CD133-Dependent Activation of Phosphoinositide 3-Kinase /AKT/Mammalian Target of Rapamycin Signaling in Melanoma Progression and Drug Resistance
Abstract
:1. Introduction
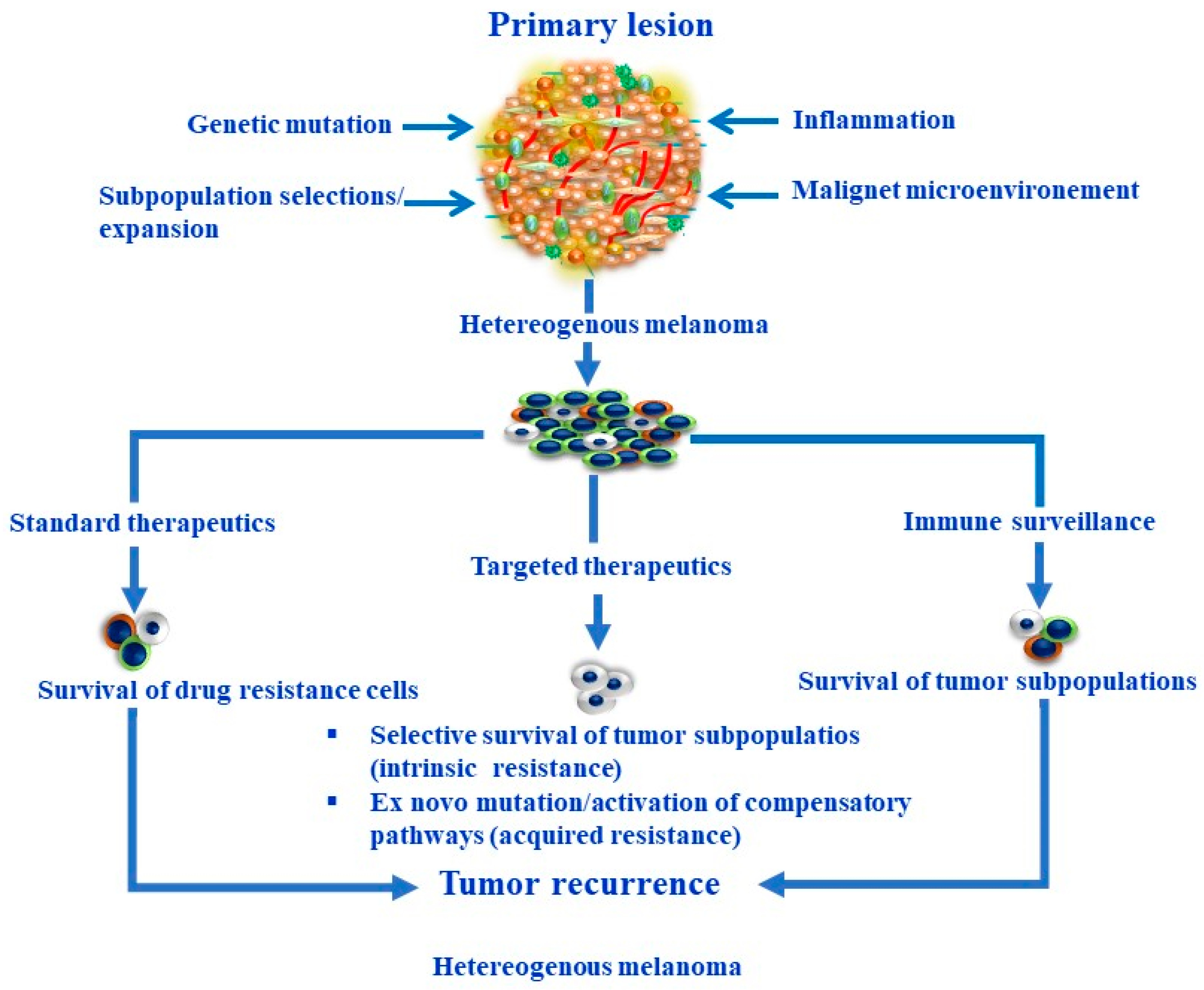
2. Melanoma Heterogeneity and Plasticity
3. Melanoma Stem Cells
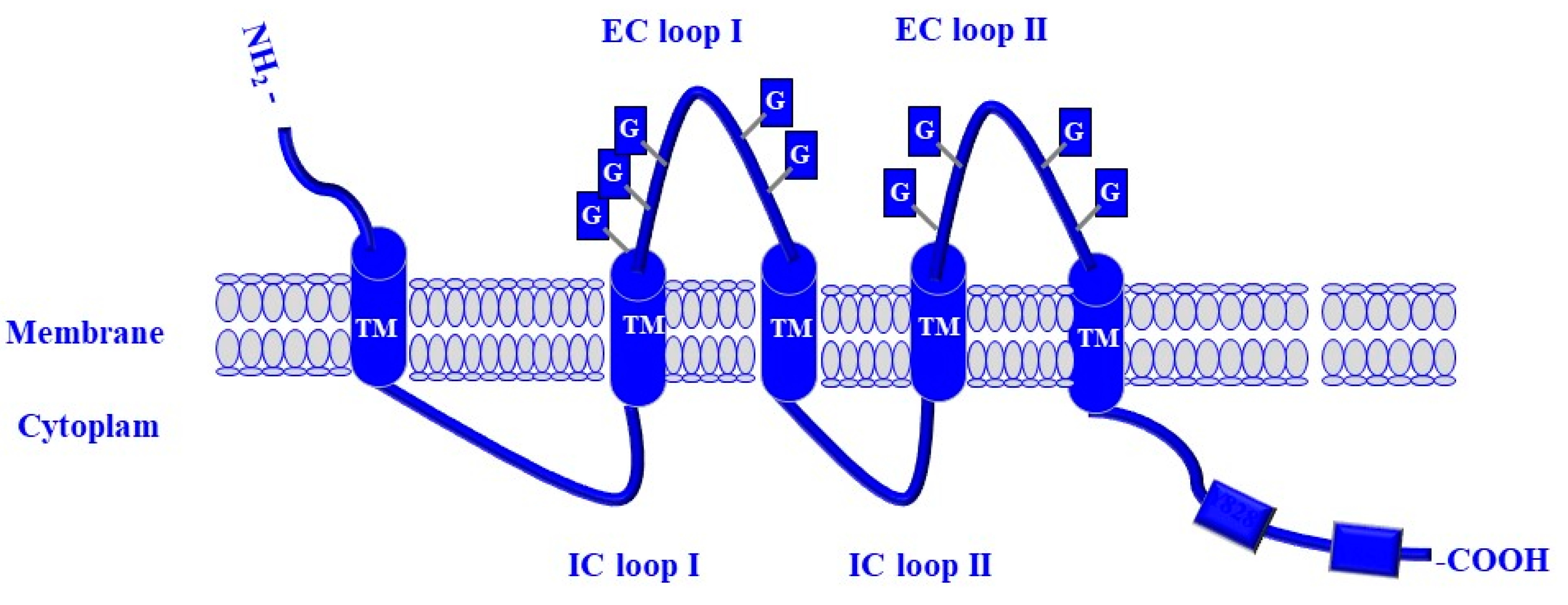
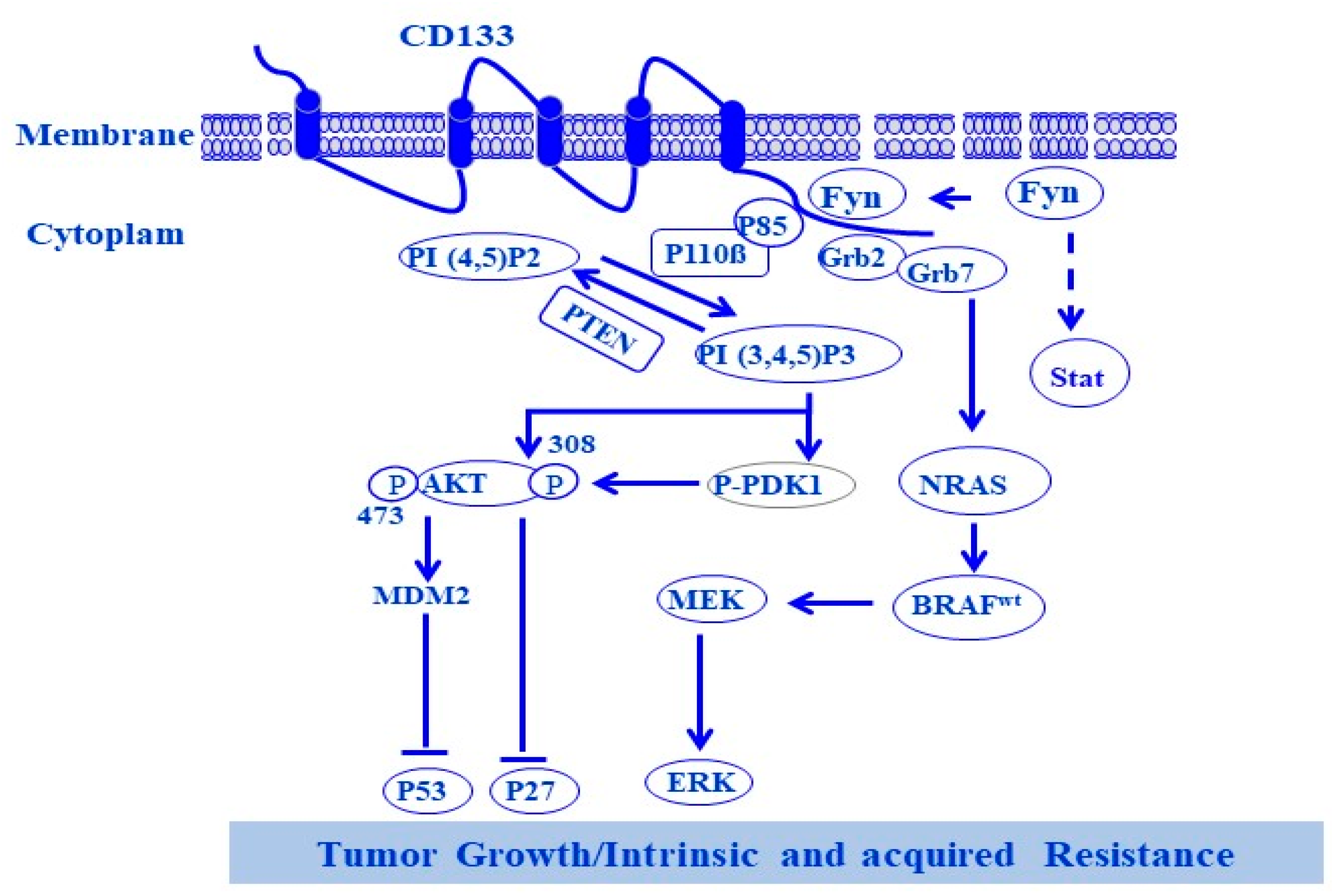
4. CD133
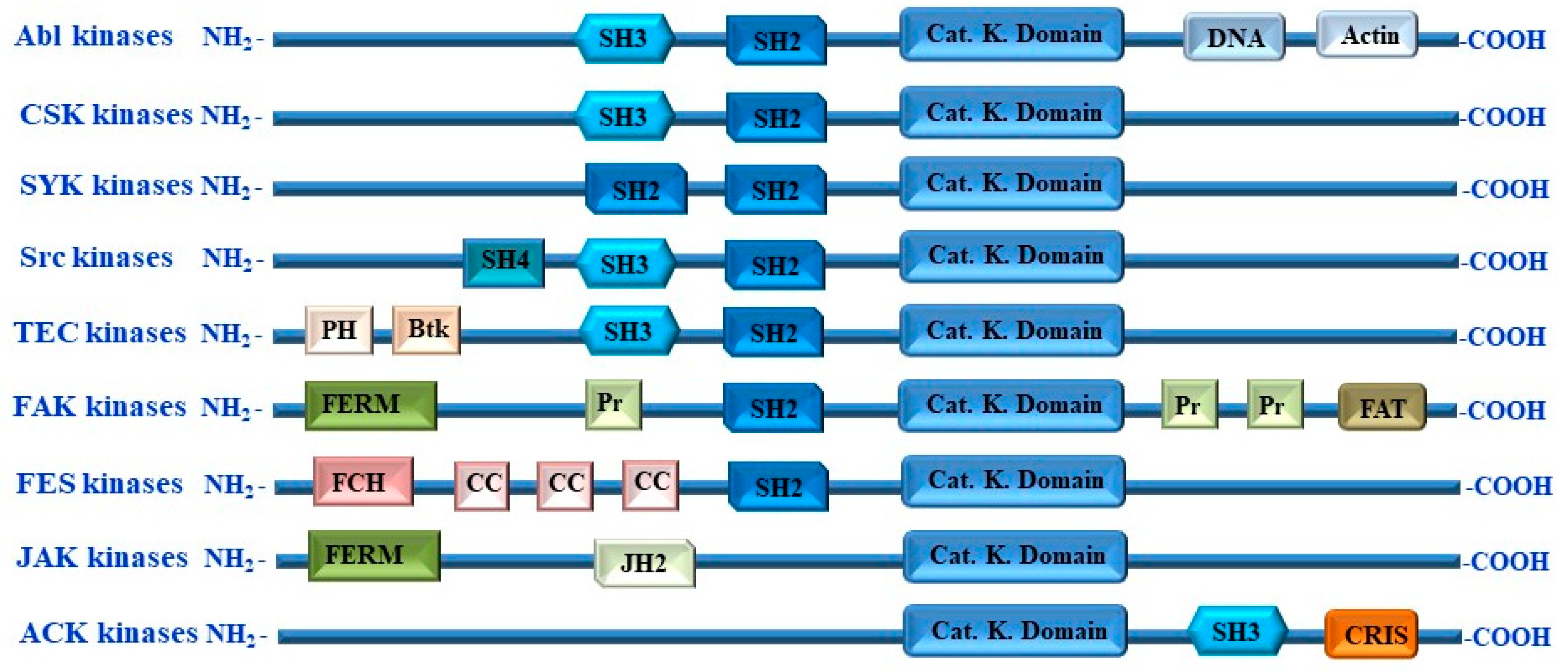
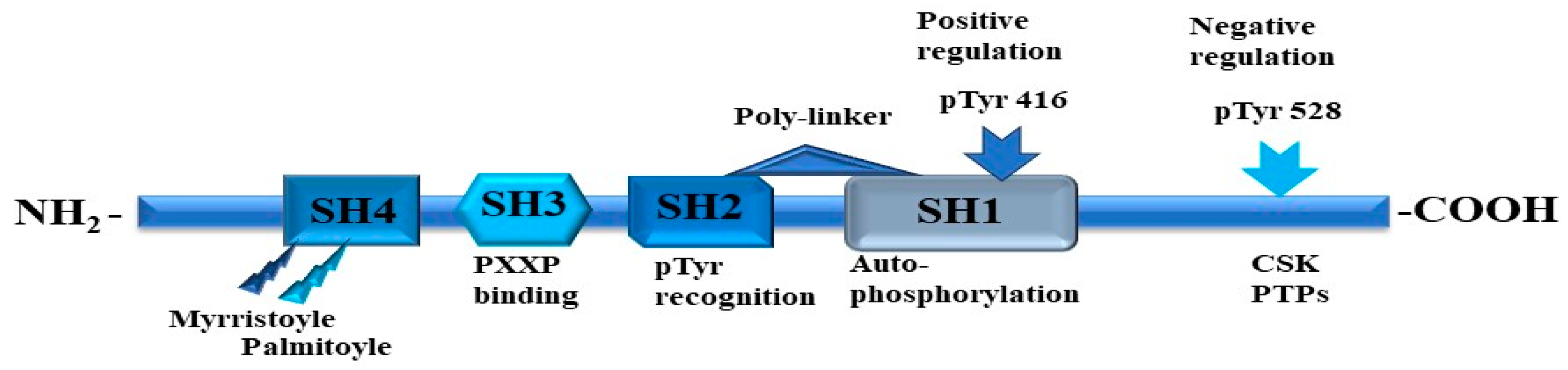
5. Non-Receptor Tyrosine Kinases
6. Non-Receptor Tyrosine Kinase Fyn
7. Receptor and Non-Receptor Tyrosine Kinase-Mediated Pathways to Melanoma Progression and Drug Resistance
8. Melanoma Progression and Drug Resistance Are Attributed to Dysregulation of PI3K/AKT/mTOR Pathways
9. PI3K/AKT/mTOR Pathway as a Therapeutic Target in Melanoma Treatment
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Boumahdi, S.; de Sauvage, F.J. The great escape: Tumour cell plasticity in resistance to targeted therapy. Nat. Rev. Drug Discov. 2020, 19, 39–56. [Google Scholar] [CrossRef]
- Li, Z.; Jia, H.; Zhang, B.; Zhang, Y.; Li, H.; Song, P. The clinical features, treatment, and prognosis of primary mediastinal malignant melanoma: A case report. Medicine 2017, 96, e6436. [Google Scholar] [CrossRef]
- Hamid, O.; Robert, C.; Daud, A.; Hodi, F.S.; Hwu, W.J.; Kefford, R.; Wolchok, J.D.; Hersey, P.; Joseph, R.; Weber, J.S.; et al. Five-year survival outcomes for patients with advanced melanoma treated with pembrolizumab in KEYNOTE-001. Ann. Oncol. 2019, 30, 582–588. [Google Scholar] [CrossRef] [PubMed]
- El-Khattouti, A.; Sheehan, N.T.; Monico, J.; Drummond, H.A.; Haikel, Y.; Brodell, R.T.; Megahed, M.; Hassan, M. CD133+ melanoma subpopulation acquired resistance to caffeic acid phenethyl ester-induced apoptosis is attributed to the elevated expression of ABCB5: Significance for melanoma treatment. Cancer Lett. 2015, 357, 83–104. [Google Scholar] [CrossRef] [PubMed]
- El-Khattouti, A.; Selimovic, D.; Haïkel, Y.; Megahed, M.; Gomez, C.R.; Hassan, M. Identification and analysis of CD133(+) melanoma stem-like cells conferring resistance to taxol: An insight into the mechanisms of their resistance and response. Cancer Lett. 2014, 343, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Barzegar Behrooz, A.; Syahir, A.; Ahmad, S. CD133: Beyond a cancer stem cell biomarker. J. Drug Target. 2019, 27, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Kumar, S.; Gorain, M.; Tomar, D.; Patil, H.S.; Radharani, N.N.V.; Kumar, T.V.S.; Patil, T.V.; Thulasiram, H.V.; Kundu, G.C. Notch1-MAPK Signaling Axis Regulates CD133. J. Investig. Dermatol. 2016, 136, 2462–2474. [Google Scholar] [CrossRef]
- Jamal, S.M.E.; Alamodi, A.; Wahl, R.U.; Grada, Z.; Shareef, M.A.; Hassan, S.Y.; Murad, F.; Hassan, S.L.; Santourlidis, S.; Gomez, C.R.; et al. Melanoma stem cell maintenance and chemo-resistance are mediated by CD133 signal to PI3K-dependent pathways. Oncogene 2020, 39, 5468–5478. [Google Scholar] [CrossRef] [PubMed]
- Inoki, K.; Ouyang, H.; Li, Y.; Guan, K.L. Signaling by target of rapamycin proteins in cell growth control. Microbiol. Mol. Biol. Rev. 2005, 69, 79–100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cao, S.; Gong, M.; Zhang, W.; Zhu, Z.; Wu, S.; Yue, Y.; Qian, W.; Ma, Q.; Wang, S.; et al. Mechanically activated ion channel Piezo1 contributes to melanoma malignant progression through AKT/mTOR signaling. Cancer Biol. Ther. 2022, 23, 336–347. [Google Scholar] [CrossRef]
- Sanchez, J.N.; Wang, T.; Cohen, M.S. BRAF and MEK Inhibitors: Use and Resistance in BRAF-Mutated Cancers. Drugs 2018, 78, 549–566. [Google Scholar] [CrossRef]
- Adashek, J.J.; Menta, A.K.; Reddy, N.K.; Desai, A.P.; Roszik, J.; Subbiah, V. Tissue-Agnostic Activity of BRAF plus MEK Inhibitor in BRAF V600-Mutant Tumors. Mol. Cancer Ther. 2022, 21, 871–878. [Google Scholar] [CrossRef]
- Mak, G.; Arkenau, H.T.; Chin, M. Resistance surveillance in a BRAF mutant melanoma patient on long-term BRAF-inhibitor treatment. Melanoma Res. 2014, 24, 408–412. [Google Scholar] [CrossRef] [PubMed]
- Savoia, P.; Zavattaro, E.; Cremona, O. Clinical Implications of Acquired BRAF Inhibitors resistance in Melanoma. Int. J. Mol. Sci. 2020, 21, 9730. [Google Scholar] [CrossRef]
- Haluska, F.; Pemberton, T.; Ibrahim, N.; Kalinsky, K. The RTK/RAS/BRAF/PI3K pathways in melanoma: Biology, small molecule inhibitors, and potential applications. Semin. Oncol. 2007, 34, 546–554. [Google Scholar] [CrossRef]
- Khaddour, K.; Maahs, L.; Avila-Rodriguez, A.M.; Maamar, Y.; Samaan, S.; Ansstas, G. Melanoma Targeted Therapies beyond. Cancers 2021, 13, 5847. [Google Scholar] [CrossRef]
- Rozengurt, E.; Soares, H.P.; Sinnet-Smith, J. Suppression of feedback loops mediated by PI3K/mTOR induces multiple overactivation of compensatory pathways: An unintended consequence leading to drug resistance. Mol. Cancer Ther. 2014, 13, 2477–2488. [Google Scholar] [CrossRef]
- Corrales, E.; Levit-Zerdoun, E.; Metzger, P.; Mertes, R.; Lehmann, A.; Münch, J.; Lemke, S.; Kowar, S.; Boerries, M. PI3K/AKT signaling allows for MAPK/ERK pathway independency mediating dedifferentiation-driven Drug resistance in melanoma. Cell Commun. Signal. 2022, 20, 187. [Google Scholar] [CrossRef]
- Yajima, I.; Kumasaka, M.Y.; Thang, N.D.; Goto, Y.; Takeda, K.; Yamanoshita, O.; Iida, M.; Ohgami, N.; Tamura, H.; Kawamoto, Y.; et al. RAS/RAF/MEK/ERK and PI3K/PTEN/AKT Signaling in Malignant Melanoma Progression and Therapy. Dermatol. Res. Pract. 2012, 2012, 354191. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Jin, L.; Wilmott, J.S.; Hu, W.L.; Yosufi, B.; Thorne, R.F.; Liu, T.; Rizos, H.; Yan, X.G.; Dong, L.; et al. PI(4,5)P2 5-phosphatase A regulates PI3K/Akt signalling and has a tumour suppressive role in human melanoma. Nat. Commun. 2013, 4, 1508. [Google Scholar] [CrossRef] [PubMed]
- De Craene, J.O.; Bertazzi, D.L.; Bär, S.; Friant, S. Phosphoinositides, Major Actors in Membrane Trafficking and Lipid Signaling Pathways. Int. J. Mol. Sci. 2017, 18, 634. [Google Scholar] [CrossRef]
- Bilanges, B.; Posor, Y.; Vanhaesebroeck, B. PI3K isoforms in cell signalling and vesicle trafficking. Nat. Rev. Mol. Cell Biol. 2019, 20, 515–534. [Google Scholar] [CrossRef]
- Jean, S.; Kiger, A.A. Classes of phosphoinositide 3-kinases at a glance. J. Cell Sci. 2014, 127 Pt 5, 923–928. [Google Scholar] [CrossRef]
- Siempelkamp, B.D.; Rathinaswamy, M.K.; Jenkins, M.L.; Burke, J.E. Molecular mechanism of activation of class IA phosphoinositide 3-kinases (PI3Ks) by membrane-localized HRas. J. Biol. Chem. 2017, 292, 12256–12266. [Google Scholar] [CrossRef]
- Vidal, S.; Bouzaher, Y.H.; El Motiam, A.; Seoane, R.; Rivas, C. Overview of the regulation of the class IA PI3K/AKT pathway by SUMO. Semin. Cell Dev. Biol. 2022, 132, 51–61. [Google Scholar] [CrossRef]
- Fox, M.; Mott, H.R.; Owen, D. Class IA PI3K regulatory subunits: p110-independent roles and structures. Biochem. Soc. Trans. 2020, 48, 1397–1417. [Google Scholar] [CrossRef] [PubMed]
- Parkman, G.L.; Foth, M.; Kircher, D.A.; Holmen, S.L.; McMahon, M. The role of PI3’-lipid signalling in melanoma initiation, progression and maintenance. Exp. Dermatol. 2022, 31, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, S.J.; Ferguson, D.T.; Mitchell, C.A.; Ooms, L.M. Regulation of PI3K effector signalling in cancer by the phosphoinositide phosphatases. Biosci. Rep. 2017, 37, BSR20160432. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Schneider, H.; Rudd, C.E. Phosphatidylinositol 3-kinase p85 adaptor function in T-cells. Co-stimulation and regulation of cytokine transcription independent of associated p110. J. Biol. Chem. 2002, 277, 912–921. [Google Scholar] [CrossRef]
- Jiang, X.; Chen, S.; Asara, J.M.; Balk, S.P. Phosphoinositide 3-kinase pathway activation in phosphate and tensin homolog (PTEN)-deficient prostate cancer cells is independent of receptor tyrosine kinases and mediated by the p110beta and p110delta catalytic subunits. J. Biol. Chem. 2010, 285, 14980–14989. [Google Scholar] [CrossRef]
- Sipeki, S.; Koprivanacz, K.; Takács, T.; Kurilla, A.; László, L.; Vas, V.; Buday, L. Novel Roles of SH2 and SH3 Domains in Lipid Binding. Cells 2021, 10, 1191. [Google Scholar] [CrossRef] [PubMed]
- Candido, S.; Salemi, R.; Piccinin, S.; Falzone, L.; Libra, M. The PIK3CA H1047R Mutation Confers Resistance to BRAF and MEK Inhibitors in A375 Melanoma Cells through the Cross-Activation of MAPK and PI3K-Akt Pathways. Pharmaceutics 2022, 14, 590. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yu, J.; Yan, J.; Dai, J.; Si, L.; Chi, Z.; Sheng, X.; Cui, C.; Ma, M.; Tang, H.; et al. PI3K/AKT/mTOR pathway inhibitors inhibit the growth of melanoma cells with mTOR H2189Y mutations in vitro. Cancer Biol. Ther. 2018, 19, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Si, L.; Li, Y.; Wu, X.; Xu, X.; Dai, J.; Tang, H.; Ma, M.; Chi, Z.; Sheng, X.; et al. Analysis of mTOR Gene Aberrations in Melanoma Patients and Evaluation of Their Sensitivity to PI3K-AKT-mTOR Pathway Inhibitors. Clin. Cancer Res. 2016, 22, 1018–1027. [Google Scholar] [CrossRef]
- Sinnberg, T.; Lasithiotakis, K.; Niessner, H.; Schittek, B.; Flaherty, K.T.; Kulms, D.; Maczey, E.; Campos, M.; Gogel, J.; Garbe, C.; et al. Inhibition of PI3K-AKT-mTOR signaling sensitizes melanoma cells to cisplatin and temozolomide. J. Investig. Dermatol. 2009, 129, 1500–1515. [Google Scholar] [CrossRef]
- Hocker, T.L.; Singh, M.K.; Tsao, H. Melanoma genetics and therapeutic approaches in the 21st century: Moving from the benchside to the bedside. J. Investig. Dermatol. 2008, 128, 2575–2595. [Google Scholar] [CrossRef]
- Pópulo, H.; Lopes, J.M.; Soares, P. The mTOR signalling pathway in human cancer. Int. J. Mol. Sci. 2012, 13, 1886–1918. [Google Scholar] [CrossRef]
- Moschetta, M.; Reale, A.; Marasco, C.; Vacca, A.; Carratù, M.R. Therapeutic targeting of the mTOR-signalling pathway in cancer: Benefits and limitations. Br. J. Pharmacol. 2014, 171, 3801–3813. [Google Scholar] [CrossRef]
- Indini, A.; Fiorilla, I.; Ponzone, L.; Calautti, E.; Audrito, V. NAD/NAMPT and mTOR Pathways in Melanoma: Drivers of Drug Resistance and Prospective Therapeutic Targets. Int. J. Mol. Sci. 2022, 23, 9985. [Google Scholar] [CrossRef] [PubMed]
- Karbowniczek, M.; Spittle, C.S.; Morrison, T.; Wu, H.; Henske, E.P. mTOR is activated in the majority of malignant melanomas. J. Investig. Dermatol. 2008, 128, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Karakousis, G.C. Melanoma of unknown primary. J. Surg. Oncol. 2019, 119, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Shain, A.H.; Yeh, I.; Kovalyshyn, I.; Sriharan, A.; Talevich, E.; Gagnon, A.; Dummer, R.; North, J.; Pincus, L.; Ruben, B.; et al. The Genetic Evolution of Melanoma from Precursor Lesions. N. Engl. J. Med. 2015, 373, 1926–1936. [Google Scholar] [CrossRef] [PubMed]
- Katzendobler, S.; Do, A.; Weller, J.; Rejeski, K.; Dorostkar, M.M.; Albert, N.L.; Forbrig, R.; Niyazi, M.; Egensperger, R.; Tonn, J.C.; et al. The value of stereotactic biopsy of primary and recurrent brain metastases in the era of precision medicine. Front. Oncol. 2022, 12, 1014711. [Google Scholar] [CrossRef] [PubMed]
- Ward-Peterson, M.; Acuña, J.M.; Alkhalifah, M.K.; Nasiri, A.M.; Al-Akeel, E.S.; Alkhaldi, T.M.; Dawari, S.A.; Aldaham, S.A. Association Between Race/Ethnicity and Survival of Melanoma Patients in the United States Over 3 Decades: A Secondary Analysis of SEER Data. Medicine 2016, 95, e3315. [Google Scholar] [CrossRef] [PubMed]
- Saginala, K.; Barsouk, A.; Aluru, J.S.; Rawla, P. Epidemiology of Melanoma. Med. Sci. 2021, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Haggar, F.A.; Boushey, R.P. Colorectal cancer epidemiology: Incidence, mortality, survival, and risk factors. Clin. Colon Rectal Surg. 2009, 22, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Volkovova, K.; Bilanicova, D.; Bartonova, A.; Letašiová, S.; Dusinska, M. Associations between environmental factors and incidence of cutaneous melanoma. Review. Environ. Health 2012, 11 (Suppl. S1), S12. [Google Scholar] [CrossRef]
- Budden, T.; Bowden, N.A. The role of altered nucleotide excision repair and UVB-induced DNA damage in melanomagenesis. Int. J. Mol. Sci. 2013, 14, 1132–1151. [Google Scholar] [CrossRef]
- Kumar, R.; Deep, G.; Agarwal, R. An Overview of Ultraviolet B Radiation-Induced Skin Cancer Chemoprevention by Silibinin. Curr. Pharmacol. Rep. 2015, 1, 206–215. [Google Scholar] [CrossRef]
- Zhang, A.; Miao, K.; Sun, H.; Deng, C.X. Tumor heterogeneity reshapes the tumor microenvironment to influence drug resistance. Int. J. Biol. Sci. 2022, 18, 3019–3033. [Google Scholar] [CrossRef]
- Yang, E.; Wang, X.; Gong, Z.; Yu, M.; Wu, H.; Zhang, D. Exosome-mediated metabolic reprogramming: The emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct. Target. Ther. 2020, 5, 242. [Google Scholar] [CrossRef]
- McQuerry, J.A.; Chang, J.T.; Bowtell, D.D.L.; Cohen, A.; Bild, A.H. Mechanisms and clinical implications of tumor heterogeneity and convergence on recurrent phenotypes. J. Mol. Med. 2017, 95, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Hida, K.; Ohga, N.; Akiyama, K.; Maishi, N.; Hida, Y. Heterogeneity of tumor endothelial cells. Cancer Sci. 2013, 104, 1391–1395. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.Y.; Menzies, A.M.; Rizos, H. Mechanisms and strategies to overcome resistance to molecularly targeted therapy for melanoma. Cancer 2017, 123, 2118–2129. [Google Scholar] [CrossRef] [PubMed]
- Tellez-Gabriel, M.; Ory, B.; Lamoureux, F.; Heymann, M.F.; Heymann, D. Tumour Heterogeneity: The Key Advantages of Single-Cell Analysis. Int. J. Mol. Sci. 2016, 17, 2142. [Google Scholar] [CrossRef]
- Sun, X.X.; Yu, Q. Intra-tumor heterogeneity of cancer cells and its implications for cancer treatment. Acta Pharmacol. Sin. 2015, 36, 1219–1227. [Google Scholar] [CrossRef]
- Sanli, Y.; Leake, J.; Odu, A.; Xi, Y.; Subramaniam, R.M. Tumor Heterogeneity on FDG PET/CT and Immunotherapy: An Imaging Biomarker for Predicting Treatment Response in Patients With Metastatic Melanoma. AJR Am. J. Roentgenol. 2019, 212, 1318–1326. [Google Scholar] [CrossRef]
- Crucitta, S.; Cucchiara, F.; Mathijssen, R.; Mateo, J.; Jager, A.; Joosse, A.; Passaro, A.; Attili, I.; Petrini, I.; van Schaik, R.; et al. Treatment-driven tumour heterogeneity and drug resistance: Lessons from solid tumours. Cancer Treat. Rev. 2022, 104, 102340. [Google Scholar] [CrossRef]
- Shannan, B.; Perego, M.; Somasundaram, R.; Herlyn, M. Heterogeneity in Melanoma. Cancer. Treat. Res. 2016, 167, 1–15. [Google Scholar] [CrossRef]
- Katenkamp, D. Cellular heterogeneity. Explanation for changing of tumor phenotype and biologic behavior in soft tissue sarcomas. Pathol. Res. Pract. 1988, 183, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Jamal-Hanjani, M.; Quezada, S.A.; Larkin, J.; Swanton, C. Translational implications of tumor heterogeneity. Clin. Cancer Res. 2015, 21, 1258–1266. [Google Scholar] [CrossRef]
- Sanna, A.; Harbst, K.; Johansson, I.; Christensen, G.; Lauss, M.; Mitra, S.; Rosengren, F.; Häkkinen, J.; Vallon-Christersson, J.; Olsson, H.; et al. Tumor genetic heterogeneity analysis of chronic sun-damaged melanoma. Pigment Cell Melanoma Res. 2020, 33, 480–489. [Google Scholar] [CrossRef]
- Testa, U.; Castelli, G.; Pelosi, E. Melanoma: Genetic Abnormalities, Tumor Progression, Clonal Evolution and Tumor Initiating Cells. Med. Sci. 2017, 5, 28. [Google Scholar] [CrossRef]
- Pipek, O.; Vizkeleti, L.; Doma, V.; Alpár, D.; Bödör, C.; Kárpáti, S.; Timar, J. The Driverless Triple-Wild-Type (BRAF, RAS, KIT) Cutaneous Melanoma: Whole Genome Sequencing Discoveries. Cancers 2023, 15, 1712. [Google Scholar] [CrossRef]
- Guo, W.; Wang, H.; Li, C. Signal pathways of melanoma and targeted therapy. Signal Transduct. Target. Ther. 2021, 6, 424. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, G.; Colombino, M.; Casula, M.; Manca, A.; Mandalà, M.; Cossu, A.; Italian Melanoma Intergroup for the Italian Melanoma Intergroup (IMI). Molecular Pathways in Melanomagenesis: What We Learned from Next-Generation Sequencing Approaches. Curr. Oncol. Rep. 2018, 20, 86. [Google Scholar] [CrossRef] [PubMed]
- Grzywa, T.M.; Paskal, W.; Włodarski, P.K. Intratumor and Intertumor Heterogeneity in Melanoma. Transl. Oncol. 2017, 10, 956–975. [Google Scholar] [CrossRef]
- Ng, M.F.; Simmons, J.L.; Boyle, G.M. Heterogeneity in Melanoma. Cancers 2022, 14, 3030. [Google Scholar] [CrossRef]
- Kumar, D.; Gorain, M.; Kundu, G.; Kundu, G.C. Therapeutic implications of cellular and molecular biology of cancer stem cells in melanoma. Mol. Cancer 2017, 16, 7. [Google Scholar] [CrossRef]
- Hass, R.; von der Ohe, J.; Ungefroren, H. Impact of the Tumor Microenvironment on Tumor Heterogeneity and Consequences for Cancer Cell Plasticity and Stemness. Cancers 2020, 12, 3716. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Shlyakhtina, Y.; Moran, K.L.; Portal, M.M. Genetic and Non-Genetic Mechanisms Underlying Cancer Evolution. Cancers 2021, 13, 1380. [Google Scholar] [CrossRef]
- Hirata, E.; Sahai, E. Tumor Microenvironment and Differential Responses to Therapy. Cold Spring Harb. Perspect. Med. 2017, 7, a026781. [Google Scholar] [CrossRef]
- Hölzel, M.; Tüting, T. Inflammation-Induced Plasticity in Melanoma Therapy and Metastasis. Trends Immunol. 2016, 37, 364–374. [Google Scholar] [CrossRef]
- Shou, Y.; Yang, L.; Yang, Y.; Zhu, X.; Li, F.; Xu, J. Determination of hypoxia signature to predict prognosis and the tumor immune microenvironment in melanoma. Mol. Omics 2021, 17, 307–316. [Google Scholar] [CrossRef]
- Umansky, V.; Sevko, A. Overcoming immunosuppression in the melanoma microenvironment induced by chronic inflammation. Cancer Immunol. Immunother. 2012, 61, 275–282. [Google Scholar] [CrossRef]
- Ahmed, F.; Haass, N.K. Microenvironment-Driven Dynamic Heterogeneity and Phenotypic Plasticity as a Mechanism of Melanoma Therapy Resistance. Front. Oncol. 2018, 8, 173. [Google Scholar] [CrossRef] [PubMed]
- Granados, K.; Poelchen, J.; Novak, D.; Utikal, J. Cellular Reprogramming-A Model for Melanoma Cellular Plasticity. Int. J. Mol. Sci. 2020, 21, 8274. [Google Scholar] [CrossRef] [PubMed]
- Bettum, I.J.; Gorad, S.S.; Barkovskaya, A.; Pettersen, S.; Moestue, S.A.; Vasiliauskaite, K.; Tenstad, E.; Øyjord, T.; Risa, Ø.; Nygaard, V.; et al. Metabolic reprogramming supports the invasive phenotype in malignant melanoma. Cancer Lett. 2015, 366, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Falletta, P.; Goding, C.R.; Vivas-García, Y. Connecting Metabolic Rewiring With Phenotype Switching in Melanoma. Front. Cell Dev. Biol. 2022, 10, 930250. [Google Scholar] [CrossRef]
- Karami Fath, M.; Ebrahimi, M.; Nourbakhsh, E.; Zia Hazara, A.; Mirzaei, A.; Shafieyari, S.; Salehi, A.; Hoseinzadeh, M.; Payandeh, Z.; Barati, G. PI3K/Akt/mTOR signaling pathway in cancer stem cells. Pathol. Res. Pract. 2022, 237, 154010. [Google Scholar] [CrossRef]
- Rambow, F.; Marine, J.C.; Goding, C.R. Melanoma plasticity and phenotypic diversity: Therapeutic barriers and opportunities. Genes Dev. 2019, 33, 1295–1318. [Google Scholar] [CrossRef]
- Pagliuca, C.; Di Leo, L.; De Zio, D. New Insights into the Phenotype Switching of Melanoma. Cancers 2022, 14, 6118. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Olbrecht, S.; Boeckx, B.; Vos, H.; Laoui, D.; Etlioglu, E.; Wauters, E.; Pomella, V.; Verbandt, S.; Busschaert, P.; et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020, 30, 745–762. [Google Scholar] [CrossRef] [PubMed]
- Hossain, S.M.; Eccles, M.R. Phenotype Switching and the Melanoma Microenvironment; Impact on Immunotherapy and Drug Resistance. Int. J. Mol. Sci. 2023, 24, 1601. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Han, F.; Du, Y.; Shi, H.; Zhou, W. Hypoxic microenvironment in cancer: Molecular mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2023, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, S.; Zuo, A.; Zhang, J.; Wen, W.; Jiang, W.; Chen, H.; Liang, D.; Sun, J.; Wang, M. HIF-1α/JMJD1A signaling regulates inflammation and oxidative stress following hyperglycemia and hypoxia-induced vascular cell injury. Cell. Mol. Biol. Lett. 2021, 26, 40. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lin, P.C. Mechanisms that drive inflammatory tumor microenvironment, tumor heterogeneity, and metastatic progression. Semin. Cancer Biol. 2017, 47, 185–195. [Google Scholar] [CrossRef]
- Benboubker, V.; Boivin, F.; Dalle, S.; Caramel, J. Cancer Cell Phenotype Plasticity as a Driver of Immune Escape in Melanoma. Front. Immunol. 2022, 13, 873116. [Google Scholar] [CrossRef]
- Knappe, N.; Novak, D.; Weina, K.; Bernhardt, M.; Reith, M.; Larribere, L.; Hölzel, M.; Tüting, T.; Gebhardt, C.; Umansky, V.; et al. Directed Dedifferentiation Using Partial Reprogramming Induces Invasive Phenotype in Melanoma Cells. Stem Cells 2016, 34, 832–846. [Google Scholar] [CrossRef]
- Diazzi, S.; Tartare-Deckert, S.; Deckert, M. The mechanical phenotypic plasticity of melanoma cell: An emerging driver of therapy cross-resistance. Oncogenesis 2023, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, N.; Berx, G. Melanoma cells revive an embryonic transcriptional network to dictate phenotypic heterogeneity. Front. Oncol. 2014, 4, 352. [Google Scholar] [CrossRef] [PubMed]
- Romano, V.; Belviso, I.; Venuta, A.; Ruocco, M.R.; Masone, S.; Aliotta, F.; Fiume, G.; Montagnani, S.; Avagliano, A.; Arcucci, A. Influence of Tumor Microenvironment and Fibroblast Population Plasticity on Melanoma Growth, Therapy Resistance and Immunoescape. Int. J. Mol. Sci. 2021, 22, 5283. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.J.; Liu, S.; Jin, P.; Stroncek, D. Mesenchymal stromal cell plasticity and the tumor microenvironment. Emerg. Top. Life Sci. 2017, 1, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Tsang, M.; Quesnel, K.; Vincent, K.; Hutchenreuther, J.; Postovit, L.M.; Leask, A. Insights into Fibroblast Plasticity: Cellular Communication Network 2 Is Required for Activation of Cancer-Associated Fibroblasts in a Murine Model of Melanoma. Am. J. Pathol. 2020, 190, 206–221. [Google Scholar] [CrossRef] [PubMed]
- Papaccio, F.; Kovacs, D.; Bellei, B.; Caputo, S.; Migliano, E.; Cota, C.; Picardo, M. Profiling Cancer-Associated Fibroblasts in Melanoma. Int. J. Mol. Sci. 2021, 22, 7255. [Google Scholar] [CrossRef]
- Yoshida, M.; Selvan, S.; McCue, P.A.; DeAngelis, T.; Baserga, R.; Fujii, A.; Rui, H.; Mastrangelo, M.J.; Sato, T. Expression of insulin-like growth factor-1 receptor in metastatic uveal melanoma and implications for potential autocrine and paracrine tumor cell growth. Pigment Cell Melanoma Res. 2014, 27, 297–308. [Google Scholar] [CrossRef]
- White, R.M.; Zon, L.I. Melanocytes in development, regeneration, and cancer. Cell Stem Cell 2008, 3, 242–252. [Google Scholar] [CrossRef]
- Centeno, P.P.; Pavet, V.; Marais, R. The journey from melanocytes to melanoma. Nat. Rev. Cancer 2023, 23, 372–390. [Google Scholar] [CrossRef]
- Cichorek, M.; Wachulska, M.; Stasiewicz, A.; Tymińska, A. Skin melanocytes: Biology and development. Postepy Dermatol. Alergol. 2013, 30, 30–41. [Google Scholar] [CrossRef]
- Li, L.; Fukunaga-Kalabis, M.; Herlyn, M. Isolation and cultivation of dermal stem cells that differentiate into functional epidermal melanocytes. Methods Mol. Biol. 2012, 806, 15–29. [Google Scholar] [CrossRef]
- Hoerter, J.D.; Bradley, P.; Casillas, A.; Chambers, D.; Weiswasser, B.; Clements, L.; Gilbert, S.; Jiao, A. Does melanoma begin in a melanocyte stem cell? J. Skin Cancer 2012, 2012, 571087. [Google Scholar] [CrossRef] [PubMed]
- Grichnik, J.M.; Ali, W.N.; Burch, J.A.; Byers, J.D.; Garcia, C.A.; Clark, R.E.; Shea, C.R. KIT expression reveals a population of precursor melanocytes in human skin. J. Investig. Dermatol. 1996, 106, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Zabierowski, S.E.; Fukunaga-Kalabis, M.; Li, L.; Herlyn, M. Dermis-derived stem cells: A source of epidermal melanocytes and melanoma? Pigment Cell Melanoma Res. 2011, 24, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Kyrgidis, A.; Tzellos, T.G.; Triaridis, S. Melanoma: Stem cells, sun exposure and hallmarks for carcinogenesis, molecular concepts and future clinical implications. J. Carcinog. 2010, 9, 3. [Google Scholar] [CrossRef]
- Valyi-Nagy, K.; Kormos, B.; Ali, M.; Shukla, D.; Valyi-Nagy, T. Stem cell marker CD271 is expressed by vasculogenic mimicry-forming uveal melanoma cells in three-dimensional cultures. Mol. Vis. 2012, 18, 588–592. [Google Scholar]
- sher, G.; Masoodi, T.; Patil, K.; Akhtar, S.; Kuttikrishnan, S.; Ahmad, A.; Uddin, S. Dysregulated FOXM1 signaling in the regulation of cancer stem cells. Semin. Cancer Biol. 2022, 86 Pt 3, 107–121. [Google Scholar] [CrossRef]
- Faião-Flores, F.; Smalley, K.S.M. Get with the Program! Stemness and Reprogramming in Melanoma Metastasis. J. Investig. Dermatol. 2018, 138, 10–13. [Google Scholar] [CrossRef]
- Pine, S.R.; Liu, W. Asymmetric cell division and template DNA co-segregation in cancer stem cells. Front. Oncol. 2014, 4, 226. [Google Scholar] [CrossRef] [PubMed]
- Gómez-López, S.; Lerner, R.G.; Petritsch, C. Asymmetric cell division of stem and progenitor cells during homeostasis and cancer. Cell. Mol. Life Sci. 2014, 71, 575–597. [Google Scholar] [CrossRef]
- Schatton, T.; Frank, M.H. Cancer stem cells and human malignant melanoma. Pigment Cell Melanoma Res. 2008, 21, 39–55. [Google Scholar] [CrossRef]
- Fargeas, C.A.; Florek, M.; Huttner, W.B.; Corbeil, D. Characterization of prominin-2, a new member of the prominin family of pentaspan membrane glycoproteins. J. Biol. Chem. 2003, 278, 8586–8596. [Google Scholar] [CrossRef]
- Corbeil, D.; Karbanová, J.; Fargeas, C.A.; Jászai, J. Prominin-1 (CD133): Molecular and Cellular Features Across Species. Adv. Exp. Med. Biol. 2013, 777, 3–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, Y.; Xing, Y.; Cao, B.; Yang, F.; Yang, T.; Ai, Z.; Wei, Y.; Jiang, J. The Interaction between Cancer Stem Cell Marker CD133 and Src Protein Promotes Focal Adhesion Kinase (FAK) Phosphorylation and Cell Migration. J. Biol. Chem. 2016, 291, 15540–15550. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Labbé, D.; Fontaine, N.; Lamy, S.; Beaulieu, E.; Gingras, D.; Béliveau, R. The stem cell marker CD133 (prominin-1) is phosphorylated on cytoplasmic tyrosine-828 and tyrosine-852 by Src and Fyn tyrosine kinases. Biochemistry 2009, 48, 3998–4007. [Google Scholar] [CrossRef]
- González-Herrero, I.; Romero-Camarero, I.; Cañueto, J.; Cardeñoso-Álvarez, E.; Fernández-López, E.; Pérez-Losada, J.; Sánchez-García, I.; Román-Curto, C. CD133+ cell content correlates with tumour growth in melanomas from skin with chronic sun-induced damage. Br. J. Dermatol. 2013, 169, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Madjd, Z.; Erfani, E.; Gheytanchi, E.; Moradi-Lakeh, M.; Shariftabrizi, A.; Asadi-Lari, M. Expression of CD133 cancer stem cell marker in melanoma: A systematic review and meta-analysis. Int. J. Biol. Markers 2016, 31, e118–e125. [Google Scholar] [CrossRef] [PubMed]
- Simbulan-Rosenthal, C.M.; Gaur, A.; Zhou, H.; AbdusSamad, M.; Qin, Q.; Dougherty, R.; Aljehane, L.; Kuo, L.W.; Vakili, S.; Karna, K.; et al. CD133 Is Associated with Increased Melanoma Cell Survival after Multikinase Inhibition. J. Oncol. 2019, 2019, 6486173. [Google Scholar] [CrossRef] [PubMed]
- Lai, I.C.; Shih, P.H.; Yao, C.J.; Yeh, C.T.; Wang-Peng, J.; Lui, T.N.; Chuang, S.E.; Hu, T.S.; Lai, T.Y.; Lai, G.M. Elimination of cancer stem-like cells and potentiation of temozolomide sensitivity by Honokiol in glioblastoma multiforme cells. PLoS ONE 2015, 10, e0114830. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.P.; Chien, Y.; Chiou, G.Y.; Cherng, J.Y.; Wang, M.L.; Lo, W.L.; Chang, Y.L.; Huang, P.I.; Chen, Y.W.; Shih, Y.H.; et al. Inhibition of cancer stem cell-like properties and reduced chemoradioresistance of glioblastoma using microRNA145 with cationic polyurethane-short branch PEI. Biomaterials 2012, 33, 1462–1476. [Google Scholar] [CrossRef] [PubMed]
- Tseng, L.M.; Huang, P.I.; Chen, Y.R.; Chen, Y.C.; Chou, Y.C.; Chen, Y.W.; Chang, Y.L.; Hsu, H.S.; Lan, Y.T.; Chen, K.H.; et al. Targeting signal transducer and activator of transcription 3 pathway by cucurbitacin I diminishes self-renewing and radiochemoresistant abilities in thyroid cancer-derived CD133+ cells. J. Pharmacol. Exp. Ther. 2012, 341, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.A. The role of the PI3K-AKT pathway in melanoma. Cancer J. 2012, 18, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Rascio, F.; Spadaccino, F.; Rocchetti, M.T.; Castellano, G.; Stallone, G.; Netti, G.S.; Ranieri, E. The Pathogenic Role of PI3K/AKT Pathway in Cancer Onset and Drug Resistance: An Updated Review. Cancers 2021, 13, 3949. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.L.; Cantley, L.C. PI3K pathway alterations in cancer: Variations on a theme. Oncogene 2008, 27, 5497–5510. [Google Scholar] [CrossRef]
- Wang, J.; Cai, S.; Xiong, Q.; Weng, D.; Wang, Q.; Ma, Z. PIK3R2 predicts poor outcomes for patients with melanoma and contributes to the malignant progression via PI3K/AKT/NF-κB axis. Clin. Transl. Oncol. 2023, 25, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Ueda, Y.; Richmond, A. NF-kappaB activation in melanoma. Pigment Cell Res. 2006, 19, 112–124. [Google Scholar] [CrossRef]
- Amiri, K.I.; Richmond, A. Role of nuclear factor-kappa B in melanoma. Cancer Metastasis Rev. 2005, 24, 301–313. [Google Scholar] [CrossRef]
- Qi, X.; Chen, Y.; Liu, S.; Liu, L.; Yu, Z.; Yin, L.; Fu, L.; Deng, M.; Liang, S.; Lü, M. Sanguinarine inhibits melanoma invasion and migration by targeting the FAK/PI3K/AKT/mTOR signalling pathway. Pharm. Biol. 2023, 61, 696–709. [Google Scholar] [CrossRef]
- Khan, K.H.; Yap, T.A.; Yan, L.; Cunningham, D. Targeting the PI3K-AKT-mTOR signaling network in cancer. Chin. J. Cancer 2013, 32, 253–265. [Google Scholar] [CrossRef]
- Babchia, N.; Calipel, A.; Mouriaux, F.; Faussat, A.M.; Mascarelli, F. The PI3K/Akt and mTOR/P70S6K signaling pathways in human uveal melanoma cells: Interaction with B-Raf/ERK. Investig. Ophthalmol. Vis. Sci. 2010, 51, 421–429. [Google Scholar] [CrossRef]
- Gil, D.; Zarzycka, M.; Pabijan, J.; Lekka, M.; Dulińska-Litewka, J. Dual targeting of melanoma translation by MNK/eIF4E and PI3K/mTOR inhibitors. Cell. Signal. 2023, 109, 110742. [Google Scholar] [CrossRef]
- Caporali, S.; Alvino, E.; Lacal, P.M.; Levati, L.; Giurato, G.; Memoli, D.; Caprini, E.; Antonini Cappellini, G.C.; D’Atri, S. Targeting the PI3K/AKT/mTOR pathway overcomes the stimulating effect of dabrafenib on the invasive behavior of melanoma cells with acquired resistance to the BRAF inhibitor. Int. J. Oncol. 2016, 49, 1164–1174. [Google Scholar] [CrossRef]
- Madhunapantula, S.V.; Robertson, G.P. The PTEN-AKT3 signaling cascade as a therapeutic target in melanoma. Pigment Cell Melanoma Res. 2009, 22, 400–419. [Google Scholar] [CrossRef]
- Gong, C.; Xia, H. Resveratrol suppresses melanoma growth by promoting autophagy through inhibiting the PI3K/AKT/mTOR signaling pathway. Exp. Ther. Med. 2020, 19, 1878–1886. [Google Scholar] [CrossRef]
- Robertson, G.P. Functional and therapeutic significance of Akt deregulation in malignant melanoma. Cancer Metastasis Rev. 2005, 24, 273–285. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.; Chang, F.; Lehmann, B.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta 2007, 1773, 1263–1284. [Google Scholar] [CrossRef]
- Gocek, E.; Moulas, A.N.; Studzinski, G.P. Non-receptor protein tyrosine kinases signaling pathways in normal and cancer cells. Crit. Rev. Clin. Lab. Sci. 2014, 51, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Chiasson, J.L. Comparative characterization of receptor and non-receptor associated protein tyrosine kinases. Biochim. Biophys. Acta 1989, 996, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Siveen, K.S.; Prabhu, K.S.; Achkar, I.W.; Kuttikrishnan, S.; Shyam, S.; Khan, A.Q.; Merhi, M.; Dermime, S.; Uddin, S. Role of Non Receptor Tyrosine Kinases in Hematological Malignances and its Targeting by Natural Products. Mol. Cancer 2018, 17, 31. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, C.D.; Rath, A.; Davidson, A.R. Recognition of non-canonical peptides by the yeast Fus1p SH3 domain: Elucidation of a common mechanism for diverse SH3 domain specificities. J. Mol. Biol. 2008, 377, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Calle, Y.; Sayed, M.A.; Kamal, J.M.; Rengaswamy, P.; Manser, E.; Meiners, S.; Nur-E-Kamal, A. Cdc42-dependent nuclear translocation of non-receptor tyrosine kinase, ACK. Biochem. Biophys. Res. Commun. 2004, 314, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Engen, J.R.; Wales, T.E.; Hochrein, J.M.; Meyn, M.A.; Banu Ozkan, S.; Bahar, I.; Smithgall, T.E. Structure and dynamic regulation of Src-family kinases. Cell. Mol. Life Sci. 2008, 65, 3058–3073. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Echagüe, V.; Miller, W.T. Regulation of ack-family nonreceptor tyrosine kinases. J. Signal Transduct. 2011, 2011, 742372. [Google Scholar] [CrossRef] [PubMed]
- Tabernero, L.; Aricescu, A.R.; Jones, E.Y.; Szedlacsek, S.E. Protein tyrosine phosphatases: Structure-function relationships. FEBS J. 2008, 275, 867–882. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Lovly, C.M. Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer 2018, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- Marholz, L.J.; Zeringo, N.A.; Lou, H.J.; Turk, B.E.; Parker, L.L. In Silico Design and in Vitro Characterization of Universal Tyrosine Kinase Peptide Substrates. Biochemistry 2018, 57, 1847–1851. [Google Scholar] [CrossRef]
- Luechapanichkul, R.; Chen, X.; Taha, H.A.; Vyas, S.; Guan, X.; Freitas, M.A.; Hadad, C.M.; Pei, D. Specificity profiling of dual specificity phosphatase vaccinia VH1-related (VHR) reveals two distinct substrate binding modes. J. Biol. Chem. 2013, 288, 6498–6510. [Google Scholar] [CrossRef]
- Songyang, Z.; Shoelson, S.E.; Chaudhuri, M.; Gish, G.; Pawson, T.; Haser, W.G.; King, F.; Roberts, T.; Ratnofsky, S.; Lechleider, R.J. SH2 domains recognize specific phosphopeptide sequences. Cell 1993, 72, 767–778. [Google Scholar] [CrossRef]
- Miller, W.T. Determinants of substrate recognition in nonreceptor tyrosine kinases. Acc. Chem. Res. 2003, 36, 393–400. [Google Scholar] [CrossRef]
- Rickles, R.J.; Botfield, M.C.; Weng, Z.; Taylor, J.A.; Green, O.M.; Brugge, J.S.; Zoller, M.J. Identification of Src, Fyn, Lyn, PI3K and Abl SH3 domain ligands using phage display libraries. EMBO J. 1994, 13, 5598–5604. [Google Scholar] [CrossRef]
- Jaber Chehayeb, R.; Boggon, T.J. SH2 Domain Binding: Diverse FLVRs of Partnership. Front. Endocrinol. 2020, 11, 575220. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.J.; Stacey, M.M.; Liu, B.A.; Pawson, T. Molecular mechanisms of SH2- and PTB-domain-containing proteins in receptor tyrosine kinase signaling. Cold Spring Harb. Perspect. Biol. 2013, 5, a008987. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, C.; Knudsen, B.S.; Ohuchi, T.; Di Fiore, P.P.; Glassman, R.H.; Hanafusa, H. The SH3 domain of Crk binds specifically to a conserved proline-rich motif in Eps15 and Eps15R. J. Biol. Chem. 1995, 270, 15341–15347. [Google Scholar] [CrossRef]
- Mehrabipour, M.; Jasemi, N.S.K.; Dvorsky, R.; Ahmadian, M.R. A Systematic Compilation of Human SH3 Domains: A Versatile Superfamily in Cellular Signaling. Cells 2023, 12, 2054. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.C.; Wierenga, R.K.; Saraste, M. Insights into Src kinase functions: Structural comparisons. Trends Biochem. Sci. 1998, 23, 179–184. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef]
- Levine, S.J. Molecular mechanisms of soluble cytokine receptor generation. J. Biol. Chem. 2008, 283, 14177–14181. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, Y.; Zhan, L.; Zhang, L.; Hu, J.; Gao, Z. The role of non-receptor protein tyrosine kinases in the excitotoxicity induced by the overactivation of NMDA receptors. Rev. Neurosci. 2016, 27, 283–289. [Google Scholar] [CrossRef]
- Chen, S.; Brier, S.; Smithgall, T.E.; Engen, J.R. The Abl SH2-kinase linker naturally adopts a conformation competent for SH3 domain binding. Protein Sci. 2007, 16, 572–581. [Google Scholar] [CrossRef]
- Donaldson, L.W.; Gish, G.; Pawson, T.; Kay, L.E.; Forman-Kay, J.D. Structure of a regulatory complex involving the Abl SH3 domain, the Crk SH2 domain, and a Crk-derived phosphopeptide. Proc. Natl. Acad. Sci. USA 2002, 99, 14053–14058. [Google Scholar] [CrossRef] [PubMed]
- Owen, L.M.; Bax, N.A.; Weis, W.I.; Dunn, A.R. The C-terminal actin-binding domain of talin forms an asymmetric catch bond with F-actin. Proc. Natl. Acad. Sci. USA 2022, 119, e2109329119. [Google Scholar] [CrossRef]
- Murayama, K.; Kato-Murayama, M.; Mishima, C.; Akasaka, R.; Shirouzu, M.; Fukui, Y.; Yokoyama, S. Crystal structure of the Bruton’s tyrosine kinase PH domain with phosphatidylinositol. Biochem. Biophys. Res. Commun. 2008, 377, 23–28. [Google Scholar] [CrossRef]
- Fukuda, M.; Kojima, T.; Kabayama, H.; Mikoshiba, K. Mutation of the pleckstrin homology domain of Bruton’s tyrosine kinase in immunodeficiency impaired inositol 1,3,4,5-tetrakisphosphate binding capacity. J. Biol. Chem. 1996, 271, 30303–30306. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Várnai, P.; Balla, A.; Jalink, K.; Rhee, S.G.; Balla, T. The pleckstrin homology domain of phosphoinositide-specific phospholipase Cdelta4 is not a critical determinant of the membrane localization of the enzyme. J. Biol. Chem. 2004, 279, 24362–24371. [Google Scholar] [CrossRef]
- Overduin, M.; Kervin, T.A. The phosphoinositide code is read by a plethora of protein domains. Expert Rev. Proteom. 2021, 18, 483–502. [Google Scholar] [CrossRef]
- Hirsch, E.; Gulluni, F.; Martini, M. Phosphoinositides in cell proliferation and metabolism. Adv. Biol. Regul. 2020, 75, 100693. [Google Scholar] [CrossRef] [PubMed]
- Bridges, D.; Saltiel, A.R. Phosphoinositides: Key modulators of energy metabolism. Biochim. Biophys. Acta 2015, 1851, 857–866. [Google Scholar] [CrossRef]
- Beziau, A.; Brand, D.; Piver, E. The Role of Phosphatidylinositol Phosphate Kinases during Viral Infection. Viruses 2020, 12, 1124. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.E. Structural Basis for Regulation of Phosphoinositide Kinases and Their Involvement in Human Disease. Mol. Cell 2018, 71, 653–673. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.L.; Walch, L.; Verbavatz, J.M. Lipids and Their Trafficking: An Integral Part of Cellular Organization. Dev. Cell 2016, 39, 139–153. [Google Scholar] [CrossRef]
- Toker, A.; Cantley, L.C. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature 1997, 387, 673–676. [Google Scholar] [CrossRef]
- Liu, P.; Cheng, H.; Roberts, T.M.; Zhao, J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009, 8, 627–644. [Google Scholar] [CrossRef]
- Desale, S.E.; Chidambaram, H.; Chinnathambi, S. G-protein coupled receptor, PI3K and Rho signaling pathways regulate the cascades of Tau and amyloid-β in Alzheimer’s disease. Mol. Biomed. 2021, 2, 17. [Google Scholar] [CrossRef]
- Taichman, R.; Merida, I.; Torigoe, T.; Gaulton, G.N.; Reed, J.C. Evidence that protein tyrosine kinase p56-Lck regulates the activity of phosphatidylinositol-3’-kinase in interleukin-2-dependent T-cells. J. Biol. Chem. 1993, 268, 20031–20036. [Google Scholar] [CrossRef]
- Karnitz, L.M.; Sutor, S.L.; Abraham, R.T. The Src-family kinase, Fyn, regulates the activation of phosphatidylinositol 3-kinase in an interleukin 2-responsive T cell line. J. Exp. Med. 1994, 179, 1799–1808. [Google Scholar] [CrossRef]
- Tang, X.; Feng, Y.; Ye, K. Src-family tyrosine kinase fyn phosphorylates phosphatidylinositol 3-kinase enhancer-activating Akt, preventing its apoptotic cleavage and promoting cell survival. Cell Death Differ. 2007, 14, 368–377. [Google Scholar] [CrossRef]
- Peng, S.; Fu, Y. FYN: Emerging biological roles and potential therapeutic targets in cancer. J. Transl. Med. 2023, 21, 84. [Google Scholar] [CrossRef]
- Matrone, C.; Petrillo, F.; Nasso, R.; Ferretti, G. Fyn Tyrosine Kinase as Harmonizing Factor in Neuronal Functions and Dysfunctions. Int. J. Mol. Sci. 2020, 21, 4444. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, J.F.; Hall, C.G.; Atkinson, T.P. Identification of an alternatively spliced isoform of the fyn tyrosine kinase. Biochem. Biophys. Res. Commun. 2002, 298, 501–504. [Google Scholar] [CrossRef]
- Vatish, M.; Yamada, E.; Pessin, J.E.; Bastie, C.C. Fyn kinase function in lipid utilization: A new upstream regulator of AMPK activity? Arch. Physiol. Biochem. 2009, 115, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Baer, A.; Colon-Moran, W.; Xiang, J.; Stapleton, J.T.; Bhattarai, N. Src-family kinases negatively regulate NFAT signaling in resting human T cells. PLoS ONE 2017, 12, e0187123. [Google Scholar] [CrossRef]
- Chong, Y.P.; Mulhern, T.D.; Zhu, H.J.; Fujita, D.J.; Bjorge, J.D.; Tantiongco, J.P.; Sotirellis, N.; Lio, D.S.; Scholz, G.; Cheng, H.C. A novel non-catalytic mechanism employed by the C-terminal Src-homologous kinase to inhibit Src-family kinase activity. J. Biol. Chem. 2004, 279, 20752–20766. [Google Scholar] [CrossRef]
- Advani, G.; Lim, Y.C.; Catimel, B.; Lio, D.S.S.; Ng, N.L.Y.; Chüeh, A.C.; Tran, M.; Anasir, M.I.; Verkade, H.; Zhu, H.J.; et al. Csk-homologous kinase (Chk) is an efficient inhibitor of Src-family kinases but a poor catalyst of phosphorylation of their C-terminal regulatory tyrosine. Cell Commun. Signal. 2017, 15, 29. [Google Scholar] [CrossRef]
- den Hertog, J.; Ostman, A.; Böhmer, F.D. Protein tyrosine phosphatases: Regulatory mechanisms. FEBS J. 2008, 275, 831–847. [Google Scholar] [CrossRef]
- Vacaresse, N.; Møller, B.; Danielsen, E.M.; Okada, M.; Sap, J. Activation of c-Src and Fyn kinases by protein-tyrosine phosphatase RPTPalpha is substrate-specific and compatible with lipid raft localization. J. Biol. Chem. 2008, 283, 35815–35824. [Google Scholar] [CrossRef]
- Liang, X.; Lu, Y.; Wilkes, M.; Neubert, T.A.; Resh, M.D. The N-terminal SH4 region of the Src family kinase Fyn is modified by methylation and heterogeneous fatty acylation: Role in membrane targeting, cell adhesion, and spreading. J. Biol. Chem. 2004, 279, 8133–8139. [Google Scholar] [CrossRef]
- Boggon, T.J.; Eck, M.J. Structure and regulation of Src family kinases. Oncogene 2004, 23, 7918–7927. [Google Scholar] [CrossRef]
- Teutschbein, J.; Schartl, M.; Meierjohann, S. Interaction of Xiphophorus and murine Fyn with focal adhesion kinase. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2009, 149, 168–174. [Google Scholar] [CrossRef]
- Zhao, X.; Guan, J.L. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv. Drug Deliv. Rev. 2011, 63, 610–615. [Google Scholar] [CrossRef] [PubMed]
- Meierjohann, S.; Wende, E.; Kraiss, A.; Wellbrock, C.; Schartl, M. The oncogenic epidermal growth factor receptor variant Xiphophorus melanoma receptor kinase induces motility in melanocytes by modulation of focal adhesions. Cancer Res. 2006, 66, 3145–3152. [Google Scholar] [CrossRef]
- Wittbrodt, J.; Adam, D.; Malitschek, B.; Mäueler, W.; Raulf, F.; Telling, A.; Robertson, S.M.; Schartl, M. Novel putative receptor tyrosine kinase encoded by the melanoma-inducing Tu locus in Xiphophorus. Nature 1989, 341, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Wittbrodt, J.; Lammers, R.; Malitschek, B.; Ullrich, A.; Schartl, M. The Xmrk receptor tyrosine kinase is activated in Xiphophorus malignant melanoma. EMBO J. 1992, 11, 4239–4246. [Google Scholar] [CrossRef] [PubMed]
- Wellbrock, C.; Schartl, M. Activation of phosphatidylinositol 3-kinase by a complex of p59fyn and the receptor tyrosine kinase Xmrk is involved in malignant transformation of pigment cells. Eur. J. Biochem. 2000, 267, 3513–3522. [Google Scholar] [CrossRef]
- Wellbrock, C.; Schartl, M. Multiple binding sites in the growth factor receptor Xmrk mediate binding to p59fyn, GRB2 and Shc. Eur. J. Biochem. 1999, 260, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Alexanian, A.; Miller, B.; Chesnik, M.; Mirza, S.; Sorokin, A. Post-translational regulation of COX2 activity by FYN in prostate cancer cells. Oncotarget 2014, 5, 4232–4243. [Google Scholar] [CrossRef] [PubMed]
- Czarnecka, A.M.; Bartnik, E.; Fiedorowicz, M.; Rutkowski, P. Targeted Therapy in Melanoma and Mechanisms of Resistance. Int. J. Mol. Sci. 2020, 21, 4576. [Google Scholar] [CrossRef]
- Denley, A.; Gymnopoulos, M.; Kang, S.; Mitchell, C.; Vogt, P.K. Requirement of phosphatidylinositol(3,4,5)trisphosphate in phosphatidylinositol 3-kinase-induced oncogenic transformation. Mol. Cancer Res. 2009, 7, 1132–1138. [Google Scholar] [CrossRef]
- Lien, E.C.; Dibble, C.C.; Toker, A. PI3K signaling in cancer: Beyond AKT. Curr. Opin. Cell Biol. 2017, 45, 62–71. [Google Scholar] [CrossRef]
- Dieterle, A.M.; Böhler, P.; Keppeler, H.; Alers, S.; Berleth, N.; Drießen, S.; Hieke, N.; Pietkiewicz, S.; Löffler, A.S.; Peter, C.; et al. PDK1 controls upstream PI3K expression and PIP3 generation. Oncogene 2014, 33, 3043–3053. [Google Scholar] [CrossRef]
- Orlacchio, A.; Ranieri, M.; Brave, M.; Arciuch, V.A.; Forde, T.; De Martino, D.; Anderson, K.E.; Hawkins, P.; Di Cristofano, A. SGK1 Is a Critical Component of an AKT-Independent Pathway Essential for PI3K-Mediated Tumor Development and Maintenance. Cancer Res. 2017, 77, 6914–6926. [Google Scholar] [CrossRef]
- Liu, R.; Chen, Y.; Liu, G.; Li, C.; Song, Y.; Cao, Z.; Li, W.; Hu, J.; Lu, C.; Liu, Y. PI3K/AKT pathway as a key link modulates the multidrug resistance of cancers. Cell Death Dis. 2020, 11, 797. [Google Scholar] [CrossRef]
- Loewith, R.; Hall, M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef]
- Thoreen, C.C.; Kang, S.A.; Chang, J.W.; Liu, Q.; Zhang, J.; Gao, Y.; Reichling, L.J.; Sim, T.; Sabatini, D.M.; Gray, N.S. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 2009, 284, 8023–8032. [Google Scholar] [CrossRef]
- Dibble, C.C.; Cantley, L.C. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015, 25, 545–555. [Google Scholar] [CrossRef]
- Kim, L.C.; Cook, R.S.; Chen, J. mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene 2017, 36, 2191–2201. [Google Scholar] [CrossRef]
- Yang, H.; Rudge, D.G.; Koos, J.D.; Vaidialingam, B.; Yang, H.J.; Pavletich, N.P. mTOR kinase structure, mechanism and regulation. Nature 2013, 497, 217–223. [Google Scholar] [CrossRef]
- Sharma, N.; Nanta, R.; Sharma, J.; Gunewardena, S.; Singh, K.P.; Shankar, S.; Srivastava, R.K. PI3K/AKT/mTOR and sonic hedgehog pathways cooperate together to inhibit human pancreatic cancer stem cell characteristics and tumor growth. Oncotarget 2015, 6, 32039–32060. [Google Scholar] [CrossRef]
- Chamcheu, J.C.; Roy, T.; Uddin, M.B.; Banang-Mbeumi, S.; Chamcheu, R.N.; Walker, A.L.; Liu, Y.Y.; Huang, S. Role and Therapeutic Targeting of the PI3K/Akt/mTOR Signaling Pathway in Skin Cancer: A Review of Current Status and Future Trends on Natural and Synthetic Agents Therapy. Cells 2019, 8, 803. [Google Scholar] [CrossRef]
- Mafi, S.; Mansoori, B.; Taeb, S.; Sadeghi, H.; Abbasi, R.; Cho, W.C.; Rostamzadeh, D. mTOR-Mediated Regulation of Immune Responses in Cancer and Tumor Microenvironment. Front. Immunol. 2021, 12, 774103. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Jiang, Y.; Zou, F.; Liu, Y.; Wang, S.; Xu, N.; Xu, W.; Cui, C.; Xing, Y.; Cao, B.; et al. Activation of PI3K/Akt pathway by CD133-p85 interaction promotes tumorigenic capacity of glioma stem cells. Proc. Natl. Acad. Sci. USA 2013, 110, 6829–6834. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Regufe da Mota, S.; Liu, R.; Moore, C.E.; Xie, J.; Lanucara, F.; Agarwala, U.; Pyr Dit Ruys, S.; Vertommen, D.; Rider, M.H.; et al. Eukaryotic elongation factor 2 kinase activity is controlled by multiple inputs from oncogenic signaling. Mol. Cell. Biol. 2014, 34, 4088–4103. [Google Scholar] [CrossRef] [PubMed]
- Hijazi, M.; Casado, P.; Akhtar, N.; Alvarez-Teijeiro, S.; Rajeeve, V.; Cutillas, P.R. eEF2K Activity Determines Synergy to Cotreatment of Cancer Cells With PI3K and MEK Inhibitors. Mol. Cell. Proteom. 2022, 21, 100240. [Google Scholar] [CrossRef] [PubMed]
- Bracho-Valdés, I.; Moreno-Alvarez, P.; Valencia-Martínez, I.; Robles-Molina, E.; Chávez-Vargas, L.; Vázquez-Prado, J. mTORC1- and mTORC2-interacting proteins keep their multifunctional partners focused. IUBMB Life 2011, 63, 896–914. [Google Scholar] [CrossRef] [PubMed]
- Origanti, S.; Nowotarski, S.L.; Carr, T.D.; Sass-Kuhn, S.; Xiao, L.; Wang, J.Y.; Shantz, L.M. Ornithine decarboxylase mRNA is stabilized in an mTORC1-dependent manner in Ras-transformed cells. Biochem. J. 2012, 442, 199–207. [Google Scholar] [CrossRef]
- Cam, H.; Easton, J.B.; High, A.; Houghton, P.J. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1α. Mol. Cell 2010, 40, 509–520. [Google Scholar] [CrossRef]
- Wong, P.M.; Feng, Y.; Wang, J.; Shi, R.; Jiang, X. Regulation of autophagy by coordinated action of mTORC1 and protein phosphatase 2A. Nat. Commun. 2015, 6, 8048. [Google Scholar] [CrossRef]
- Peterson, T.R.; Sengupta, S.S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011, 146, 408–420. [Google Scholar] [CrossRef]
- Muscella, A.; Vetrugno, C.; Calabriso, N.; Cossa, L.G.; De Pascali, S.A.; Fanizzi, F.P.; Marsigliante, S. [Pt(O,O’-acac)(γ-acac)(DMS)] alters SH-SY5Y cell migration and invasion by the inhibition of Na+/H+ exchanger isoform 1 occurring through a PKC-ε/ERK/mTOR Pathway. PLoS ONE 2014, 9, e112186. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Liu, L.N.; Hosoi, H.; Dilling, M.B.; Shikata, T.; Houghton, P.J. p53/p21(CIP1) cooperate in enforcing rapamycin-induced G(1) arrest and determine the cellular response to rapamycin. Cancer Res. 2001, 61, 3373–3381. [Google Scholar] [PubMed]
- Nourse, J.; Firpo, E.; Flanagan, W.M.; Coats, S.; Polyak, K.; Lee, M.H.; Massague, J.; Crabtree, G.R.; Roberts, J.M. Interleukin-2-mediated elimination of the p27Kip1 cyclin-dependent kinase inhibitor prevented by rapamycin. Nature 1994, 372, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, K.; Liu, P.; Geng, Y.; Wang, B.; Gan, W.; Guo, J.; Wu, F.; Chin, Y.R.; Berrios, C.; et al. Inhibition of Rb Phosphorylation Leads to mTORC2-Mediated Activation of Akt. Mol. Cell 2016, 62, 929–942. [Google Scholar] [CrossRef]
- Massi, D.; Mihic-Probst, D.; Schadendorf, D.; Dummer, R.; Mandalà, M. Dedifferentiated melanomas: Morpho-phenotypic profile, genetic reprogramming and clinical implications. Cancer Treat. Rev. 2020, 88, 102060. [Google Scholar] [CrossRef]
- Rodrik-Outmezguine, V.S.; Chandarlapaty, S.; Pagano, N.C.; Poulikakos, P.I.; Scaltriti, M.; Moskatel, E.; Baselga, J.; Guichard, S.; Rosen, N. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov. 2011, 1, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Mercurio, L.; Albanesi, C.; Madonna, S. Recent Updates on the Involvement of PI3K/AKT/mTOR Molecular Cascade in the Pathogenesis of Hyperproliferative Skin Disorders. Front. Med. 2021, 8, 665647. [Google Scholar] [CrossRef]
- Urso, C. Are growth phases exclusive to cutaneous melanoma? J. Clin. Pathol. 2004, 57, 560. [Google Scholar] [CrossRef]
- Ciarletta, P.; Foret, L.; Ben Amar, M. The radial growth phase of malignant melanoma: Multi-phase modelling, numerical simulations and linear stability analysis. J. R. Soc. Interface 2011, 8, 345–368. [Google Scholar] [CrossRef]
- Sinha, S.; Singh, S.K.; Jangde, N.; Ray, R.; Rai, V. p32 promotes melanoma progression and metastasis by targeting EMT markers, Akt/PKB pathway, and tumor microenvironment. Cell Death Dis. 2021, 12, 1012. [Google Scholar] [CrossRef] [PubMed]
- Zbytek, B.; Carlson, J.A.; Granese, J.; Ross, J.; Mihm, M.C.; Slominski, A. Current concepts of metastasis in melanoma. Expert Rev. Dermatol. 2008, 3, 569–585. [Google Scholar] [CrossRef]
- Lo, J.A.; Fisher, D.E. The melanoma revolution: From UV carcinogenesis to a new era in therapeutics. Science 2014, 346, 945–949. [Google Scholar] [CrossRef]
- Sample, A.; He, Y.Y. Mechanisms and prevention of UV-induced melanoma. Photodermatol. Photoimmunol. Photomed. 2018, 34, 13–24. [Google Scholar] [CrossRef]
- Piérard, G.E. Cell proliferation in cutaneous malignant melanoma: Relationship with neoplastic progression. Int. Sch. Res. Not. Dermatol. 2012, 2012, 828146. [Google Scholar] [CrossRef]
- Leonardi, G.C.; Falzone, L.; Salemi, R.; Zanghì, A.; Spandidos, D.A.; Mccubrey, J.A.; Candido, S.; Libra, M. Cutaneous melanoma: From pathogenesis to therapy (Review). Int. J. Oncol. 2018, 52, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; He, S.M.; He, Z.X.; Li, M.; Yang, Y.; Pang, J.X.; Zhang, X.; Chow, K.; Zhou, Q.; Duan, W.; et al. Plumbagin induces apoptotic and autophagic cell death through inhibition of the PI3K/Akt/mTOR pathway in human non-small cell lung cancer cells. Cancer Lett. 2014, 344, 239–259. [Google Scholar] [CrossRef]
- Xie, X.; White, E.P.; Mehnert, J.M. Coordinate autophagy and mTOR pathway inhibition enhances cell death in melanoma. PLoS ONE 2013, 8, e55096. [Google Scholar] [CrossRef] [PubMed]
- Shtivelman, E.; Davies, M.Q.; Hwu, P.; Yang, J.; Lotem, M.; Oren, M.; Flaherty, K.T.; Fisher, D.E. Pathways and therapeutic targets in melanoma. Oncotarget 2014, 5, 1701–1752. [Google Scholar] [CrossRef] [PubMed]
- Tentori, L.; Lacal, P.M.; Graziani, G. Challenging resistance mechanisms to therapies for metastatic melanoma. Trends Pharmacol. Sci. 2013, 34, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, W.; Zhang, G.; Kwong, L.; Lu, H.; Tan, J.; Sadek, N.; Xiao, M.; Zhang, J.; Labrie, M.; et al. Targeting mTOR signaling overcomes acquired resistance to combined BRAF and MEK inhibition in BRAF-mutant melanoma. Oncogene 2021, 40, 5590–5599. [Google Scholar] [CrossRef] [PubMed]
- Fenton, S.E.; Sosman, J.A.; Chandra, S. Resistance mechanisms in melanoma to immuneoncologic therapy with checkpoint inhibitors. Cancer Drug Resist. 2019, 2, 744–761. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Eckburg, A.; Gantiwala, S.; Hart, Z.; Dein, J.; Lam, K.; Puri, N. Resistance to Molecularly Targeted Therapies in Melanoma. Cancers 2021, 13, 1115. [Google Scholar] [CrossRef]
- Kearney, A.L.; Norris, D.M.; Ghomlaghi, M.; Kin Lok Wong, M.; Humphrey, S.J.; Carroll, L.; Yang, G.; Cooke, K.C.; Yang, P.; Geddes, T.A.; et al. Akt phosphorylates insulin receptor substrate to limit PI3K-mediated PIP3 synthesis. Elife 2021, 10, e66942. [Google Scholar] [CrossRef] [PubMed]
- Suleymanova, N.; Crudden, C.; Worrall, C.; Dricu, A.; Girnita, A.; Girnita, L. Enhanced response of melanoma cells to MEK inhibitors following unbiased IGF-1R down-regulation. Oncotarget 2017, 8, 82256–82267. [Google Scholar] [CrossRef] [PubMed]
- Leroy, C.; Ramos, P.; Cornille, K.; Bonenfant, D.; Fritsch, C.; Voshol, H.; Bentires-Alj, M. Activation of IGF1R/p110β/AKT/mTOR confers resistance to α-specific PI3K inhibition. Breast Cancer Res. 2016, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Hemmings, B.A.; Restuccia, D.F. PI3K-PKB/Akt pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011189. [Google Scholar] [CrossRef]
- Glaviano, A.; Foo, A.S.C.; Lam, H.Y.; Yap, K.C.H.; Jacot, W.; Jones, R.H.; Eng, H.; Nair, M.G.; Makvandi, P.; Geoerger, B.; et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol. Cancer 2023, 22, 138. [Google Scholar] [CrossRef] [PubMed]
- Chiarini, F.; Evangelisti, C.; Lattanzi, G.; McCubrey, J.A.; Martelli, A.M. Advances in understanding the mechanisms of evasive and innate resistance to mTOR inhibition in cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 1322–1337. [Google Scholar] [CrossRef]
- Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Kempf, R.C.; Long, J.; Laidler, P.; Mijatovic, S.; Maksimovic-Ivanic, D.; Stivala, F.; Mazzarino, M.C.; et al. Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in controlling growth and sensitivity to therapy-implications for cancer and aging. Aging 2011, 3, 192–222. [Google Scholar] [CrossRef]
- Li, Q.; Li, Z.; Luo, T.; Shi, H. Targeting the PI3K/AKT/mTOR and RAF/MEK/ERK pathways for cancer therapy. Mol. Biomed. 2022, 3, 47. [Google Scholar] [CrossRef]
- Palušová, V.; Renzová, T.; Verlande, A.; Vaclová, T.; Medková, M.; Cetlová, L.; Sedláčková, M.; Hříbková, H.; Slaninová, I.; Krutá, M.; et al. Dual Targeting of BRAF and mTOR Signaling in Melanoma Cells with Pyridinyl Imidazole Compounds. Cancers 2020, 12, 1516. [Google Scholar] [CrossRef]
- Carlino, M.S.; Larkin, J.; Long, G.V. Immune checkpoint inhibitors in melanoma. Lancet 2021, 398, 1002–1014. [Google Scholar] [CrossRef]
- Fukuda, K.; Okamura, K.; Riding, R.L.; Fan, X.; Afshari, K.; Haddadi, N.S.; McCauley, S.M.; Guney, M.H.; Luban, J.; Funakoshi, T.; et al. AIM2 regulates anti-tumor immunity and is a viable therapeutic target for melanoma. J. Exp. Med. 2021, 218, e20200962. [Google Scholar] [CrossRef]
- Weber, R.; Fleming, V.; Hu, X.; Nagibin, V.; Groth, C.; Altevogt, P.; Utikal, J.; Umansky, V. Myeloid-Derived Suppressor Cells Hinder the Anti-Cancer Activity of Immune Checkpoint Inhibitors. Front. Immunol. 2018, 9, 1310. [Google Scholar] [CrossRef]
- Simiczyjew, A.; Dratkiewicz, E.; Mazurkiewicz, J.; Ziętek, M.; Matkowski, R.; Nowak, D. The Influence of Tumor Microenvironment on Immune Escape of Melanoma. Int. J. Mol. Sci. 2020, 21, 8359. [Google Scholar] [CrossRef] [PubMed]
- Deken, M.A.; Gadiot, J.; Jordanova, E.S.; Lacroix, R.; van Gool, M.; Kroon, P.; Pineda, C.; Geukes Foppen, M.H.; Scolyer, R.; Song, J.Y.; et al. Targeting the MAPK and PI3K pathways in combination with PD1 blockade in melanoma. Oncoimmunology 2016, 5, e1238557. [Google Scholar] [CrossRef] [PubMed]
- Rager, T.; Eckburg, A.; Patel, M.; Qiu, R.; Gantiwala, S.; Dovalovsky, K.; Fan, K.; Lam, K.; Roesler, C.; Rastogi, A.; et al. Treatment of Metastatic Melanoma with a Combination of Immunotherapies and Molecularly Targeted Therapies. Cancers 2022, 14, 3779. [Google Scholar] [CrossRef] [PubMed]
- Aasen, S.N.; Parajuli, H.; Hoang, T.; Feng, Z.; Stokke, K.; Wang, J.; Roy, K.; Bjerkvig, R.; Knappskog, S.; Thorsen, F. Effective Treatment of Metastatic Melanoma by Combining MAPK and PI3K Signaling Pathway Inhibitors. Int. J. Mol. Sci. 2019, 20, 4235. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Long, J.; Li, K.; Zhang, X.; Chen, X.; Peng, C. A novel chalcone derivative suppresses melanoma cell growth through targeting Fyn/Stat3 pathway. Cancer Cell Int. 2020, 20, 256. [Google Scholar] [CrossRef]
- Gangadhar, T.C.; Clark, J.I.; Karrison, T.; Gajewski, T.F. Phase II study of the Src kinase inhibitor saracatinib 260-Gangadhar, T.C.; Clark, J.I.; Karrison, T.; Gajewski, T.F. Phase II study of the Src kinase inhibitor saracatinib (AZD0530) in metastatic melanoma. Investig. New Drugs 2013, 31, 769–773. [Google Scholar] [CrossRef]
- Schneider, P.; Schön, M.; Pletz, N.; Seitz, C.S.; Liu, N.; Ziegelbauer, K.; Zachmann, K.; Emmert, S.; Schön, M.P. The novel PI3 kinase inhibitor, BAY 80-6946, impairs melanoma growth in vivo and in vitro. Exp. Dermatol. 2014, 23, 579–584. [Google Scholar] [CrossRef]
- Amaral, T.; Niessner, H.; Sinnberg, T.; Thomas, I.; Meiwes, A.; Garbe, C.; Garzarolli, M.; Rauschenberg, R.; Eigentler, T.; Meier, F. An open-label, single-arm, phase II trial of buparlisib in patients with melanoma brain metastases not eligible for surgery or radiosurgery-the BUMPER study. Neurooncol. Adv. 2020, 2, vdaa140. [Google Scholar] [CrossRef]
- Tran, K.B.; Kolekar, S.; Jabed, A.; Jaynes, P.; Shih, J.H.; Wang, Q.; Flanagan, J.U.; Rewcastle, G.W.; Baguley, B.C.; Shepherd, P.R. Diverse mechanisms activate the PI 3-kinase/mTOR pathway in melanomas: Implications for the use of PI 3-kinase inhibitors to overcome resistance to inhibitors of BRAF and MEK. BMC Cancer 2021, 21, 136. [Google Scholar] [CrossRef] [PubMed]
- Ernst, D.S.; Eisenhauer, E.; Wainman, N.; Davis, M.; Lohmann, R.; Baetz, T.; Belanger, K.; Smylie, M. Phase II study of perifosine in previously untreated patients with metastatic melanoma. Investig. New Drugs 2005, 23, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Rebecca, V.W.; Massaro, R.R.; Fedorenko, I.V.; Sondak, V.K.; Anderson, A.R.; Kim, E.; Amaravadi, R.K.; Maria-Engler, S.S.; Messina, J.L.; Gibney, G.T.; et al. Inhibition of autophagy enhances the effects of the AKT inhibitor MK-2206 when combined with paclitaxel and carboplatin in BRAF wild-type melanoma. Pigment Cell Melanoma Res. 2014, 27, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Sznol, J.A.; Jilaveanu, L.B.; Kluger, H.M. Studies of NVP-BEZ235 in melanoma. Curr. Cancer Drug Targets 2013, 13, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Bernard, S.; Poon, A.C.; Tam, P.M.; Mutsaers, A.J. Investigation of the effects of mTOR inhibitors rapamycin and everolimus in combination with carboplatin on canine malignant melanoma cells. BMC Vet. Res. 2021, 17, 382. [Google Scholar] [CrossRef]
- Slingluff, C.L.; Petroni, G.R.; Molhoek, K.R.; Brautigan, D.L.; Chianese-Bullock, K.A.; Shada, A.L.; Smolkin, M.E.; Olson, W.C.; Gaucher, A.; Chase, C.M.; et al. Clinical activity and safety of combination therapy with temsirolimus and bevacizumab for advanced melanoma: A phase II trial (CTEP 7190/Mel47). Clin. Cancer Res. 2013, 19, 3611–3620. [Google Scholar] [CrossRef]
- Gopal, Y.N.; Rizos, H.; Chen, G.; Deng, W.; Frederick, D.T.; Cooper, Z.A.; Scolyer, R.A.; Pupo, G.; Komurov, K.; Sehgal, V.; et al. Inhibition of mTORC1/2 overcomes resistance to MAPK pathway inhibitors mediated by PGC1α and oxidative phosphorylation in melanoma. Cancer Res. 2014, 74, 7037–7047. [Google Scholar] [CrossRef]
- Espona-Fiedler, M.; Soto-Cerrato, V.; Hosseini, A.; Lizcano, J.M.; Guallar, V.; Quesada, R.; Gao, T.; Pérez-Tomás, R. Identification of dual mTORC1 and mTORC2 inhibitors in melanoma cells: Prodigiosin vs. obatoclax. Biochem. Pharmacol. 2012, 83, 489–496. [Google Scholar] [CrossRef]





| Description of the Study | References |
|---|---|
| Melanoma progression and treatment resistance are mediated by CD133 signaling to the PI3K pathway | Jamal et al., 2020 [8] |
| Inhibition of melanoma growth in an autophagy-dependent manner through inhibition of PI3K/AKT/mTOR signaling | Gong et al., 2020 [135] |
| Inhibition of the PI3K/AKT/mTOR pathway can efficiently counteract dabrafenib-induced stimulation of the invasive capacity of melanoma cells with required resistance. | Caporali et al., 2014 [133] |
| Description of the Study | Therapeutic Target | References |
|---|---|---|
| Phase II study of the Src kinase inhibitor saracatinib (AZD0530) in metastatic melanoma | NRTK, Fyn | Tang et al., 2020; Gangadhar, et al., 2013 [257,258] |
| Clinical reliability of PI3K pathway as therapeutic target for melanoma treatment | PI3K | Amaral et al., 2020; Schneider et al., 2014; Tran et al., 2021 [259,260,261] |
| Clinical relevance of AKT as therapeutic in melanoma treatment | AKT | Ernst et al., 2005; Rebecca et al., 2014 [262,263] |
| Target both mTORC1 and mTORC2 based on their clinical reliability in melanoma treatment | mTORC1/mTORC2 | Sznol et al., 2013; Bernard et al., 2021; Slingluff et al., 2013; Gopal et al., 2014; Espona-Fiedler et al., 2012 [264,265,266,267,268] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kharouf, N.; Flanagan, T.W.; Alamodi, A.A.; Al Hmada, Y.; Hassan, S.-Y.; Shalaby, H.; Santourlidis, S.; Hassan, S.-L.; Haikel, Y.; Megahed, M.; et al. CD133-Dependent Activation of Phosphoinositide 3-Kinase /AKT/Mammalian Target of Rapamycin Signaling in Melanoma Progression and Drug Resistance. Cells 2024, 13, 240. https://doi.org/10.3390/cells13030240
Kharouf N, Flanagan TW, Alamodi AA, Al Hmada Y, Hassan S-Y, Shalaby H, Santourlidis S, Hassan S-L, Haikel Y, Megahed M, et al. CD133-Dependent Activation of Phosphoinositide 3-Kinase /AKT/Mammalian Target of Rapamycin Signaling in Melanoma Progression and Drug Resistance. Cells. 2024; 13(3):240. https://doi.org/10.3390/cells13030240
Chicago/Turabian StyleKharouf, Naji, Thomas W. Flanagan, Abdulhadi A. Alamodi, Youssef Al Hmada, Sofie-Yasmin Hassan, Hosam Shalaby, Simeon Santourlidis, Sarah-Lilly Hassan, Youssef Haikel, Mossad Megahed, and et al. 2024. "CD133-Dependent Activation of Phosphoinositide 3-Kinase /AKT/Mammalian Target of Rapamycin Signaling in Melanoma Progression and Drug Resistance" Cells 13, no. 3: 240. https://doi.org/10.3390/cells13030240
APA StyleKharouf, N., Flanagan, T. W., Alamodi, A. A., Al Hmada, Y., Hassan, S.-Y., Shalaby, H., Santourlidis, S., Hassan, S.-L., Haikel, Y., Megahed, M., Brodell, R. T., & Hassan, M. (2024). CD133-Dependent Activation of Phosphoinositide 3-Kinase /AKT/Mammalian Target of Rapamycin Signaling in Melanoma Progression and Drug Resistance. Cells, 13(3), 240. https://doi.org/10.3390/cells13030240










