Pathogenesis of DJ-1/PARK7-Mediated Parkinson’s Disease
Abstract
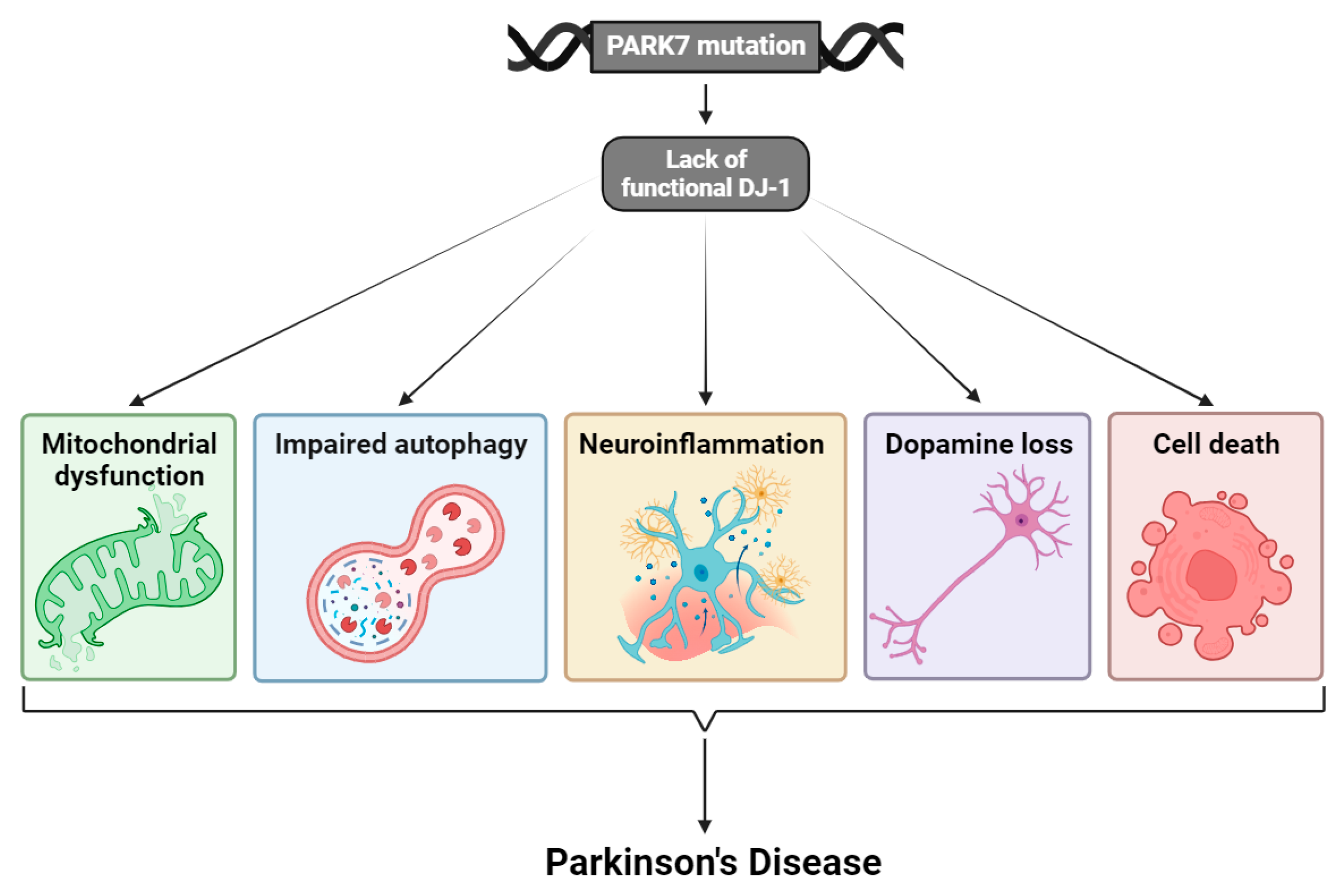
:1. Introduction
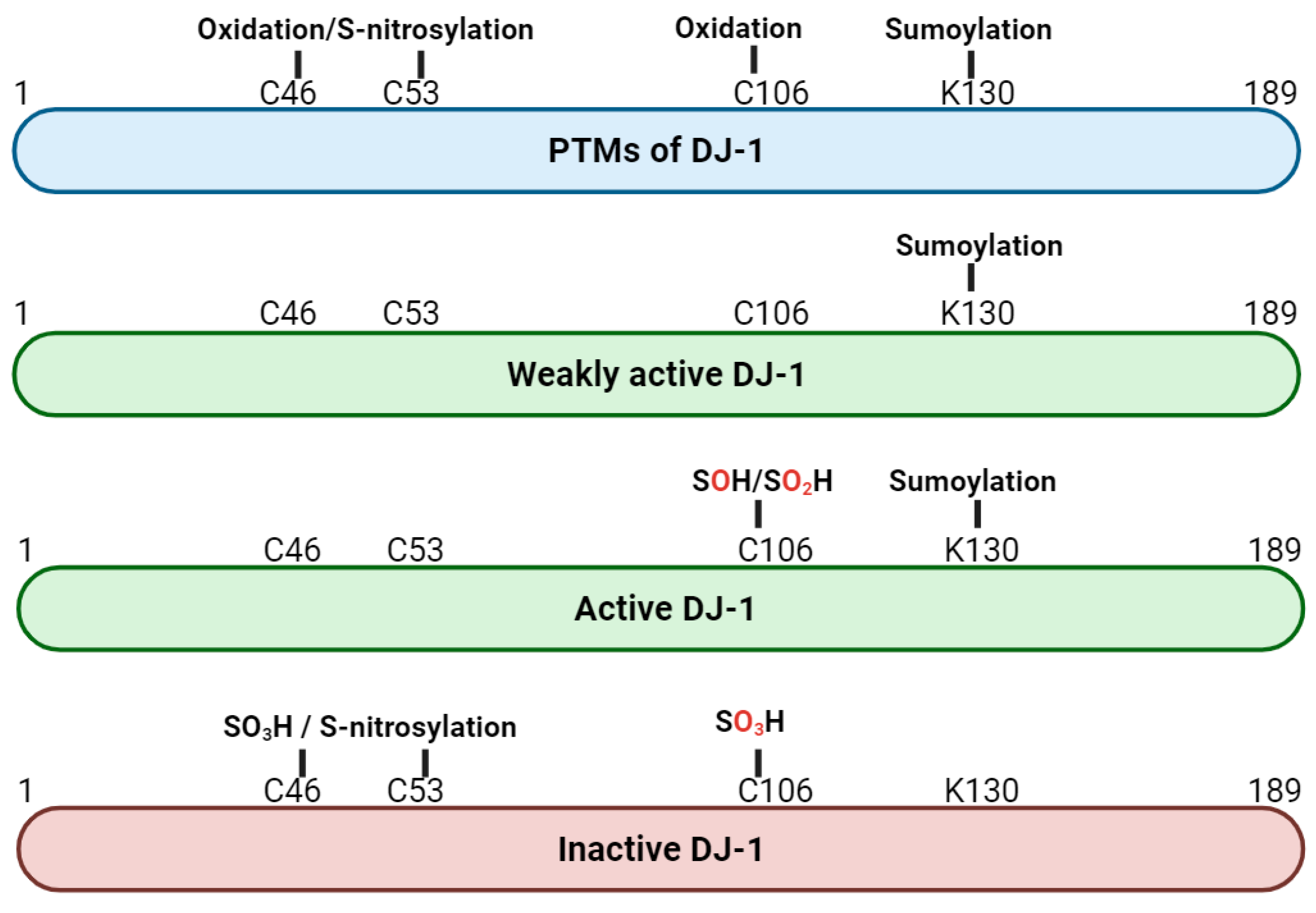
2. Structure and Post-Translational Modification of DJ-1 Protein
3. Mutations of DJ-1
4. The Multiple Cellular and Molecular Functions of DJ-1
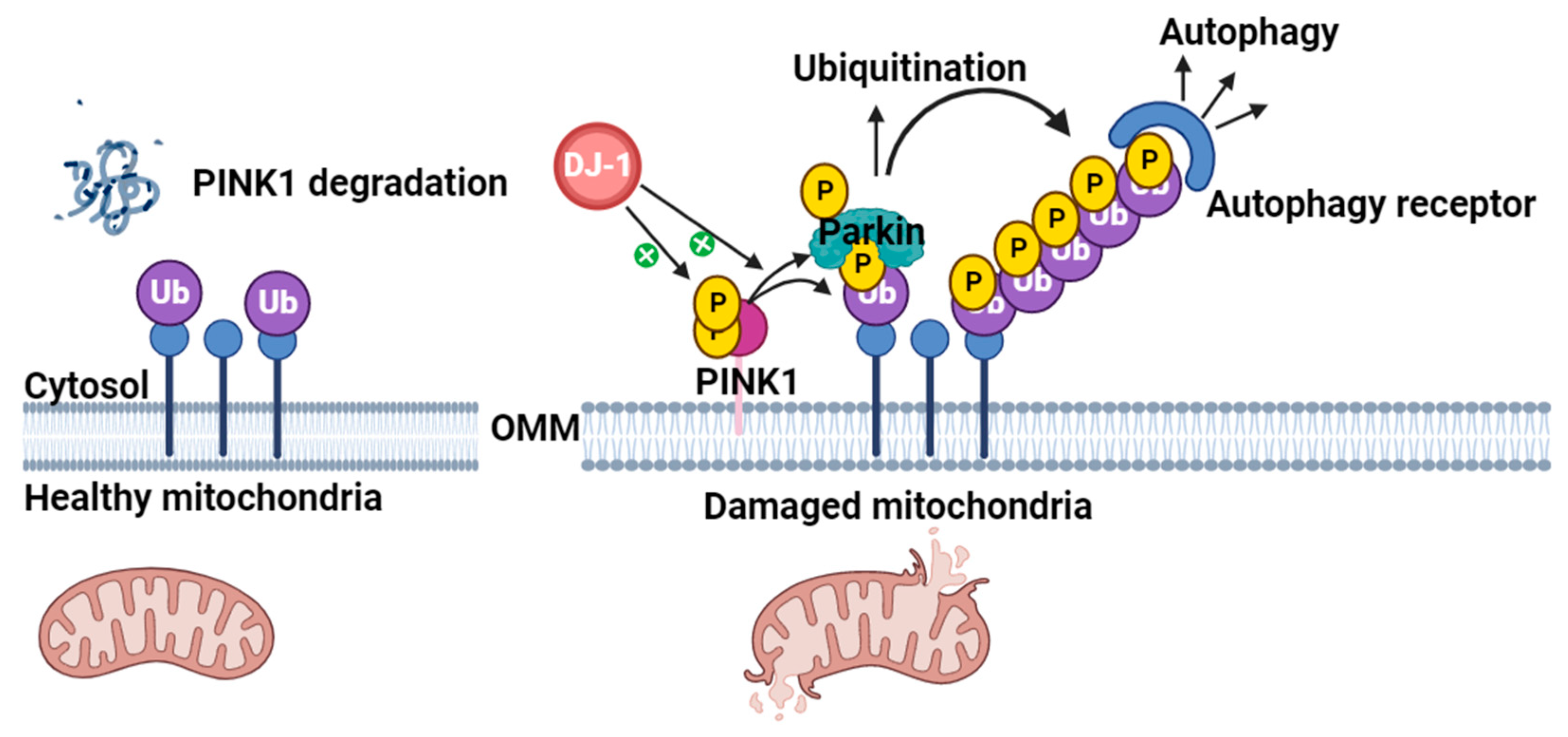
4.1. Role of DJ-1 in Mitochondrial Homeostasis
Interplay of Parkin, PINK1, and DJ-1
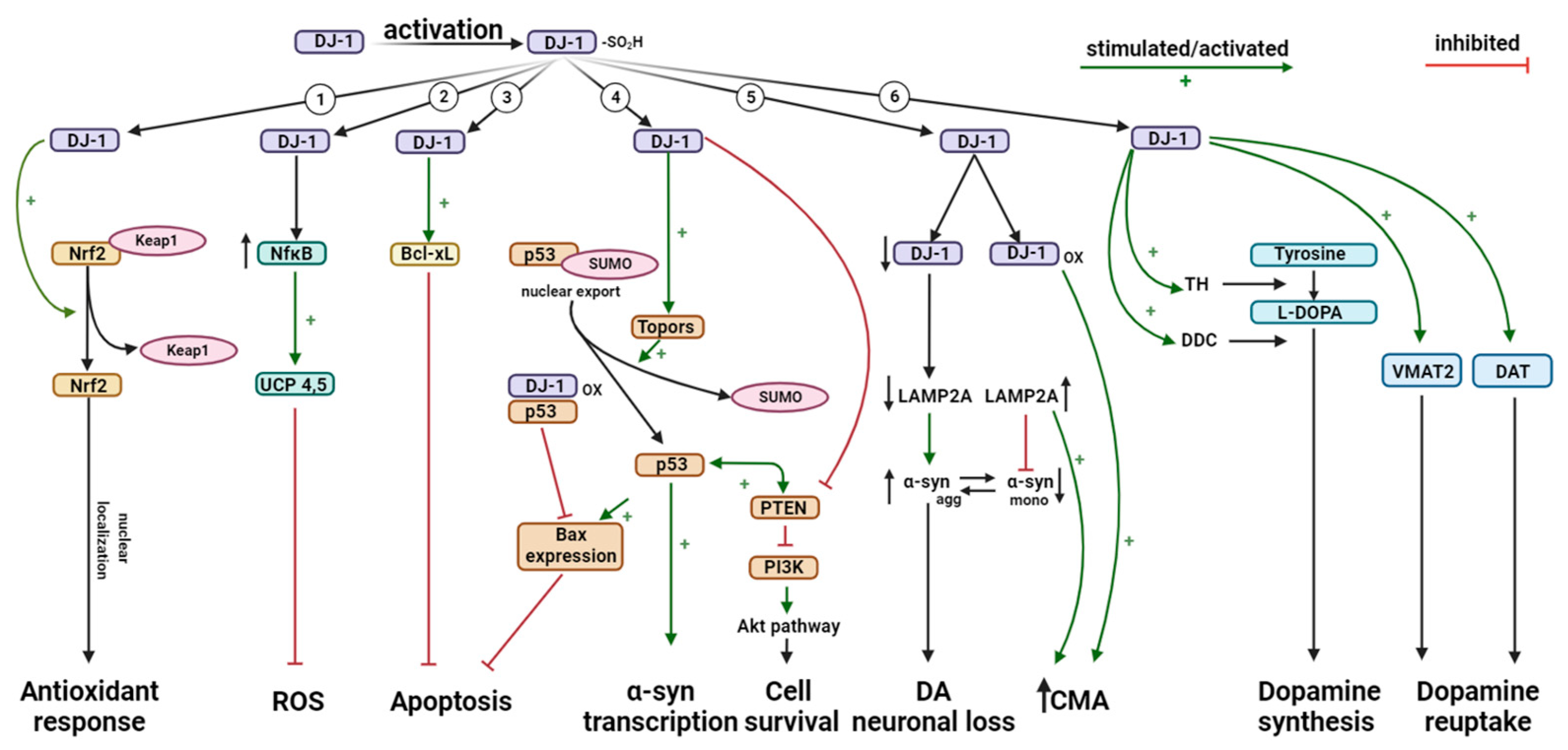
4.2. Antioxidant Activity of DJ-1
4.3. Role of DJ-1 in Inflammatory Responses
4.4. Role of DJ-1 in Apoptosis
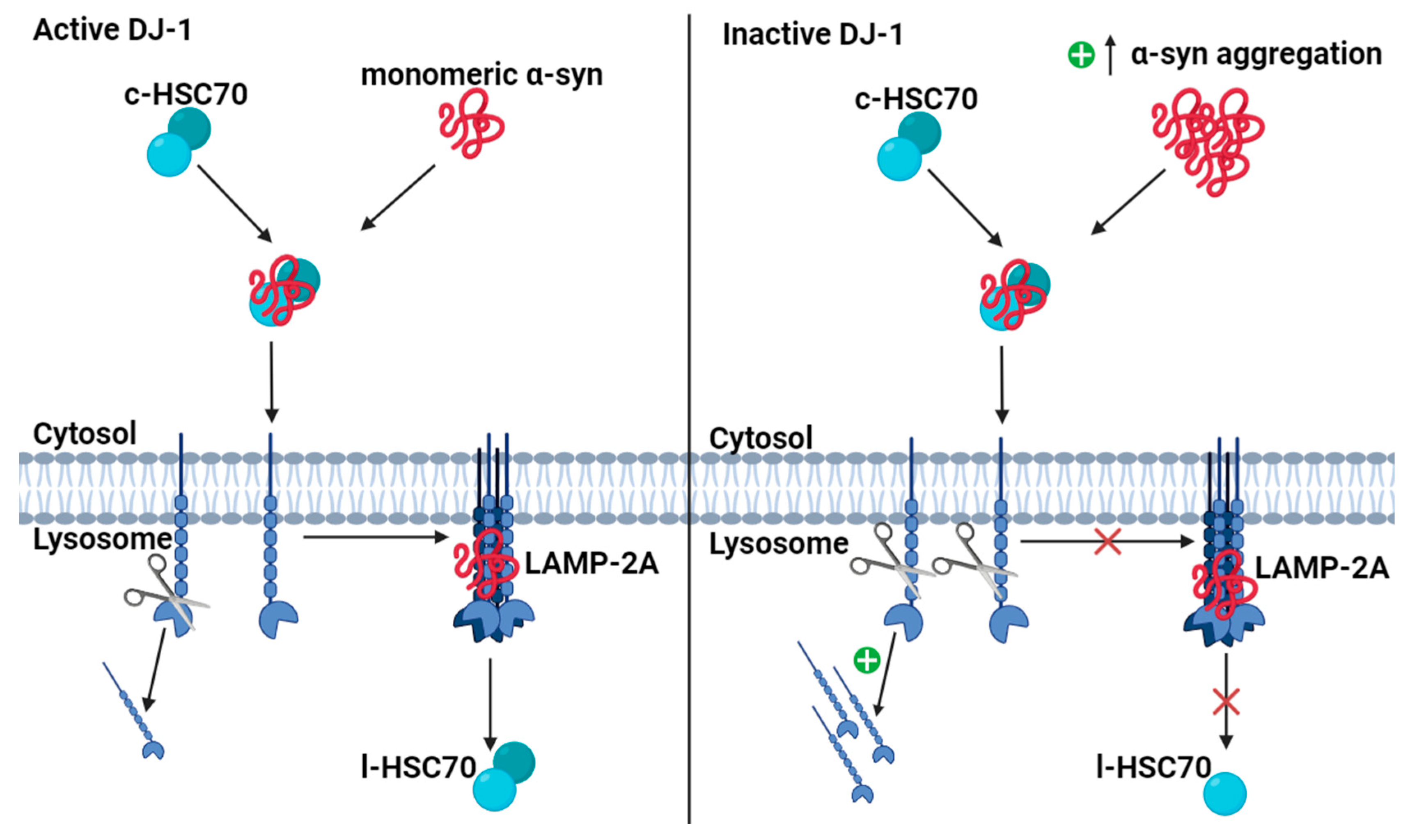
4.5. DJ-1 Regulates Degradation of Monomeric α-Synuclein (α-syn) via Chaperone-Mediated Autophagy (CMA)
4.6. DJ-1 Regulates Dopamine Homeostasis
5. DJ-1 as a Biomarker and Therapeutic Agent
6. Discussion
7. Conclusions—Many Functions, but One Goal: Neuroprotectivity
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goetz, C.G. The history of Parkinson’s disease: Early clinical descriptions and neurological therapies. Cold Spring Harb. Perspect. Med. 2011, 1, a008862. [Google Scholar] [CrossRef]
- Jankovic, J. Parkinson’s disease: Clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 2008, 79, 368–376. [Google Scholar] [CrossRef]
- Sveinbjornsdottir, S. The clinical symptoms of Parkinson’s disease. J. Neurochem. 2016, 139 (Suppl. S1), 318–324. [Google Scholar] [CrossRef]
- MacMahon Copas, A.N.; McComish, S.F.; Fletcher, J.M.; Caldwell, M.A. The Pathogenesis of Parkinson’s Disease: A Complex Interplay Between Astrocytes, Microglia, and T Lymphocytes? Front. Neurol. 2021, 12, 666737. [Google Scholar] [CrossRef]
- Balestrino, R.; Schapira, A.H.V. Parkinson disease. Eur. J. Neurol. 2020, 27, 27–42. [Google Scholar] [CrossRef]
- Madsen, D.A.; Schmidt, S.I.; Blaabjerg, M.; Meyer, M. Interaction between Parkin and alpha-Synuclein in PARK2-Mediated Parkinson’s Disease. Cells 2021, 10, 283. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Schmidt, M.L.; Lee, V.M.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. Alpha-synuclein in Lewy bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef] [PubMed]
- Schaser, A.J.; Osterberg, V.R.; Dent, S.E.; Stackhouse, T.L.; Wakeham, C.M.; Boutros, S.W.; Weston, L.J.; Owen, N.; Weissman, T.A.; Luna, E.; et al. Alpha-synuclein is a DNA binding protein that modulates DNA repair with implications for Lewy body disorders. Sci. Rep. 2019, 9, 10919. [Google Scholar] [CrossRef] [PubMed]
- Ascherio, A.; Schwarzschild, M.A. The epidemiology of Parkinson’s disease: Risk factors and prevention. Lancet Neurol. 2016, 15, 1257–1272. [Google Scholar] [CrossRef] [PubMed]
- Emamzadeh, F.N.; Surguchov, A. Parkinson’s Disease: Biomarkers, Treatment, and Risk Factors. Front. Neurosci. 2018, 12, 612. [Google Scholar] [CrossRef] [PubMed]
- Lunati, A.; Lesage, S.; Brice, A. The genetic landscape of Parkinson’s disease. Rev. Neurol. 2018, 174, 628–643. [Google Scholar] [CrossRef]
- Kouli, A.; Torsney, K.M.; Kuan, W.L. Parkinson’s Disease: Etiology, Neuropathology, and Pathogenesis. In Parkinson’s Disease: Pathogenesis and Clinical Aspects; Stoker, T.B., Greenland, J.C., Eds.; Exon Publications: Brisbane, Australia, 2018. [Google Scholar]
- Inzelberg, R.; Samuels, Y.; Azizi, E.; Qutob, N.; Inzelberg, L.; Domany, E.; Schechtman, E.; Friedman, E. Parkinson disease (PARK) genes are somatically mutated in cutaneous melanoma. Neurol. Genet. 2016, 2, e70. [Google Scholar] [CrossRef]
- Kitada, T.; Asakawa, S.; Hattori, N.; Matsumine, H.; Yamamura, Y.; Minoshima, S.; Yokochi, M.; Mizuno, Y.; Shimizu, N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 1998, 392, 605–608. [Google Scholar] [CrossRef]
- Li, W.; Fu, Y.; Halliday, G.M.; Sue, C.M. PARK Genes Link Mitochondrial Dysfunction and Alpha-Synuclein Pathology in Sporadic Parkinson’s Disease. Front. Cell Dev. Biol. 2021, 9, 612476. [Google Scholar] [CrossRef]
- Wang, X.L.; Feng, S.T.; Wang, Y.T.; Yuan, Y.H.; Li, Z.P.; Chen, N.H.; Wang, Z.-Z.; Zhang, Y. Mitophagy, a Form of Selective Autophagy, Plays an Essential Role in Mitochondrial Dynamics of Parkinson’s Disease. Cell Mol. Neurobiol. 2022, 42, 1321–1339. [Google Scholar] [CrossRef]
- Nagakubo, D.; Taira, T.; Kitaura, H.; Ikeda, M.; Tamai, K.; Iguchi-Ariga, S.M.; Ariga, H. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem. Biophys. Res. Commun. 1997, 231, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Bonifati, V.; Rizzu, P.; van Baren, M.J.; Schaap, O.; Breedveld, G.J.; Krieger, E.; Dekker, M.C.J.; Squitieri, F.; Ibanez, P.; Joosse, M.; et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 2003, 299, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Macedo, M.G.; Anar, B.; Bronner, I.F.; Cannella, M.; Squitieri, F.; Bonifati, V.; Hoogeveen, A.; Heutink, P.; Rizzu, P. The DJ-1L166P mutant protein associated with early onset Parkinson’s disease is unstable and forms higher-order protein complexes. Hum. Mol. Genet. 2003, 12, 2807–2816. [Google Scholar] [CrossRef] [PubMed]
- Repici, M.; Giorgini, F. DJ-1 in Parkinson’s Disease: Clinical Insights and Therapeutic Perspectives. J. Clin. Med. 2019, 8, 1377. [Google Scholar] [CrossRef] [PubMed]
- Dias, V.; Junn, E.; Mouradian, M.M. The role of oxidative stress in Parkinson’s disease. J. Parkinsons. Dis. 2013, 3, 461–491. [Google Scholar] [CrossRef] [PubMed]
- Lind-Holm Mogensen, F.; Scafidi, A.; Poli, A.; Michelucci, A. PARK7/DJ-1 in microglia: Implications in Parkinson’s disease and relevance as a therapeutic target. J. Neuroinflammation. 2023, 20, 95. [Google Scholar] [CrossRef] [PubMed]
- Dolgacheva, L.P.; Berezhnov, A.V.; Fedotova, E.I.; Zinchenko, V.P.; Abramov, A.Y. Role of DJ-1 in the mechanism of pathogenesis of Parkinson’s disease. J. Bioenerg. Biomembr. 2019, 51, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Di Nottia, M.; Masciullo, M.; Verrigni, D.; Petrillo, S.; Modoni, A.; Rizzo, V.; Di Giuda, D.; Rizza, T.; Niceta, M.; Torraco, A.; et al. DJ-1 modulates mitochondrial response to oxidative stress: Clues from a novel diagnosis of PARK7. Clin. Genet. 2017, 92, 18–25. [Google Scholar] [CrossRef]
- Raninga, P.V.; Di Trapani, G.; Tonissen, K.F. The Multifaceted Roles of DJ-1 as an Antioxidant. Adv. Exp. Med. Biol. 2017, 1037, 67–87. [Google Scholar] [PubMed]
- van Duijn, C.M.; Dekker, M.C.; Bonifati, V.; Galjaard, R.J.; Houwing-Duistermaat, J.J.; Snijders, P.J.; Testers, L.; Breedveld, G.J.; Horstink, M.; Sandkuijl, L.A.; et al. Park7, a novel locus for autosomal recessive early-onset parkinsonism, on chromosome 1p36. Am. J. Hum. Genet. 2001, 69, 629–634. [Google Scholar] [CrossRef]
- Olzmann, J.A.; Brown, K.; Wilkinson, K.D.; Rees, H.D.; Huai, Q.; Ke, H.; Levey, A.I.; Li, L.; Chin, L.S. Familial Parkinson’s disease-associated L166P mutation disrupts DJ-1 protein folding and function. J. Biol. Chem. 2004, 279, 8506–8515. [Google Scholar] [CrossRef]
- Huai, Q.; Sun, Y.; Wang, H.; Chin, L.S.; Li, L.; Robinson, H.; Ke, H. Crystal structure of DJ-1/RS and implication on familial Parkinson’s disease. FEBS Lett. 2003, 549, 171–175. [Google Scholar] [CrossRef]
- Tao, X.; Tong, L. Crystal structure of human DJ-1, a protein associated with early onset Parkinson’s disease. J. Biol. Chem. 2003, 278, 31372–31379. [Google Scholar] [CrossRef]
- Zhang, L.; Shimoji, M.; Thomas, B.; Moore, D.J.; Yu, S.W.; Marupudi, N.I.; Torp, R.; Torgner, I.A.; Ottersen, O.P.; Dawson, T.M.; et al. Mitochondrial localization of the Parkinson’s disease related protein DJ-1: Implications for pathogenesis. Hum. Mol. Genet. 2005, 14, 2063–2073. [Google Scholar] [CrossRef] [PubMed]
- Bandopadhyay, R.; Kingsbury, A.E.; Cookson, M.R.; Reid, A.R.; Evans, I.M.; Hope, A.D.; Pittman, A.M.; Lashley, T.; Canet-Aviles, R.; Miller, D.W.; et al. The expression of DJ-1 (PARK7) in normal human CNS and idiopathic Parkinson’s disease. Brain 2004, 127 Pt 2, 420–430. [Google Scholar] [CrossRef]
- Junn, E.; Jang, W.H.; Zhao, X.; Jeong, B.S.; Mouradian, M.M. Mitochondrial localization of DJ-1 leads to enhanced neuroprotection. J. Neurosci. Res. 2009, 87, 123–129. [Google Scholar] [CrossRef]
- Kinumi, T.; Kimata, J.; Taira, T.; Ariga, H.; Niki, E. Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2004, 317, 722–728. [Google Scholar] [CrossRef]
- Kiss, R.; Zhu, M.; Jojart, B.; Czajlik, A.; Solti, K.; Forizs, B.; Nagy, É.; Zsila, F.; Beke-Somfai, T.; Tóth, G.; et al. Structural features of human DJ-1 in distinct Cys106 oxidative states and their relevance to its loss of function in disease. Biochim. Biophys. Acta Gen. Subj. 2017, 1861 Pt A, 2619–2629. [Google Scholar] [CrossRef]
- Mussakhmetov, A.; Shumilin, I.A.; Nugmanova, R.; Shabalin, I.G.; Baizhumanov, T.; Toibazar, D.; Khassenov, B.; Minor, W.; Utepbergenov, D. A transient post-translational modification of active site cysteine alters binding properties of the parkinsonism protein DJ-1. Biochem. Biophys. Res. Commun. 2018, 504, 328–333. [Google Scholar] [CrossRef]
- Ariga, H.; Takahashi-Niki, K.; Kato, I.; Maita, H.; Niki, T.; Iguchi-Ariga, S.M. Neuroprotective function of DJ-1 in Parkinson’s disease. Oxid. Med. Cell Longev. 2013, 2013, 683920. [Google Scholar] [CrossRef]
- Shinbo, Y.; Niki, T.; Taira, T.; Ooe, H.; Takahashi-Niki, K.; Maita, C.; Seino, C.; Iguchi-Ariga, S.M.M.; Ariga, H. Proper SUMO-1 conjugation is essential to DJ-1 to exert its full activities. Cell Death Differ. 2006, 13, 96–108. [Google Scholar] [CrossRef]
- Ito, G.; Ariga, H.; Nakagawa, Y.; Iwatsubo, T. Roles of distinct cysteine residues in S-nitrosylation and dimerization of DJ-1. Biochem. Biophys. Res. Commun. 2006, 339, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.A.; Collins, J.L.; Hod, Y.; Ringe, D.; Petsko, G.A. The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2003, 100, 9256–9261. [Google Scholar] [CrossRef] [PubMed]
- Abou-Sleiman, P.M.; Healy, D.G.; Quinn, N.; Lees, A.J.; Wood, N.W. The role of pathogenic DJ-1 mutations in Parkinson’s disease. Ann. Neurol. 2003, 54, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Kahle, P.J.; Waak, J.; Gasser, T. DJ-1 and prevention of oxidative stress in Parkinson’s disease and other age-related disorders. Free Radic. Biol. Med. 2009, 47, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Blackinton, J.; Lakshminarasimhan, M.; Thomas, K.J.; Ahmad, R.; Greggio, E.; Raza, A.S.; Cookson, M.R.; Wilson, M.A. Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1. J. Biol. Chem. 2009, 284, 6476–6485. [Google Scholar] [CrossRef] [PubMed]
- Piston, D.; Alvarez-Erviti, L.; Bansal, V.; Gargano, D.; Yao, Z.; Szabadkai, G.; Odell, M.; Puno, M.R.; Björkblom, B.; Maple-Grødem, J.; et al. DJ-1 is a redox sensitive adapter protein for high molecular weight complexes involved in regulation of catecholamine homeostasis. Hum. Mol. Genet. 2018, 27, 576. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, H.; Zhang, L.; Liu, X.; Zhang, C.; Wang, Y.; He, Q.; Zhang, Y.; Li, Y.; Chen, Q.; et al. DJ-1 promotes colorectal cancer progression through activating PLAGL2/Wnt/BMP4 axis. Cell Death Dis. 2018, 9, 865. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.N.; Afridi, S.; Mejia-Santana, H.; Harris, J.; Louis, E.D.; Cote, L.J.; Andrews, H.; Singleton, A.; Wavrant De-Vrieze, F.; Hardy, J.; et al. Analysis of an early-onset Parkinson’s disease cohort for DJ-1 mutations. Mov. Disord. 2004, 19, 796–800. [Google Scholar] [CrossRef]
- Taira, T.; Saito, Y.; Niki, T.; Iguchi-Ariga, S.M.; Takahashi, K.; Ariga, H. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004, 5, 213–218. [Google Scholar] [CrossRef]
- Zhou, W.; Zhu, M.; Wilson, M.A.; Petsko, G.A.; Fink, A.L. The oxidation state of DJ-1 regulates its chaperone activity toward alpha-synuclein. J. Mol. Biol. 2006, 356, 1036–1048. [Google Scholar] [CrossRef]
- Irrcher, I.; Aleyasin, H.; Seifert, E.L.; Hewitt, S.J.; Chhabra, S.; Phillips, M.; Lutz, A.K.; Rousseaux, M.W.C.; Bevilacqua, L.; Jahani-Asl, A.; et al. Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum. Mol. Genet. 2010, 19, 3734–3746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, J.; Wang, J.; Yang, B.; He, Q.; Weng, Q. Role of DJ-1 in Immune and Inflammatory Diseases. Front. Immunol. 2020, 11, 994. [Google Scholar] [CrossRef]
- Palikaras, K.; Tavernarakis, N. Mitochondrial homeostasis: The interplay between mitophagy and mitochondrial biogenesis. Exp. Gerontol. 2014, 56, 182–188. [Google Scholar] [CrossRef]
- Scorziello, A.; Borzacchiello, D.; Sisalli, M.J.; Di Martino, R.; Morelli, M.; Feliciello, A. Mitochondrial Homeostasis and Signaling in Parkinson’s Disease. Front. Aging Neurosci. 2020, 12, 100. [Google Scholar] [CrossRef]
- Sheng, Z.H.; Cai, Q. Mitochondrial transport in neurons: Impact on synaptic homeostasis and neurodegeneration. Nat. Rev. Neurosci. 2012, 13, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Kim, W.I.; Park, T.H.; Bae, J.H.; Nam, H.S.; Cho, S.W.; Choi, Y.J.; Lee, S.H.; Cho, M.K. Upregulation of DJ-1 expression in melanoma regulates PTEN/AKT pathway for cell survival and migration. Arch. Dermatol. Res. 2021, 313, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Canet-Aviles, R.M.; Wilson, M.A.; Miller, D.W.; Ahmad, R.; McLendon, C.; Bandyopadhyay, S.; Baptista, M.J.; Ringe, D.; Petsko, G.A.; Cookson, M.R. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA 2004, 101, 9103–9108. [Google Scholar] [CrossRef] [PubMed]
- Krebiehl, G.; Ruckerbauer, S.; Burbulla, L.F.; Kieper, N.; Maurer, B.; Waak, J.; Wolburg, H.; Gizatullina, Z.; Gellerich, F.N.; Woitalla, D.; et al. Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson’s disease-associated protein DJ-1. PLoS ONE 2010, 5, e9367. [Google Scholar] [CrossRef]
- Hayashi, T.; Ishimori, C.; Takahashi-Niki, K.; Taira, T.; Kim, Y.C.; Maita, H.; Maita, C.; Ariga, H.; Iguchi-Ariga, S.M. DJ-1 binds to mitochondrial complex I and maintains its activity. Biochem. Biophys. Res. Commun. 2009, 390, 667–672. [Google Scholar] [CrossRef]
- Chu, C.; He, N.; Zeljic, K.; Zhang, Z.; Wang, J.; Li, J.; Liu, Y.; Zhang, Y.; Sun, B.; Li, D.; et al. Subthalamic and pallidal stimulation in Parkinson’s disease induce distinct brain topological reconstruction. Neuroimage 2022, 255, 119196. [Google Scholar] [CrossRef]
- Lanciego, J.L.; Luquin, N.; Obeso, J.A. Functional neuroanatomy of the basal ganglia. Cold Spring Harb. Perspect. Med. 2012, 2, a009621. [Google Scholar] [CrossRef]
- Caligiore, D.; Helmich, R.C.; Hallett, M.; Moustafa, A.A.; Timmermann, L.; Toni, I.; Baldassarre, G. Parkinson’s disease as a system-level disorder. NPJ Park. Dis. 2016, 2, 16025. [Google Scholar] [CrossRef]
- Picca, A.; Mankowski, R.T.; Burman, J.L.; Donisi, L.; Kim, J.S.; Marzetti, E.; Leeuwenburgh, C. Mitochondrial quality control mechanisms as molecular targets in cardiac ageing. Nat. Rev. Cardiol. 2018, 15, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Shefa, U.; Jeong, N.Y.; Song, I.O.; Chung, H.J.; Kim, D.; Jung, J.; Huh, Y. Mitophagy links oxidative stress conditions and neurodegenerative diseases. Neural. Regen. Res. 2019, 14, 749–756. [Google Scholar] [PubMed]
- Jin, S.M.; Youle, R.J. PINK1- and Parkin-mediated mitophagy at a glance. J. Cell Sci. 2012, 125 Pt 4, 795–799. [Google Scholar] [CrossRef]
- Killackey, S.A.; Philpott, D.J.; Girardin, S.E. Mitophagy pathways in health and disease. J. Cell Biol. 2020, 219, e202004029. [Google Scholar] [CrossRef]
- Dagda, R.K.; Pien, I.; Wang, R.; Zhu, J.; Wang, K.Z.; Callio, J.; Das Banerjee, T.; Dagda, R.Y.; Chu, C.T. Beyond the mitochondrion: Cytosolic PINK1 remodels dendrites through protein kinase A. J. Neurochem. 2014, 128, 864–877. [Google Scholar] [CrossRef]
- Matsuda, S.; Kitagishi, Y.; Kobayashi, M. Function and characteristics of PINK1 in mitochondria. Oxid. Med. Cell Longev. 2013, 2013, 601587. [Google Scholar] [CrossRef]
- Youle, R.J.; van der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef] [PubMed]
- Sekine, S.; Youle, R.J. PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol. 2018, 16, 2. [Google Scholar] [CrossRef]
- Julienne, H.; Buhl, E.; Leslie, D.S.; Hodge, J.J.L. Drosophila PINK1 and parkin loss-of-function mutants display a range of non-motor Parkinson’s disease phenotypes. Neurobiol. Dis. 2017, 104, 15–23. [Google Scholar] [CrossRef]
- Durcan, T.M.; Fon, E.A. The three ‘P’s of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev. 2015, 29, 989–999. [Google Scholar] [CrossRef]
- Geisler, S.; Holmström, K.M.; Skujat, D.; Fiesel, F.C.; Rothfuss, O.C.; Kahle, P.J.; Springer, W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010, 12, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, N.; Sato, S.; Shiba, K.; Okatsu, K.; Saisho, K.; Gautier, C.A.; Sou, Y.-S.; Saiki, S.; Kawajiri, S.; Sato, F.; et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010, 189, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.P.; Jin, S.M.; Tanaka, A.; Suen, D.F.; Gautier, C.A.; Shen, J.; Cookson, M.R.; Youle, R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010, 8, e1000298. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Xiong, H.; Sun, P.; Zhang, Y.; Wang, D.; Hu, Z.; Zhu, Z.; Ma, H.; Pan, Q.; Xia, J.H.; et al. Association of PINK1 and DJ-1 confers digenic inheritance of early-onset Parkinson’s disease. Hum. Mol. Genet. 2006, 15, 1816–1825. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.A. The role of cysteine oxidation in DJ-1 function and dysfunction. Antioxid. Redox Signal. 2011, 15, 111–122. [Google Scholar] [CrossRef]
- Girotto, S.; Cendron, L.; Bisaglia, M.; Tessari, I.; Mammi, S.; Zanotti, G.; Bubacco, L. DJ-1 is a copper chaperone acting on SOD1 activation. J. Biol. Chem. 2014, 289, 10887–10899. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef]
- Clements, C.M.; McNally, R.S.; Conti, B.J.; Mak, T.W.; Ting, J.P. DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc. Natl. Acad. Sci. USA 2006, 103, 15091–15096. [Google Scholar] [CrossRef] [PubMed]
- Im, J.Y.; Lee, K.W.; Woo, J.M.; Junn, E.; Mouradian, M.M. DJ-1 induces thioredoxin 1 expression through the Nrf2 pathway. Hum. Mol. Genet. 2012, 21, 3013–3024. [Google Scholar] [CrossRef]
- Xu, J.; Zhong, N.; Wang, H.; Elias, J.E.; Kim, C.Y.; Woldman, I.; Pifl, C.; Gygi, S.P.; Geula, C.; Yankner, B.A. The Parkinson’s disease-associated DJ-1 protein is a transcriptional co-activator that protects against neuronal apoptosis. Hum. Mol. Genet. 2005, 14, 1231–1241. [Google Scholar] [CrossRef]
- Gan, L.; Johnson, D.A.; Johnson, J.A. Keap1-Nrf2 activation in the presence and absence of DJ-1. Eur. J. Neurosci. 2010, 31, 967–977. [Google Scholar] [CrossRef]
- Cuevas, S.; Yang, Y.; Konkalmatt, P.; Asico, L.D.; Feranil, J.; Jones, J.; Villar, V.A.; Armando, I.; Jose, P.A. Role of nuclear factor erythroid 2-related factor 2 in the oxidative stress-dependent hypertension associated with the depletion of DJ-1. Hypertension 2015, 65, 1251–1257. [Google Scholar] [CrossRef]
- Guzman, J.N.; Sanchez-Padilla, J.; Wokosin, D.; Kondapalli, J.; Ilijic, E.; Schumacker, P.T.; Surmeier, D.J. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature 2010, 468, 696–700. [Google Scholar] [CrossRef] [PubMed]
- Ramsden, D.B.; Ho, P.W.; Ho, J.W.; Liu, H.F.; So, D.H.; Tse, H.M.; Chan, K.; Ho, S. Human neuronal uncoupling proteins 4 and 5 (UCP4 and UCP5): Structural properties, regulation, and physiological role in protection against oxidative stress and mitochondrial dysfunction. Brain Behav. 2012, 2, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Yang, X.; Qian, Y.; Xiao, Q. Parkinson’s disease-related DJ-1 modulates the expression of uncoupling protein 4 against oxidative stress. J. Neurochem. 2018, 145, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.W.; Ho, P.W.; Liu, H.F.; So, D.H.; Chan, K.H.; Tse, Z.H.; Kung, M.H.; Ramsden, D.B.; Ho, S.L. UCP4 is a target effector of the NF-kappaB c-Rel prosurvival pathway against oxidative stress. Free Radic. Biol. Med. 2012, 53, 383–394. [Google Scholar] [PubMed]
- Mittal, M.; Siddiqui, M.R.; Tran, K.; Reddy, S.P.; Malik, A.B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 2014, 20, 1126–1167. [Google Scholar] [CrossRef]
- Rotblat, B.; Grunewald, T.G.; Leprivier, G.; Melino, G.; Knight, R.A. Anti-oxidative stress response genes: Bioinformatic analysis of their expression and relevance in multiple cancers. Oncotarget 2013, 4, 2577–2590. [Google Scholar] [CrossRef]
- Kim, S.J.; Park, Y.J.; Hwang, I.Y.; Youdim, M.B.; Park, K.S.; Oh, Y.J. Nuclear translocation of DJ-1 during oxidative stress-induced neuronal cell death. Free Radic. Biol. Med. 2012, 53, 936–950. [Google Scholar] [CrossRef]
- Peng, L.; Zhou, Y.; Jiang, N.; Wang, T.; Zhu, J.; Chen, Y.; Li, L.; Zhang, J.; Yu, S.; Zhao, Y. DJ-1 exerts anti-inflammatory effects and regulates NLRX1-TRAF6 via SHP-1 in stroke. J. Neuroinflammation 2020, 17, 81. [Google Scholar] [CrossRef]
- Yang, L.; Mao, K.; Yu, H.; Chen, J. Neuroinflammatory Responses and Parkinson’ Disease: Pathogenic Mechanisms and Therapeutic Targets. J. Neuroimmune Pharmacol. 2020, 15, 830–837. [Google Scholar] [CrossRef]
- Froyset, A.K.; Edson, A.J.; Gharbi, N.; Khan, E.A.; Dondorp, D.; Bai, Q.; Tiraboschi, E.; Suster, M.L.; Connolly, J.B.; Burton, E.A.; et al. Astroglial DJ-1 over-expression up-regulates proteins involved in redox regulation and is neuroprotective in vivo. Redox Biol. 2018, 16, 237–247. [Google Scholar] [CrossRef]
- Choi, D.J.; An, J.; Jou, I.; Park, S.M.; Joe, E.H. A Parkinson’s disease gene, DJ-1, regulates anti-inflammatory roles of astrocytes through prostaglandin D(2) synthase expression. Neurobiol. Dis. 2019, 127, 482–491. [Google Scholar] [CrossRef]
- Tiwari, P.C.; Pal, R. The potential role of neuroinflammation and transcription factors in Parkinson disease. Dialogues Clin. Neurosci. 2017, 19, 71–80. [Google Scholar] [CrossRef]
- Ren, H.; Fu, K.; Wang, D.; Mu, C.; Wang, G. Oxidized DJ-1 interacts with the mitochondrial protein BCL-XL. J. Biol. Chem. 2011, 286, 35308–35317. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.K.; Lee, M.S.; Bae, D.W.; Lee, D.H.; Cha, S.S.; Chi, S.W. Structural basis for the interaction between DJ-1 and Bcl-X(L). Biochem. Biophys. Res. Commun. 2018, 495, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Shinbo, Y.; Taira, T.; Niki, T.; Iguchi-Ariga, S.M.; Ariga, H. DJ-1 restores p53 transcription activity inhibited by Topors/p53BP3. Int. J. Oncol. 2005, 26, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Haldar, S. The relationship between BcI2, Bax and p53: Consequences for cell cycle progression and cell death. Mol. Hum. Reprod. 1998, 4, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Ren, H.; Fei, E.; Jia, N.; Ying, Z.; Jiang, P.; Wu, M.; Wang, G. Sumoylation is critical for DJ-1 to repress p53 transcriptional activity. FEBS Lett. 2008, 582, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Kato, I.; Maita, H.; Takahashi-Niki, K.; Saito, Y.; Noguchi, N.; Iguchi-Ariga, S.M.; Ariga, H. Oxidized DJ-1 inhibits p53 by sequestering p53 from promoters in a DNA-binding affinity-dependent manner. Mol. Cell Biol. 2013, 33, 340–359. [Google Scholar] [CrossRef]
- Takahashi-Niki, K.; Ganaha, Y.; Niki, T.; Nakagawa, S.; Kato-Ose, I.; Iguchi-Ariga, S.M.M.; Ariga, H. DJ-1 activates SIRT1 through its direct binding to SIRT1. Biochem. Biophys. Res. Commun. 2016, 474, 131–136. [Google Scholar] [CrossRef]
- Duplan, E.; Giordano, C.; Checler, F.; Alves da Costa, C. Direct alpha-synuclein promoter transactivation by the tumor suppressor p53. Mol. Neurodegener. 2016, 11, 13. [Google Scholar] [CrossRef]
- Choi, M.S.; Nakamura, T.; Cho, S.J.; Han, X.; Holland, E.A.; Qu, J.; Petsko, G.A.; Yates, J.R., 3rd; Liddington, R.C.; Lipton, S.A. Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in parkinson’s disease models. J. Neurosci. 2014, 34, 15123–15131. [Google Scholar] [CrossRef]
- Yang, Y.; Gehrke, S.; Haque, M.E.; Imai, Y.; Kosek, J.; Yang, L.; Beal, M.F.; Nishimura, I.; Wakamatsu, K.; Ito, S.; et al. Inactivation of Drosophila DJ-1 leads to impairments of oxidative stress response and phosphatidylinositol 3-kinase/Akt signaling. Proc. Natl. Acad. Sci. USA 2005, 102, 13670–13675. [Google Scholar] [CrossRef]
- Yadav, S.K.; Pandey, A.; Sarkar, S.; Yadav, S.S.; Parmar, D.; Yadav, S. Identification of Altered Blood MicroRNAs and Plasma Proteins in a Rat Model of Parkinson’s Disease. Mol. Neurobiol. 2022, 59, 1781–1798. [Google Scholar] [CrossRef]
- Stein, C.S.; McLendon, J.M.; Witmer, N.H.; Boudreau, R.L. Modulation of miR-181 influences dopaminergic neuronal degeneration in a mouse model of Parkinson’s disease. Mol. Ther. Nucleic Acids 2022, 28, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.E.; Park, H.J.; He, L.; Skibiel, C.; Junn, E.; Mouradian, M.M. The Parkinson’s disease gene product DJ-1 modulates miR-221 to promote neuronal survival against oxidative stress. Redox Biol. 2018, 19, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Stefanis, L. alpha-Synuclein in Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012, 2, a009399. [Google Scholar] [CrossRef] [PubMed]
- Bendor, J.T.; Logan, T.P.; Edwards, R.H. The function of alpha-synuclein. Neuron 2013, 79, 1044–1066. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Cuervo, A.M. Selective autophagy: Talking with the UPS. Cell Biochem. Biophys. 2013, 67, 3–13. [Google Scholar] [CrossRef]
- Bisi, N.; Feni, L.; Peqini, K.; Perez-Pena, H.; Ongeri, S.; Pieraccini, S.; Pellegrino, S. alpha-Synuclein: An All-Inclusive Trip Around its Structure, Influencing Factors and Applied Techniques. Front. Chem. 2021, 9, 666585. [Google Scholar] [CrossRef]
- Fauvet, B.; Mbefo, M.K.; Fares, M.B.; Desobry, C.; Michael, S.; Ardah, M.T.; Tsika, E.; Coune, P.; Prudent, M.; Lion, N.; et al. alpha-Synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. J. Biol. Chem. 2012, 287, 15345–15364. [Google Scholar] [CrossRef]
- Alfaro, I.E.; Albornoz, A.; Molina, A.; Moreno, J.; Cordero, K.; Criollo, A.; Budini, M. Chaperone Mediated Autophagy in the Crosstalk of Neurodegenerative Diseases and Metabolic Disorders. Front. Endocrinol. 2018, 9, 778. [Google Scholar] [CrossRef]
- Xu, C.Y.; Kang, W.Y.; Chen, Y.M.; Jiang, T.F.; Zhang, J.; Zhang, L.N.; Ding, J.Q.; Liu, J.; Chen, S.D. DJ-1 Inhibits alpha-Synuclein Aggregation by Regulating Chaperone-Mediated Autophagy. Front. Aging Neurosci. 2017, 9, 308. [Google Scholar] [CrossRef]
- Patel, B.; Cuervo, A.M. Methods to study chaperone-mediated autophagy. Methods 2015, 75, 133–140. [Google Scholar] [CrossRef]
- Schneider, J.L.; Cuervo, A.M. Chaperone-mediated autophagy: Dedicated saviour and unfortunate victim in the neurodegeneration arena. Biochem. Soc. Trans. 2013, 41, 1483–1488. [Google Scholar] [CrossRef]
- Richarme, G.; Dairou, J. Parkinsonism-associated protein DJ-1 is a bona fide deglycase. Biochem. Biophys. Res. Commun. 2017, 483, 387–391. [Google Scholar] [CrossRef]
- Allaman, I.; Belanger, M.; Magistretti, P.J. Methylglyoxal, the dark side of glycolysis. Front. Neurosci. 2015, 9, 23. [Google Scholar] [CrossRef]
- Desai, K.M.; Chang, T.; Wang, H.; Banigesh, A.; Dhar, A.; Liu, J.; Untereiner, A.; Wu, L. Oxidative stress and aging: Is methylglyoxal the hidden enemy? Can. J. Physiol. Pharmacol. 2010, 88, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Chegao, A.; Miranda, H.V. Unveiling new secrets in Parkinson’s disease: The glycatome. Behav. Brain Res. 2023, 442, 114309. [Google Scholar] [CrossRef] [PubMed]
- Semenyuk, P.; Barinova, K.; Muronetz, V. Glycation of alpha-synuclein amplifies the binding with glyceraldehyde-3-phosphate dehydrogenase. Int. J. Biol. Macromol. 2019, 127, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.; Vogt Weisenhorn, D.M.; Wurst, W. Chapter 5—“Parkinson’s disease—A role of non-enzymatic posttranslational modifications in disease onset and progression?”. Mol. Aspects Med. 2022, 86, 101096. [Google Scholar] [CrossRef]
- Atieh, T.B.; Roth, J.; Yang, X.; Hoop, C.L.; Baum, J. DJ-1 Acts as a Scavenger of alpha-Synuclein Oligomers and Restores Monomeric Glycated alpha-Synuclein. Biomolecules 2021, 11, 1466. [Google Scholar] [CrossRef] [PubMed]
- Jun, Y.W.; Kool, E.T. Small Substrate or Large? Debate Over the Mechanism of Glycation Adduct Repair by DJ-1. Cell Chem. Biol. 2020, 27, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Daubner, S.C.; Le, T.; Wang, S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 2011, 508, 1–12. [Google Scholar] [PubMed]
- Takahashi-Niki, K.; Niki, T.; Iguchi-Ariga, S.M.M.; Ariga, H. Transcriptional Regulation of DJ-1. Adv. Exp. Med. Biol. 2017, 1037, 89–95. [Google Scholar] [PubMed]
- Ishikawa, S.; Taira, T.; Niki, T.; Takahashi-Niki, K.; Maita, C.; Maita, H.; Ariga, H.; Iguchi-Ariga, S.M. Oxidative status of DJ-1-dependent activation of dopamine synthesis through interaction of tyrosine hydroxylase and 4-dihydroxy-L-phenylalanine (L-DOPA) decarboxylase with DJ-1. J. Biol. Chem. 2009, 284, 28832–28844. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Taira, T.; Takahashi-Niki, K.; Niki, T.; Ariga, H.; Iguchi-Ariga, S.M. Human DJ-1-specific transcriptional activation of tyrosine hydroxylase gene. J. Biol. Chem. 2010, 285, 39718–39731. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Tanaka, Y.; Takahashi-Niki, K.; Niki, T.; Ariga, H.; Iguchi-Ariga, S.M. Stimulation of vesicular monoamine transporter 2 activity by DJ-1 in SH-SY5Y cells. Biochem. Biophys. Res. Commun. 2012, 421, 813–818. [Google Scholar] [CrossRef]
- Choi, J.; Sullards, M.C.; Olzmann, J.A.; Rees, H.D.; Weintraub, S.T.; Bostwick, D.E.; Gearing, M.; Levey, A.I.; Chin, L.-S.; Li, L. Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J. Biol. Chem. 2006, 281, 10816–10824. [Google Scholar] [CrossRef]
- Saito, Y.; Hamakubo, T.; Yoshida, Y.; Ogawa, Y.; Hara, Y.; Fujimura, H.; Imai, Y.; Iwanari, H.; Mochizuki, Y.; Shichiri, M.; et al. Preparation and application of monoclonal antibodies against oxidized DJ-1. Significant elevation of oxidized DJ-1 in erythrocytes of early-stage Parkinson disease patients. Neurosci. Lett. 2009, 465, 1–5. [Google Scholar] [CrossRef]
- Jang, J.; Jeong, S.; Lee, S.I.; Seol, W.; Seo, H.; Son, I.; Ho, D.H. Oxidized DJ-1 Levels in Urine Samples as a Putative Biomarker for Parkinson’s Disease. Park. Dis. 2018, 2018, 1241757. [Google Scholar] [CrossRef]
- Drechsel, J.; Mandl, F.A.; Sieber, S.A. Chemical Probe To Monitor the Parkinsonism-Associated Protein DJ-1 in Live Cells. ACS Chem. Biol. 2018, 13, 2016–2019. [Google Scholar] [CrossRef]
- Gao, H.; Yang, W.; Qi, Z.; Lu, L.; Duan, C.; Zhao, C.; Yang, H. DJ-1 protects dopaminergic neurons against rotenone-induced apoptosis by enhancing ERK-dependent mitophagy. J. Mol. Biol. 2012, 423, 232–248. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.Y.; An, C.N.; Pu, X.P. DJ-1 protein protects dopaminergic neurons against 6-OHDA/MG-132-induced neurotoxicity in rats. Brain Res. Bull. 2012, 88, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Schwarze, S.R.; Ho, A.; Vocero-Akbani, A.; Dowdy, S.F. In vivo protein transduction: Delivery of a biologically active protein into the mouse. Science 1999, 285, 1569–1572. [Google Scholar] [CrossRef]
- Batelli, S.; Invernizzi, R.W.; Negro, A.; Calcagno, E.; Rodilossi, S.; Forloni, G.; Albani, D. The Parkinson’s disease-related protein DJ-1 protects dopaminergic neurons in vivo and cultured cells from alpha-synuclein and 6-hydroxydopamine toxicity. Neurodegener. Dis. 2015, 15, 13–23. [Google Scholar] [CrossRef]
- Miyazaki, S.; Yanagida, T.; Nunome, K.; Ishikawa, S.; Inden, M.; Kitamura, Y.; Nakagawa, S.; Taira, T.; Hirota, K.; Niwa, M.; et al. DJ-1-binding compounds prevent oxidative stress-induced cell death and movement defect in Parkinson’s disease model rats. J. Neurochem. 2008, 105, 2418–2434. [Google Scholar] [CrossRef]
- Takahashi-Niki, K.; Inafune, A.; Michitani, N.; Hatakeyama, Y.; Suzuki, K.; Sasaki, M.; Kitamura, Y.; Niki, T.; Iguchi-Ariga, S.M.; Ariga, H. DJ-1-dependent protective activity of DJ-1-binding compound no. 23 against neuronal cell death in MPTP-treated mouse model of Parkinson’s disease. J. Pharmacol. Sci. 2015, 127, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Andres-Mateos, E.; Perier, C.; Zhang, L.; Blanchard-Fillion, B.; Greco, T.M.; Thomas, B.; Ko, H.S.; Sasaki, M.; Ischiropoulos, H.; Przedborski, S.; et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc. Natl. Acad. Sci. USA 2007, 104, 14807–14812. [Google Scholar] [CrossRef]
- Junn, E.; Taniguchi, H.; Jeong, B.S.; Zhao, X.; Ichijo, H.; Mouradian, M.M. Interaction of DJ-1 with Daxx inhibits apoptosis signal-regulating kinase 1 activity and cell death. Proc. Natl. Acad. Sci. USA 2005, 102, 9691–9696. [Google Scholar] [CrossRef]
- Konovalova, J.; Gerasymchuk, D.; Parkkinen, I.; Chmielarz, P.; Domanskyi, A. Interplay between MicroRNAs and Oxidative Stress in Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 6055. [Google Scholar] [CrossRef]
- Jeffery, C.J. Moonlighting proteins: Old proteins learning new tricks. Trends Genet. 2003, 19, 415–417. [Google Scholar] [CrossRef]
- Aslam, K.; Hazbun, T.R. Hsp31, a member of the DJ-1 superfamily, is a multitasking stress responder with chaperone activity. Prion 2016, 10, 103–111. [Google Scholar] [CrossRef]
- Hasim, S.; Hussin, N.A.; Alomar, F.; Bidasee, K.R.; Nickerson, K.W.; Wilson, M.A. A glutathione-independent glyoxalase of the DJ-1 superfamily plays an important role in managing metabolically generated methylglyoxal in Candida albicans. J. Biol. Chem. 2014, 289, 1662–1674. [Google Scholar] [CrossRef] [PubMed]
- Panicker, N.; Ge, P.; Dawson, V.L.; Dawson, T.M. The cell biology of Parkinson’s disease. J. Cell Biol. 2021, 220, e202012095. [Google Scholar] [CrossRef] [PubMed]
- Mencke, P.; Boussaad, I.; Romano, C.D.; Kitami, T.; Linster, C.L.; Kruger, R. The Role of DJ-1 in Cellular Metabolism and Pathophysiological Implications for Parkinson’s Disease. Cells 2021, 10, 347. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Rao, S.P.; Kalivendi, S.V. The deglycase activity of DJ-1 mitigates alpha-synuclein glycation and aggregation in dopaminergic cells: Role of oxidative stress mediated downregulation of DJ-1 in Parkinson’s disease. Free Radic. Biol. Med. 2019, 135, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Mita, Y.; Kataoka, Y.; Saito, Y.; Kashi, T.; Hayashi, K.; Iwasaki, A.; Imanishi, T.; Miyasaka, T.; Noguchi, N. Distribution of oxidized DJ-1 in Parkinson’s disease-related sites in the brain and in the peripheral tissues: Effects of aging and a neurotoxin. Sci. Rep. 2018, 8, 12056. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skou, L.D.; Johansen, S.K.; Okarmus, J.; Meyer, M. Pathogenesis of DJ-1/PARK7-Mediated Parkinson’s Disease. Cells 2024, 13, 296. https://doi.org/10.3390/cells13040296
Skou LD, Johansen SK, Okarmus J, Meyer M. Pathogenesis of DJ-1/PARK7-Mediated Parkinson’s Disease. Cells. 2024; 13(4):296. https://doi.org/10.3390/cells13040296
Chicago/Turabian StyleSkou, Line Duborg, Steffi Krudt Johansen, Justyna Okarmus, and Morten Meyer. 2024. "Pathogenesis of DJ-1/PARK7-Mediated Parkinson’s Disease" Cells 13, no. 4: 296. https://doi.org/10.3390/cells13040296




