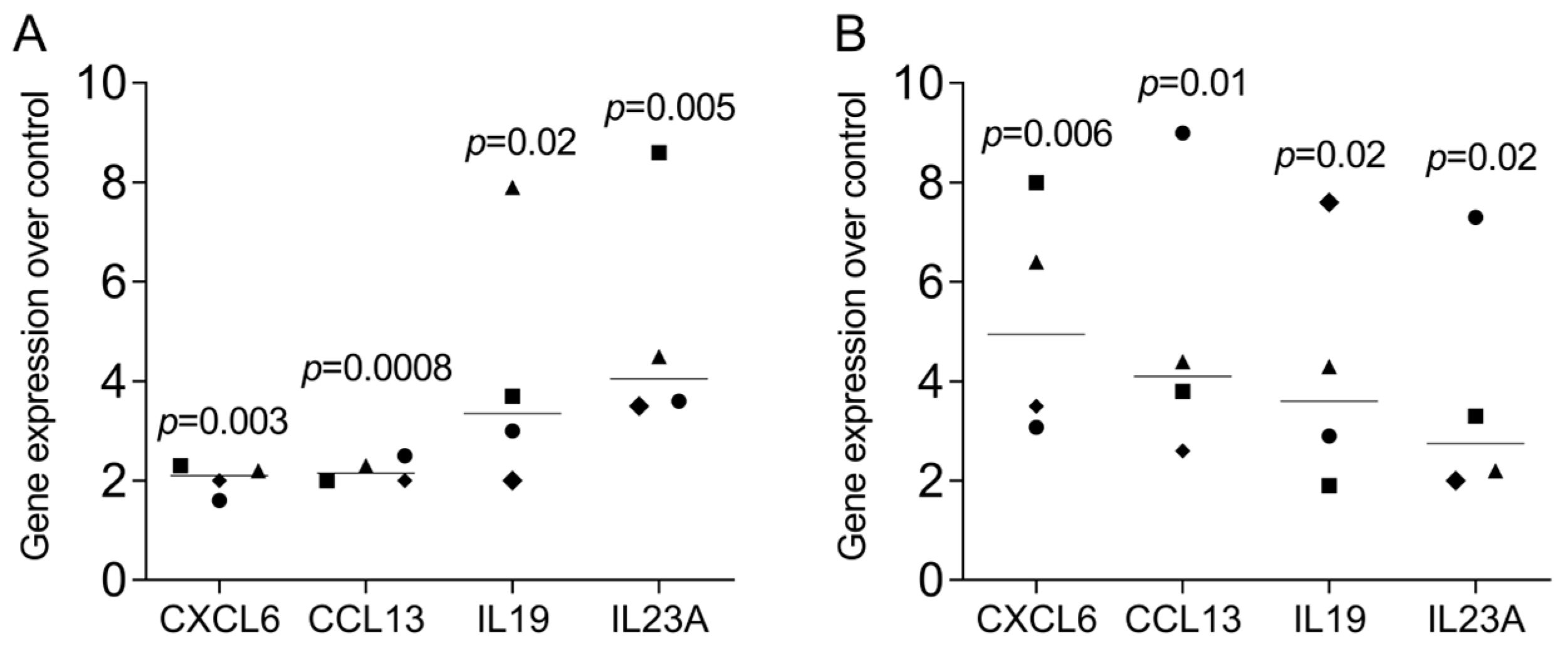
3.7. RT-PCR Gene Expression Changes in THP1 and U937 Cells by LPS and IFNG
4. Discussions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pena, O.A.; Martin, P. Cellular and molecular mechanisms of skin wound healing. Nat. Rev. Mol. Cell Biol. 2024, 25, 599–616. [Google Scholar] [CrossRef]
- Gruber, R. Osteoimmunology: Inflammatory osteolysis and regeneration of the alveolar bone. J. Clin. Periodontol. 2019, 46 (Suppl. 21), 52–69. [Google Scholar] [CrossRef] [PubMed]
- Mbalaviele, G.; Novack, D.V.; Schett, G.; Teitelbaum, S.L. Inflammatory osteolysis: A conspiracy against bone. J. Clin. Investig. 2017, 127, 2030–2039. [Google Scholar] [CrossRef]
- Kinane, D.F.; Stathopoulou, P.G.; Papapanou, P.N. Periodontal diseases. Nat. Rev. Dis. Prim. 2017, 3, 17038. [Google Scholar] [CrossRef] [PubMed]
- Neurath, M.F. Strategies for targeting cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 2024, 24, 559–576. [Google Scholar] [CrossRef]
- Chen, Z.; Bozec, A.; Ramming, A.; Schett, G. Anti-inflammatory and immune-regulatory cytokines in rheumatoid arthritis. Nat. Rev. Rheumatol. 2019, 15, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Pamer, E.G. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 2011, 11, 762–774. [Google Scholar] [CrossRef]
- Guilliams, M.; Svedberg, F.R. Does tissue imprinting restrict macrophage plasticity? Nat. Immunol. 2021, 22, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Okabe, Y.; Medzhitov, R. Tissue biology perspective on macrophages. Nat. Immunol. 2016, 17, 9–17. [Google Scholar] [CrossRef]
- Berdel, W.E.; Fink, U.; Thiel, E.; Stunkel, K.; Greiner, E.; Schwarzkopf, G.; Reichert, A.; Rastetter, J. Purification of human monocytes by adherence to polymeric fluorocarbon. Characterization of the monocyte-enriched cell fraction. Immunobiology 1982, 163, 511–520. [Google Scholar] [CrossRef]
- Bhattacharjee, J.; Das, B.; Mishra, A.; Sahay, P.; Upadhyay, P. Monocytes isolated by positive and negative magnetic sorting techniques show different molecular characteristics and immunophenotypic behaviour. F1000Res 2017, 6, 2045. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chakravarty, S.D.; Ivashkiv, L.B. Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunol. Rev. 2008, 226, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Sweet, M.J.; Hume, D.A. Signal integration between IFNgamma and TLR signalling pathways in macrophages. Immunobiology 2006, 211, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Pickl, W.F.; Majdic, O.; Kohl, P.; Stockl, J.; Riedl, E.; Scheinecker, C.; Bello-Fernandez, C.; Knapp, W. Molecular and functional characteristics of dendritic cells generated from highly purified CD14+ peripheral blood monocytes. J. Immunol. 1996, 157, 3850–3859. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, S.; Yamabe, M.; Yamaguchi, Y.; Kobayashi, Y.; Konno, T.; Tada, K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 1980, 26, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Sundstrom, C.; Nilsson, K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 1976, 17, 565–577. [Google Scholar] [CrossRef]
- Baxter, E.W.; Graham, A.E.; Re, N.A.; Carr, I.M.; Robinson, J.I.; Mackie, S.L.; Morgan, A.W. Standardized protocols for differentiation of THP-1 cells to macrophages with distinct M(IFNgamma+LPS), M(IL-4) and M(IL-10) phenotypes. J. Immunol. Methods 2020, 478, 112721. [Google Scholar] [CrossRef] [PubMed]
- Tamai, R.; Sugawara, S.; Takeuchi, O.; Akira, S.; Takada, H. Synergistic effects of lipopolysaccharide and interferon-gamma in inducing interleukin-8 production in human monocytic THP-1 cells is accompanied by up-regulation of CD14, Toll-like receptor 4, MD-2 and MyD88 expression. J. Endotoxin Res. 2003, 9, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Liu, C.; Wu, B.; Lin, Y.; Ma, T.; Xiong, H.; Wang, Q.; Li, Z.; Ma, C.; Tu, Z. Effects of IRF1 and IFN-beta interaction on the M1 polarization of macrophages and its antitumor function. Int. J. Mol. Med. 2016, 38, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Ucla, C.; Roux-Lombard, P.; Fey, S.; Dayer, J.M.; Mach, B. Interferon gamma drastically modifies the regulation of interleukin 1 genes by endotoxin in U937 cells. J. Clin. Investig. 1990, 85, 185–191. [Google Scholar] [CrossRef]
- Nascimento, C.R.; Rodrigues Fernandes, N.A.; Gonzalez Maldonado, L.A.; Rossa Junior, C. Comparison of monocytic cell lines U937 and THP-1 as macrophage models for in vitro studies. Biochem. Biophys. Rep. 2022, 32, 101383. [Google Scholar] [CrossRef]
- Chanput, W.; Peters, V.; Wichers, H. THP-1 and U937 Cells. In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., Lopez-Exposito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Cham, Switzerland, 2015; pp. 147–159. [Google Scholar]
- Ueno, T.; Yamamoto, Y.; Kawasaki, K. Phagocytosis of microparticles increases responsiveness of macrophage-like cell lines U937 and THP-1 to bacterial lipopolysaccharide and lipopeptide. Sci. Rep. 2021, 11, 6782. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, M.P.; Jones Taggart, H.; Strap, J.L.; Edun, G.; Green-Johnson, J.M. Milk fermented with Lactobacillus rhamnosus R0011 induces a regulatory cytokine profile in LPS-challenged U937 and THP-1 macrophages. Curr. Res. Food Sci. 2020, 3, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Shiratori, H.; Feinweber, C.; Luckhardt, S.; Linke, B.; Resch, E.; Geisslinger, G.; Weigert, A.; Parnham, M.J. THP-1 and human peripheral blood mononuclear cell-derived macrophages differ in their capacity to polarize in vitro. Mol. Immunol. 2017, 88, 58–68. [Google Scholar] [CrossRef]
- Genin, M.; Clement, F.; Fattaccioli, A.; Raes, M.; Michiels, C. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer 2015, 15, 577. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Li, Y.; Chen, J.; Wang, N. Azilsartan Suppressed LPS-Induced Inflammation in U937 Macrophages through Suppressing Oxidative Stress and Inhibiting the TLR2/MyD88 Signal Pathway. ACS Omega 2021, 6, 113–118. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Gerst, R.; Hölzer, M. PCAGO: An interactive tool to analyze RNA-Seq data with principal component analysis. bioRxiv 2019. [Google Scholar] [CrossRef]
- Goedhart, J.; Luijsterburg, M.S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep. 2020, 10, 20560. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Alasoo, K.; Martinez, F.O.; Hale, C.; Gordon, S.; Powrie, F.; Dougan, G.; Mukhopadhyay, S.; Gaffney, D.J. Transcriptional profiling of macrophages derived from monocytes and iPS cells identifies a conserved response to LPS and novel alternative transcription. Sci. Rep. 2015, 5, 12524. [Google Scholar] [CrossRef] [PubMed]
- Baek, Y.S.; Haas, S.; Hackstein, H.; Bein, G.; Hernandez-Santana, M.; Lehrach, H.; Sauer, S.; Seitz, H. Identification of novel transcriptional regulators involved in macrophage differentiation and activation in U937 cells. BMC Immunol. 2009, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shi, B.; Huang, C.C.; Eksarko, P.; Pope, R.M. Transcriptional diversity during monocyte to macrophage differentiation. Immunol. Lett. 2008, 117, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhu, Y.; Zhao, Z.; Yan, L.; Xu, T.; Wang, X.; He, H.; Xia, X.; Zheng, W.; Xue, X. RNA sequencing analyses of gene expressions in a canine macrophages cell line DH82 infected with canine distemper virus. Infect. Genet. Evol. 2020, 80, 104206. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, Y.; Zhang, K.; Xiao, Y.; Lu, J.; Fan, R. Multiplexed, Sequential Secretion Analysis of the Same Single Cells Reveals Distinct Effector Response Dynamics Dependent on the Initial Basal State. Adv. Sci. 2019, 6, 1801361. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.; Lu, Y.; Eisele, M.R.; Sulistijo, E.S.; Khan, N.; Fan, R.; Miller-Jensen, K. Analysis of single-cell cytokine secretion reveals a role for paracrine signaling in coordinating macrophage responses to TLR4 stimulation. Sci. Signal. 2015, 8, ra59. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.; Metsger, M.; Bauch, S.; Vidal, R.; Bottcher, M.; Grote, P.; Kliem, M.; Sauer, S. Signals trigger state-specific transcriptional programs to support diversity and homeostasis in immune cells. Sci. Signal. 2019, 12, eaao5820. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Yang, C.S.; Arifuzzaman, S.; Kim, S.; Kim, S.Y.; Jung, K.H.; Lee, Y.S.; Chai, Y.G. High-Resolution Mapping and Dynamics of the Transcriptome, Transcription Factors, and Transcription Co-Factor Networks in Classically and Alternatively Activated Macrophages. Front. Immunol. 2018, 9, 22. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panahipour, L.; Micucci, C.; Gruber, R. Inflammatory Response of THP1 and U937 Cells: The RNAseq Approach. Cells 2024, 13, 2062. https://doi.org/10.3390/cells13242062
Panahipour L, Micucci C, Gruber R. Inflammatory Response of THP1 and U937 Cells: The RNAseq Approach. Cells. 2024; 13(24):2062. https://doi.org/10.3390/cells13242062
Chicago/Turabian StylePanahipour, Layla, Chiara Micucci, and Reinhard Gruber. 2024. "Inflammatory Response of THP1 and U937 Cells: The RNAseq Approach" Cells 13, no. 24: 2062. https://doi.org/10.3390/cells13242062
APA StylePanahipour, L., Micucci, C., & Gruber, R. (2024). Inflammatory Response of THP1 and U937 Cells: The RNAseq Approach. Cells, 13(24), 2062. https://doi.org/10.3390/cells13242062






