Gene Signature of Regulatory T Cells Isolated from Children with Selective IgA Deficiency and Common Variable Immunodeficiency
Abstract
:1. Introduction
2. Materials and Methods
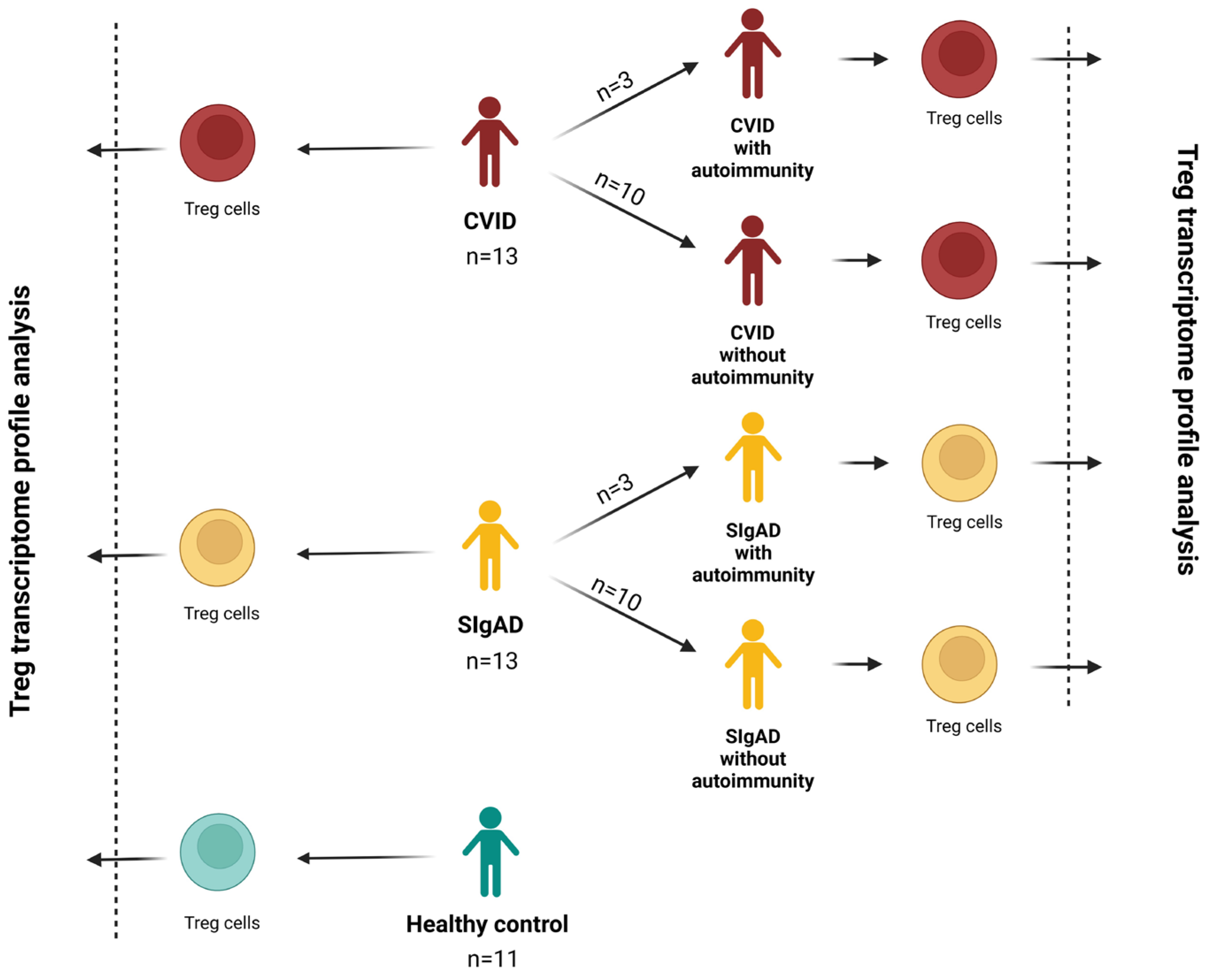
2.1. Patients
2.2. Regulatory T Cells Number Evaluation
2.3. Isolation of Regulatory T Cells
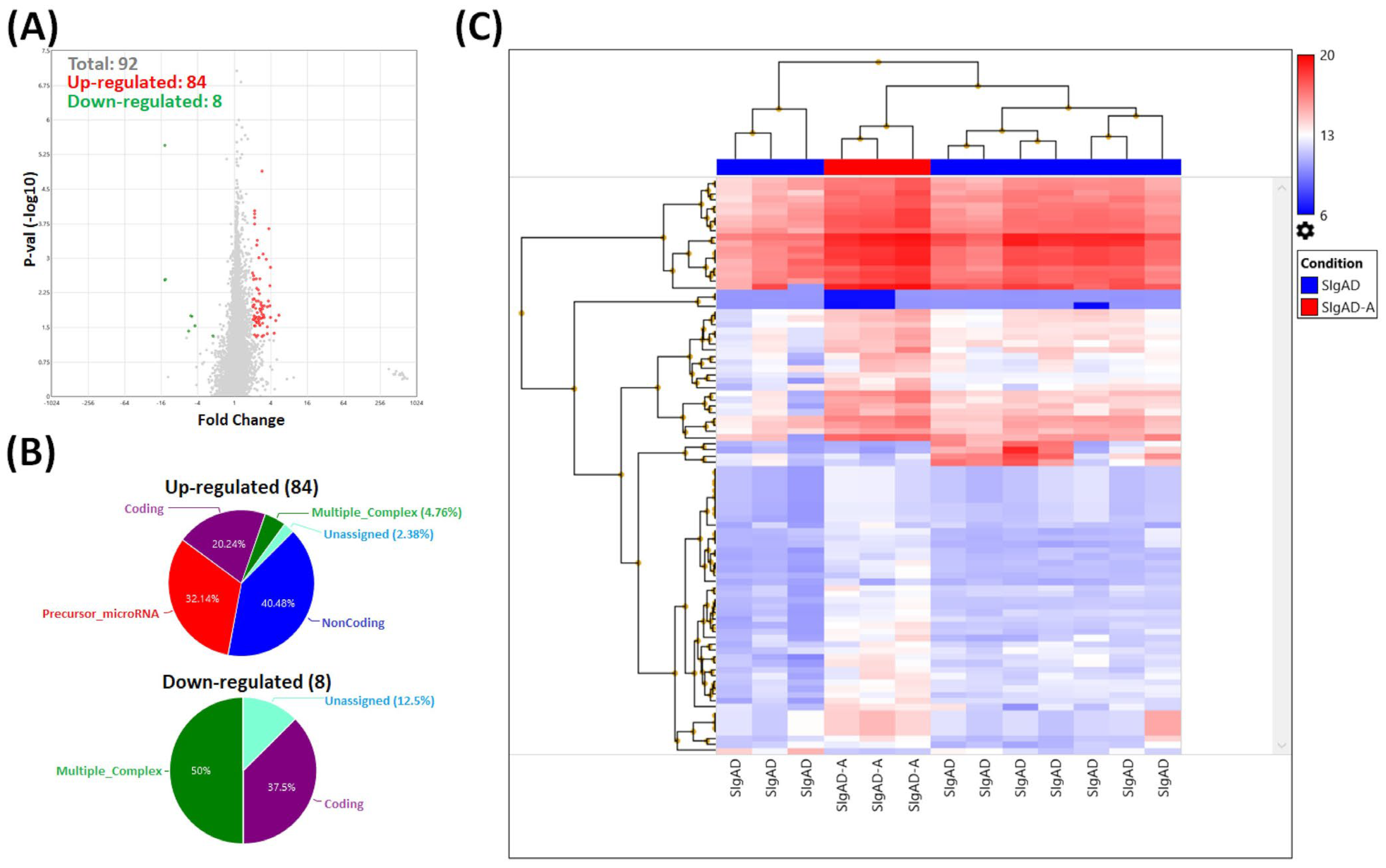
2.4. Gene Expression Analysis
2.5. Quantitative Real-Time PCR (RT-qPCR) Analysis
2.6. Statistical Analysis
3. Results
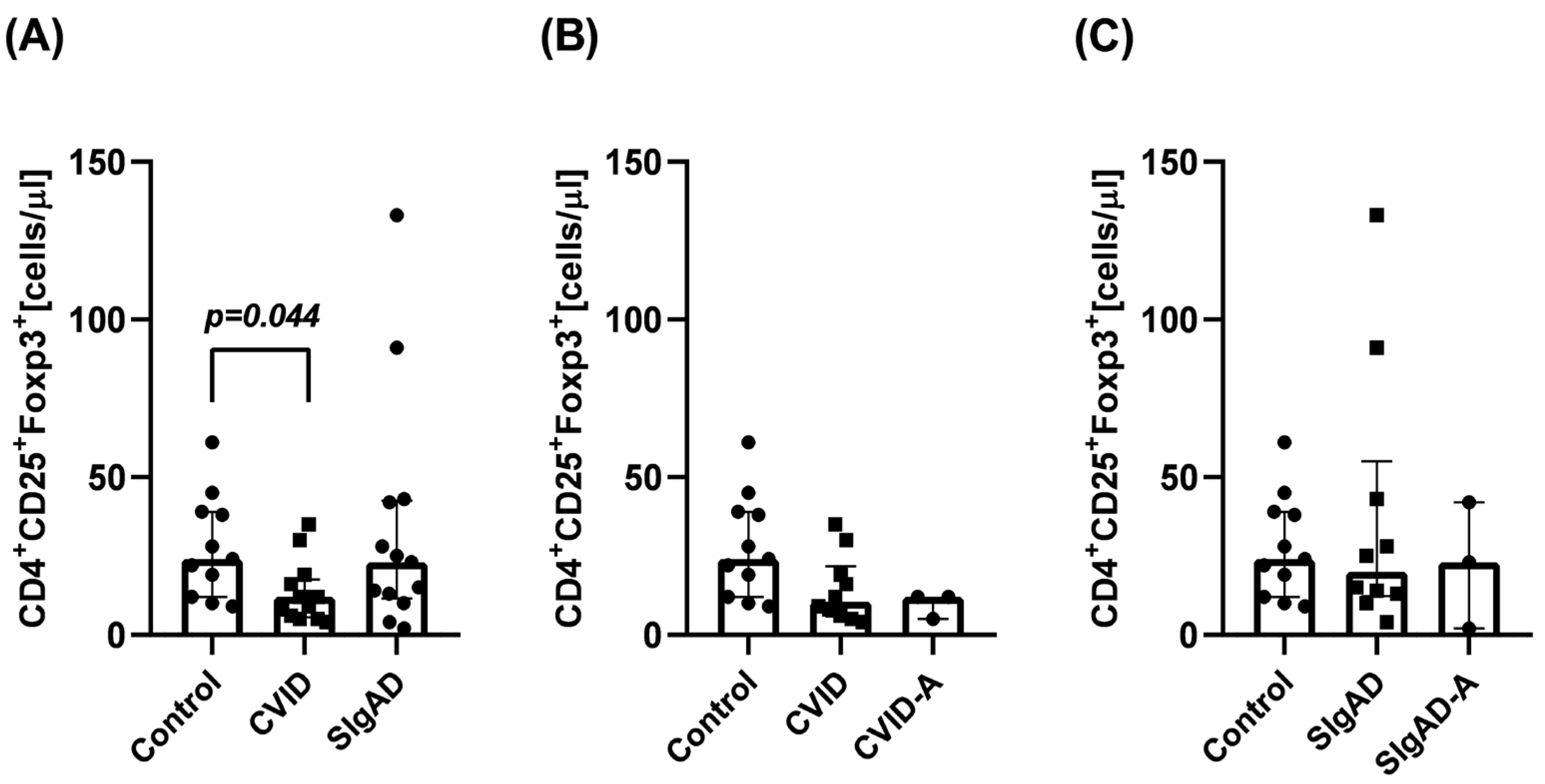
3.1. Regulatory T Cells Number
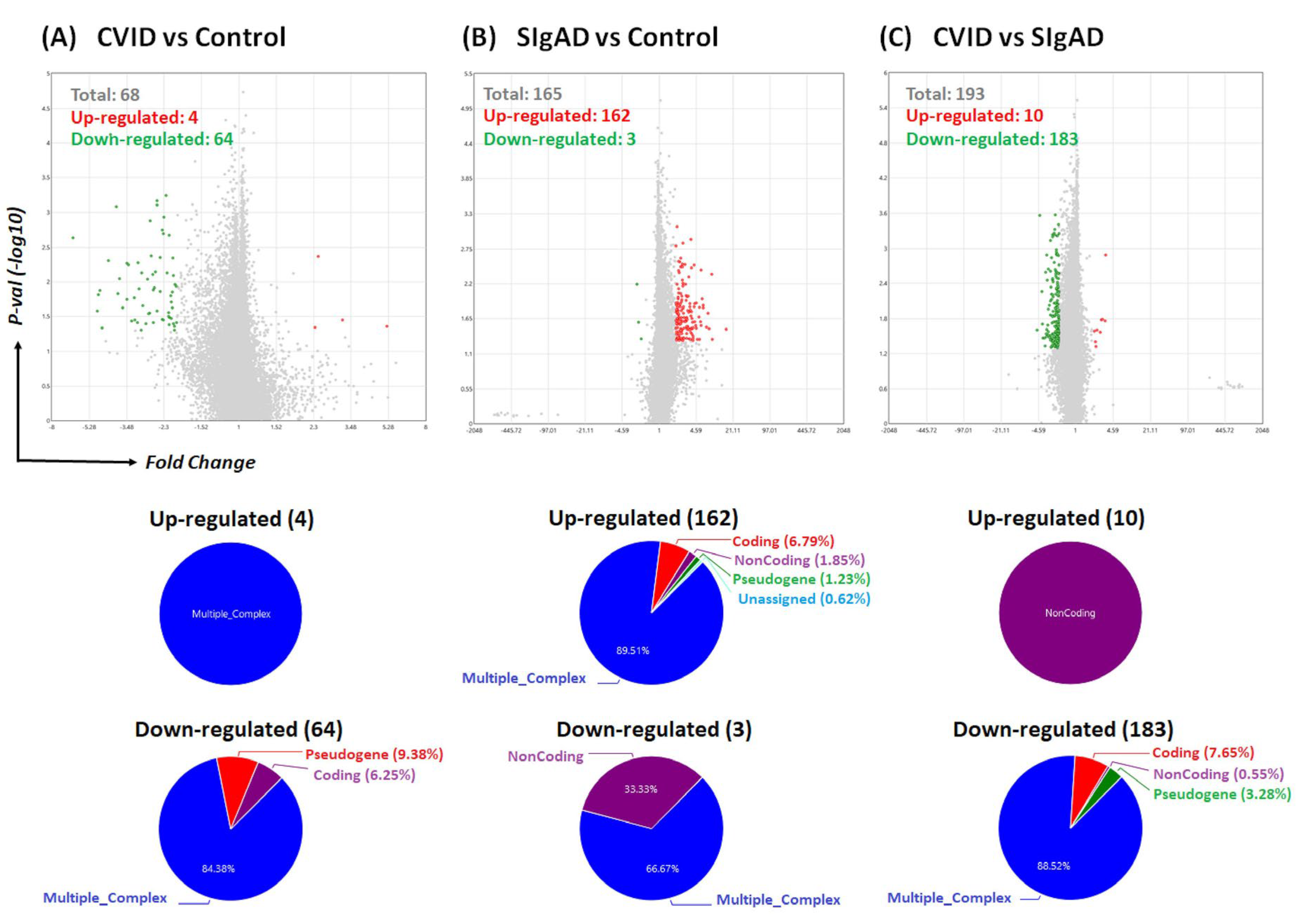
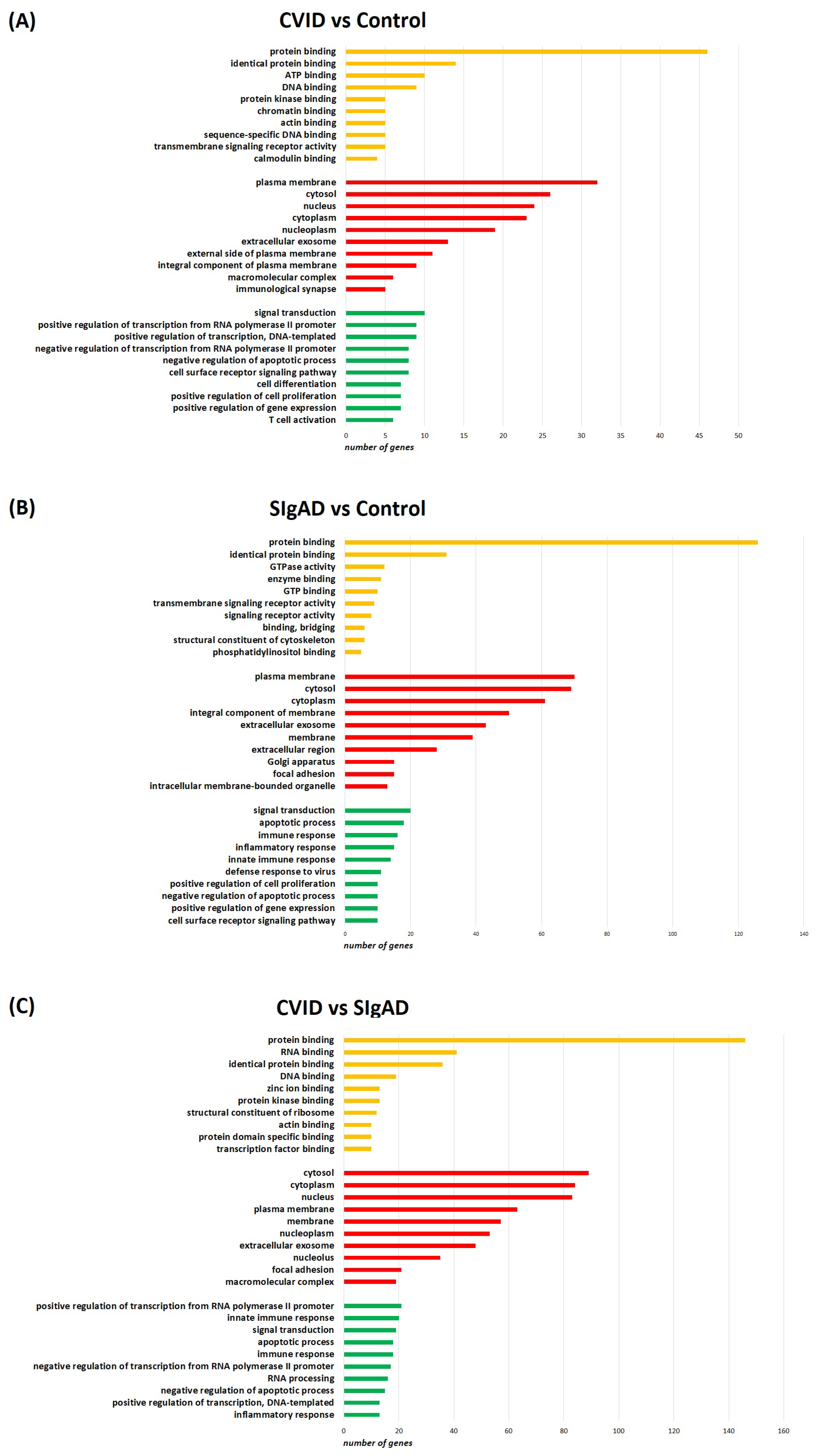
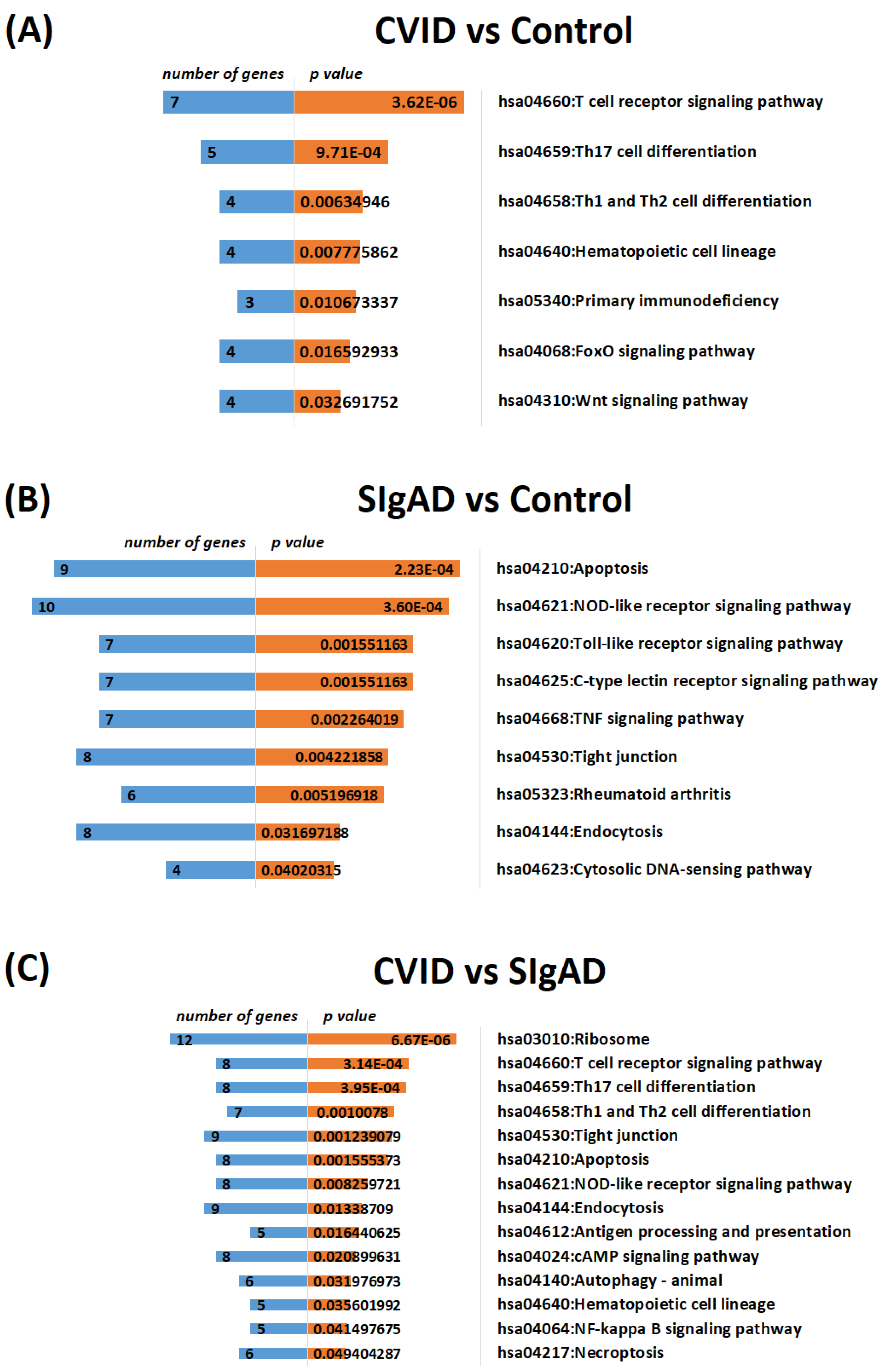
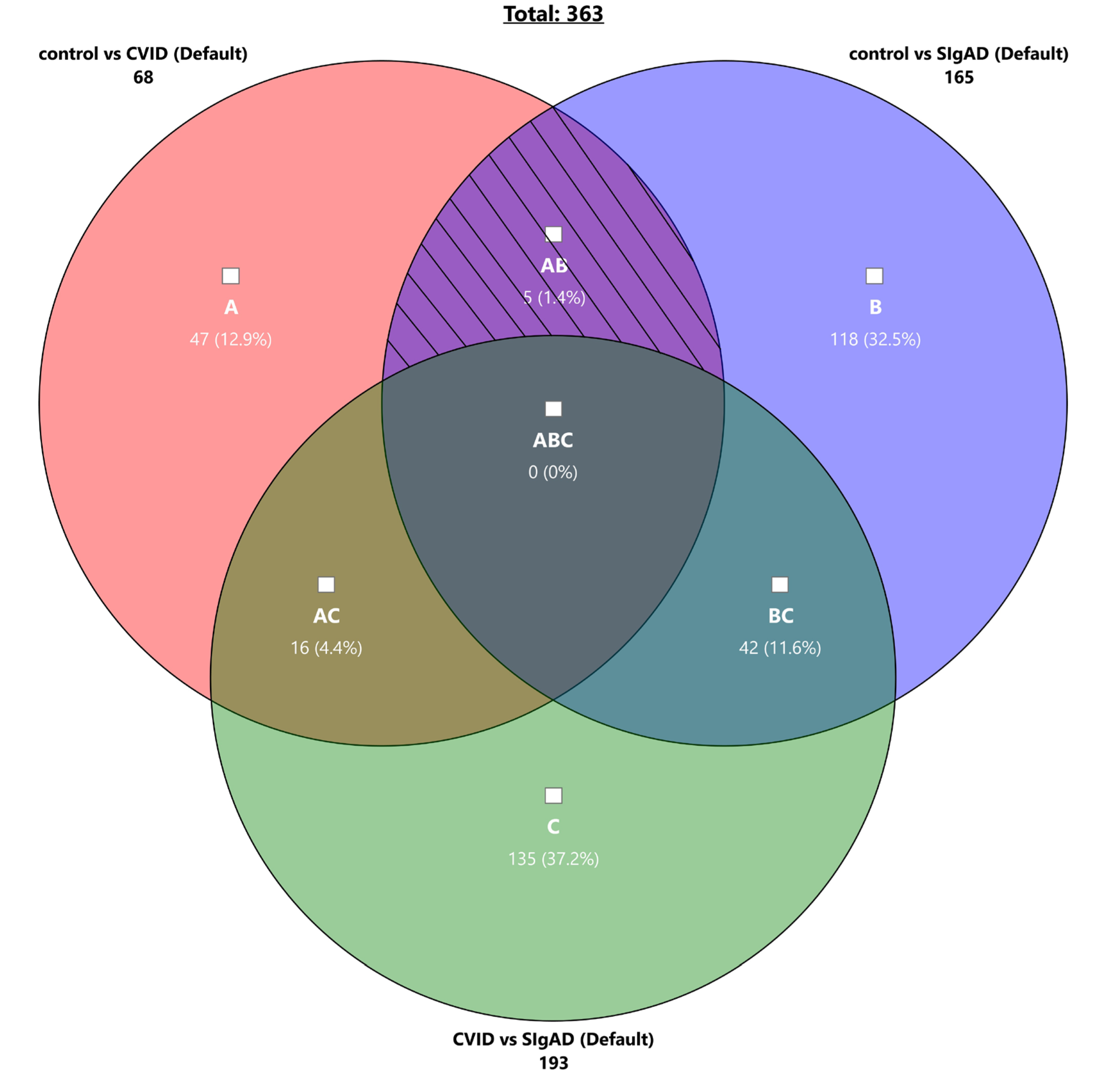
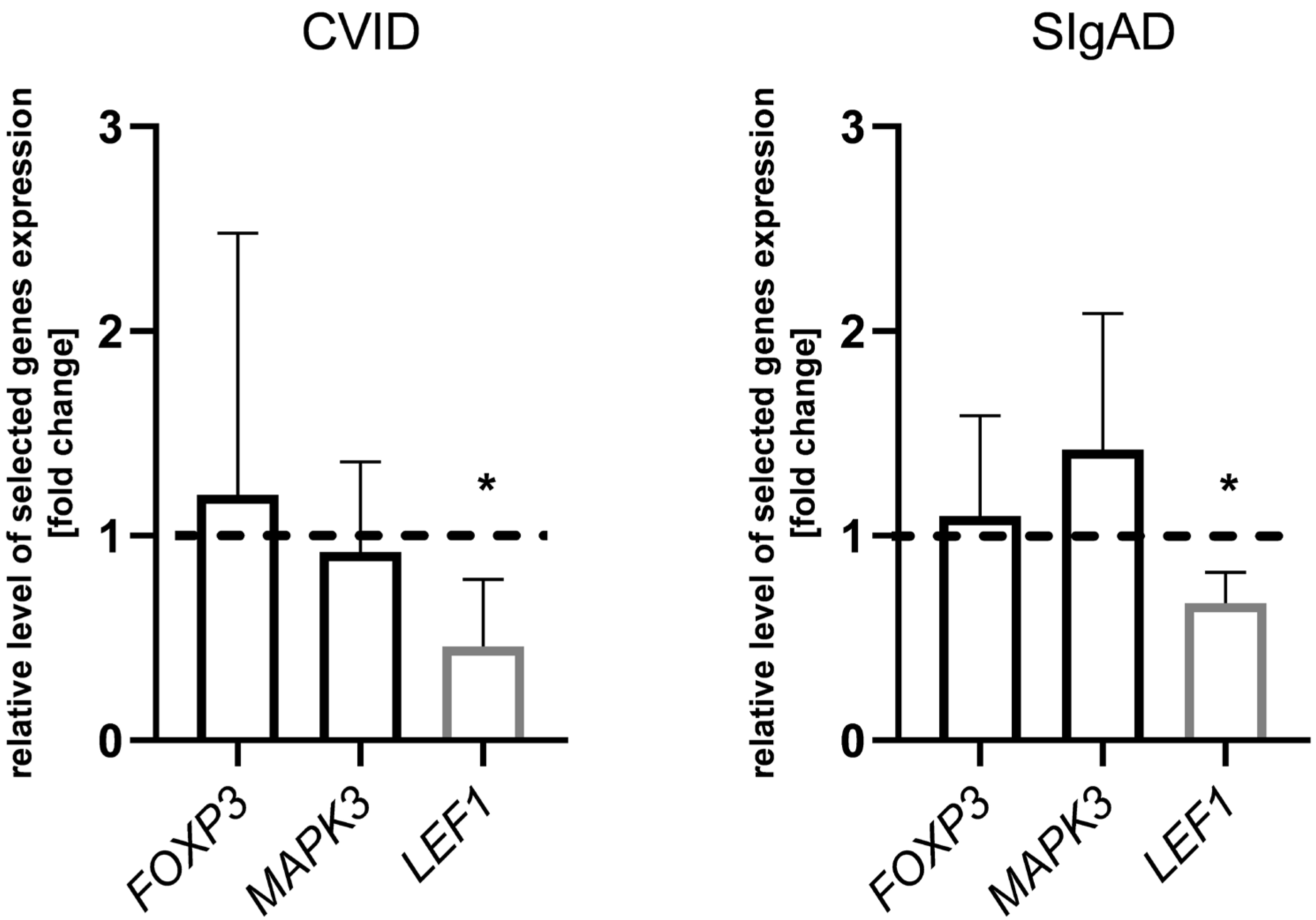
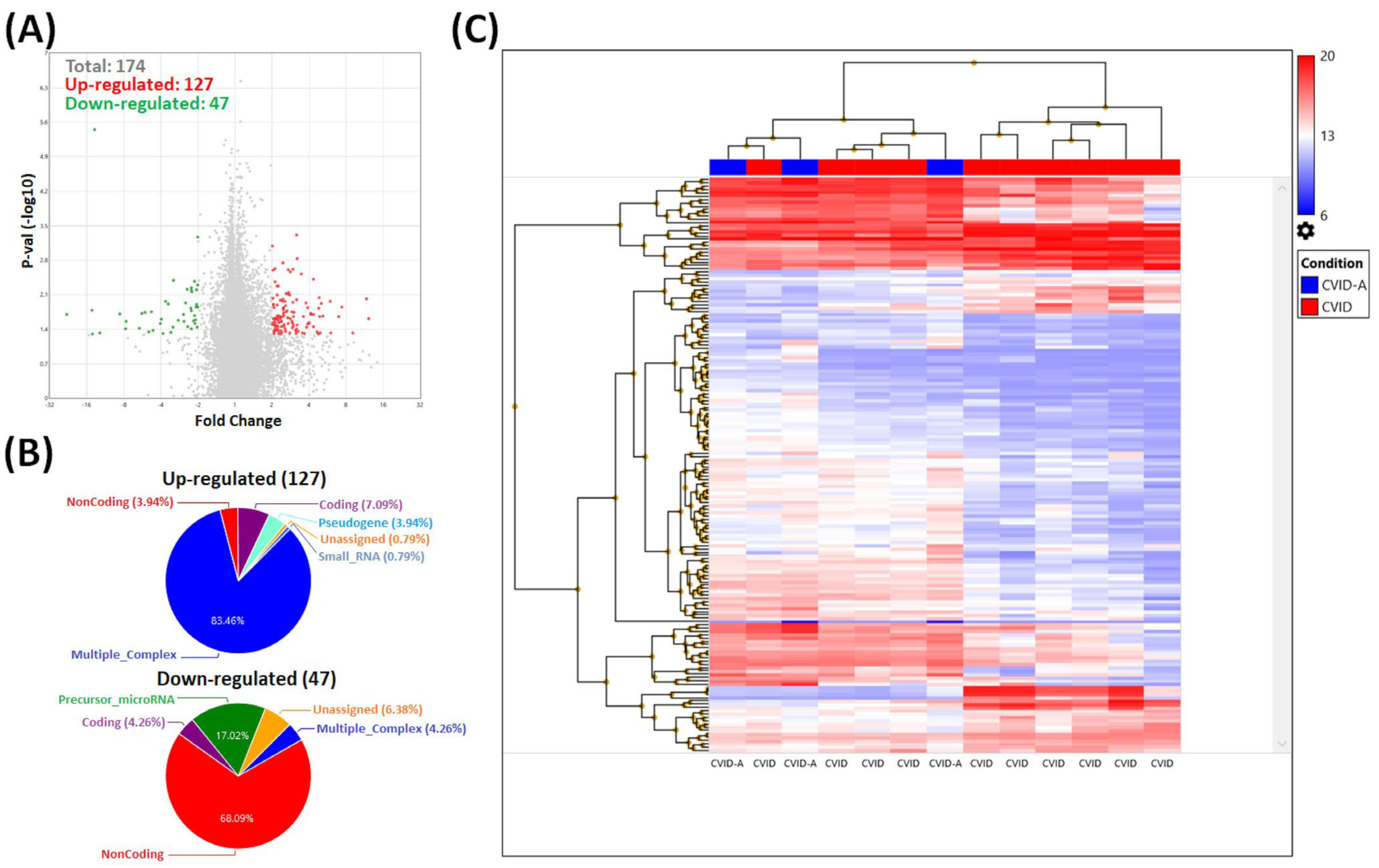
3.2. Gene Expression Analysis in Treg Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hammarström, L.; Vorechovsky, I.; Webster, D. Selective IgA deficiency (sIgAD) and common variable immunodeficiency (CVID). Clin. Exp. Immunol. 2000, 120, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Abolhassani, H.; Gharib, B.; Shahinpour, S.; Masoom, S.N.; Havaei, A.; Mirminachi, B.; Arandi, N.; Torabi-Sagvand, B.; Khazaei, H.A.; Mohammadi, J.; et al. Autoimmunity in patients with selective IgA deficiency. J. Investig. Allergol. Clin. Immunol. 2015, 25, 112–119. [Google Scholar] [PubMed]
- International Union of Immunological Societies Expert Committee on Primary Immunodeficiencies; Notarangelo, L.D.; Fischer, A.; Geha, R.S.; Casanova, J.L.; Chapel, H.; Conley, M.E.; Cunningham-Rundles, C.; Etzioni, A.; Hammartröm, L.; et al. Primary immunodeficiencies: 2009 update. J. Allergy Clin. Immunol. 2009, 124, 1161–1178, Erratum in J. Allergy Clin. Immunol. 2010, 125, 771–773. [Google Scholar] [CrossRef] [PubMed]
- Yel, L. Selective IgA deficiency. J. Clin. Immunol. 2010, 30, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Latiff, A.H.; Kerr, M.A. The clinical significance of immunoglobulin A deficiency. Ann. Clin. Biochem. 2007, 44 Pt 2, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Yazdani, R.; Azizi, G.; Abolhassani, H.; Aghamohammadi, A. Selective IgA Deficiency: Epidemiology, Pathogenesis, Clinical Phenotype, Diagnosis, Prognosis and Management. Scand. J. Immunol. 2017, 85, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, G.H.; Gardulf, A.; Sigurdsson, M.I.; Sigurdardottir, S.T.; Thorsteinsdottir, I.; Gudmundsson, S.; Hammarström, L.; Ludviksson, B.R. Clinical symptoms in adults with selective IgA deficiency: A case-control study. J. Clin. Immunol. 2013, 33, 742–747. [Google Scholar] [CrossRef]
- Resnick, E.S.; Moshier, E.L.; Godbold, J.H.; Cunningham-Rundles, C. Morbidity and mortality in common variable immune deficiency over 4 decades. Blood 2012, 119, 1650–1657. [Google Scholar] [CrossRef]
- Seidel, M.G.; Kindle, G.; Gathmann, B.; Quinti, I.; Buckland, M.; van Montfrans, J.; Scheible, R.; Rusch, S.; Gasteiger, L.M.; Grimbacher, B.; et al. The European Society for Immunodeficiencies (ESID) Registry Working Definitions for the Clinical Diagnosis of Inborn Errors of Immunity. J. Allergy Clin. Immunol. Pract. 2019, 7, 1763–1770. [Google Scholar] [CrossRef]
- Cunningham-Rundles, C.; Warnatz, K. Hypogammaglobulinemia and common variable immune deficiency. In Stiehm’s Immune Deficiencies Inborn Errors of Immunity; Academic Press: Cambridge, MA, USA, 2020; ISBN 978-0-12-816768-7. [Google Scholar]
- Abolhassani, H.; Hammarström, L.; Cunningham-Rundles, C. Current genetic landscape in common variable immune deficiency. Blood 2020, 135, 656–667. [Google Scholar] [CrossRef]
- Agarwal, S.; Cunningham-Rundles, C. Autoimmunity in common variable immunodeficiency. Ann. Allergy Asthma Immunol. 2019, 123, 454–460. [Google Scholar] [CrossRef]
- Ni Choileain, N.; Redmond, H.P. Regulatory T-cells and autoimmunity. J. Surg. Res. 2006, 130, 124–135. [Google Scholar] [CrossRef]
- Georgiev, P.; Charbonnier, L.M.; Chatila, T.A. Regulatory T Cells: The Many Faces of Foxp3. J. Clin. Immunol. 2019, 39, 623–640. [Google Scholar] [CrossRef]
- Verbsky, J.W.; Chatila, T.A. T-regulatory cells in primary immune deficiencies. Curr. Opin. Allergy Clin. Immunol. 2011, 11, 539–544. [Google Scholar] [CrossRef]
- Rutkowska-Zapała, M.; Grabowska, A.; Lenart, M.; Kluczewska, A.; Szaflarska, A.; Kobylarz, K.; Pituch-Noworolska, A.; Siedlar, M. Transcriptome profiling of regulatory T cells from children with transient hypogammaglobulinemia of infancy. Clin. Exp. Immunol. 2023, 214, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Fevang, B.; Yndestad, A.; Sandberg, W.J.; Holm, A.M.; Müller, F.; Aukrust, P.; Frøland, S.S. Low numbers of regulatory T cells in common variable immunodeficiency: Association with chronic inflammation in vivo. Clin. Exp. Immunol. 2007, 147, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Azizi, G.; Hafezi, N.; Mohammadi, H.; Yazdani, R.; Alinia, T.; Tavakol, M.; Aghamohammadi, A.; Mirshafiey, A. Abnormality of regulatory T cells in common variable immunodeficiency. Cell Immunol. 2017, 315, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Arandi, N.; Mirshafiey, A.; Jeddi-Tehrani, M.; Abolhassani, H.; Sadeghi, B.; Mirminachi, B.; Shaghaghi, M.; Aghamohammadi, A. Evaluation of CD4+CD25+FOXP3+ regulatory T cells function in patients with common variable immunodeficiency. Cell Immunol. 2013, 281, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Melo, K.M.; Carvalho, K.I.; Bruno, F.R.; Ndhlovu, L.C.; Ballan, W.M.; Nixon, D.F.; Kallas, E.G.; Costa-Carvalho, B.T. A decreased frequency of regulatory T cells in patients with common variable immunodeficiency. PLoS ONE 2009, 4, e6269. [Google Scholar] [CrossRef]
- Horn, J.; Manguiat, A.; Berglund, L.J.; Knerr, V.; Tahami, F.; Grimbacher, B.; Fulcher, D.A. Decrease in phenotypic regulatory T cells in subsets of patients with common variable immunodeficiency. Clin. Exp. Immunol. 2009, 156, 446–454. [Google Scholar] [CrossRef]
- Azizi, G.; Mirshafiey, A.; Abolhassani, H.; Yazdani, R.; Jafarnezhad-Ansariha, F.; Shaghaghi, M.; Mortazavi-Jahromi, S.S.; Noorbakhsh, F.; Rezaei, N.; Aghamohammadi, A. Circulating Helper T-Cell Subsets and Regulatory T Cells in Patients with Common Variable Immunodeficiency without Known Monogenic Disease. J. Investig. Allergol. Clin. Immunol. 2018, 28, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Genre, J.; Errante, P.R.; Kokron, C.M.; Toledo-Barros, M.; Câmara, N.O.; Rizzo, L.V. Reduced frequency of CD4+CD25HIGHFOXP3+ cells and diminished FOXP3 expression in patients with Common Variable Immunodeficiency: A link to autoimmunity? Clin. Immunol. 2009, 132, 215–221. [Google Scholar] [CrossRef]
- Yu, G.P.; Chiang, D.; Song, S.J.; Hoyte, E.G.; Huang, J.; Vanishsarn, C.; Nadeau, K.C. Regulatory T cell dysfunction in subjects with common variable immunodeficiency complicated by autoimmune disease. Clin. Immunol. 2009, 131, 240–253. [Google Scholar] [CrossRef]
- Więsik-Szewczyk, E.; Rutkowska, E.; Kwiecień, I.; Korzeniowska, M.; Sołdacki, D.; Jahnz-Różyk, K. Patients with Common Variable Immunodeficiency Complicated by Autoimmune Phenomena Have Lymphopenia and Reduced Treg, Th17, and NK Cells. J. Clin. Med. 2021, 10, 3356. [Google Scholar] [CrossRef] [PubMed]
- Tofighi Zavareh, F.; Mirshafiey, A.; Yazdani, R.; Keshtkar, A.A.; Abolhassani, H.; Bagheri, Y.; Rezaei, A.; Delavari, S.; Rezaei, N.; Aghamohammadi, A. Lymphocytes subsets in correlation with clinical profile in CVID patients without monogenic defects. Expert. Rev. Clin. Immunol. 2021, 17, 1041–1051. [Google Scholar] [CrossRef]
- Azizi, G.; Abolhassani, H.; Kiaee, F.; Tavakolinia, N.; Rafiemanesh, H.; Yazdani, R.; Mahdaviani, S.A.; Mohammadikhajehdehi, S.; Tavakol, M.; Ziaee, V.; et al. Autoimmunity and its association with regulatory T cells and B cell subsets in patients with common variable immunodeficiency. Allergol. Immunopathol. 2018, 46, 127–135. [Google Scholar] [CrossRef]
- ESID Database. Available online: http://esid.org/Working-Parties/Registry/Diagnosis-criteria (accessed on 1 December 2023).
- The Database for Annotation, Visualization and Integrated Discovery, Version 6.8. Available online: https://david.ncifcrf.gov (accessed on 1 December 2023).
- UniProt Database. Available online: https://www.uniprot.org/ (accessed on 1 December 2023).
- Calonga-Solís, V.; Amorim, L.M.; Farias, T.D.J.; Petzl-Erler, M.L.; Malheiros, D.; Augusto, D.G. Variation in genes implicated in B-cell development and antibody production affects susceptibility to pemphigus. Immunology 2021, 162, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Wang, Y.; Xiao, Y.; Zheng, J.; Liu, R.; He, X.; Yu, J.; Tang, B.; Qiu, X.; Tang, R.; et al. Overexpression of OASL upregulates TET1 to induce aberrant activation of CD4+ T cells in systemic sclerosis via IRF1 signaling. Arthritis Res. Ther. 2022, 24, 50. [Google Scholar] [CrossRef]
- Erlandsson, M.C.; Andersson, K.M.E.; Oparina, N.Y.; Chandrasekaran, V.; Saghy, T.; Damdimopoulos, A.; Garcia-Bonete, M.J.; Einbeigi, Z.; Silfverswärd, S.T.; Pekna, M.; et al. Survivin promotes a glycolytic switch in CD4+ T cells by suppressing the transcription of PFKFB3 in rheumatoid arthritis. iScience 2022, 25, 105526. [Google Scholar] [CrossRef]
- Werner, G.; Sanyal, A.; Mirizio, E.; Hutchins, T.; Tabib, T.; Lafyatis, R.; Jacobe, H.; Torok, K.S. Single-Cell Transcriptome Analysis Identifies Subclusters with Inflammatory Fibroblast Responses in Localized Scleroderma. Int. J. Mol. Sci. 2023, 24, 9796. [Google Scholar] [CrossRef]
- Gallucci, S.; Meka, S.; Gamero, A.M. Abnormalities of the type I interferon signaling pathway in lupus autoimmunity. Cytokine 2021, 146, 155633. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Zhang, W.; Liu, D.; Zeng, Z.; Liao, S.; Cai, W.; Liu, J.; Li, L.; Hong, X.; Tang, D.; et al. Screening biomarkers for Sjogren’s Syndrome by computer analysis and evaluating the expression correlations with the levels of immune cells. Front. Immunol. 2023, 14, 1023248. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Guo, Y.; Chang, L.; Zhang, J. Long noncoding RNA H19 synergizes with STAT1 to regulate SNX10 in rheumatoid arthritis. Mol. Immunol. 2023, 153, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, T.; Li, B.; Huang, T.; Hai, Y.; Huang, C.; Xiang, W. Bioinformatic analysis of immune-related transcriptome affected by IFIT1 gene in childhood systemic lupus erythematosus. Transl. Pediatr. 2023, 12, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, H.; Wang, Z.; Gong, B.; Al-Ward, H.; Deng, Y.; Fan, O.; Wang, J.; Zhu, W.; Sun, Y.E. Exploring the shared molecular mechanisms between systemic lupus erythematosus and primary Sjögren’s syndrome based on integrated bioinformatics and single-cell RNA-seq analysis. Front. Immunol. 2023, 14, 1212330. [Google Scholar] [CrossRef] [PubMed]
- Karathanasis, D.K.; Rapti, A.; Nezos, A.; Skarlis, C.; Kilidireas, C.; Mavragani, C.P.; Evangelopoulos, M.E. Differentiating central nervous system demyelinating disorders: The role of clinical, laboratory, imaging characteristics and peripheral blood type I interferon activity. Front. Pharmacol. 2022, 13, 898049. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, X.; Zheng, Y. Screening for key genes and transcription factors in ankylosing spondylitis by RNA-Seq. Exp. Ther. Med. 2018, 15, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Batten, I.; Robinson, M.W.; White, A.; Walsh, C.; Fazekas, B.; Wyse, J.; Buettner, A.; D’Arcy, S.; Greenan, E.; Murphy, C.C.; et al. Investigation of type I interferon responses in ANCA-associated vasculitis. Sci. Rep. 2021, 11, 8272. [Google Scholar] [CrossRef]
- Kim, H.; Gunter-Rahman, F.; McGrath, J.A.; Lee, E.; de Jesus, A.A.; Targoff, I.N.; Huang, Y.; O’Hanlon, T.P.; Tsai, W.L.; Gadina, M.; et al. Expression of interferon-regulated genes in juvenile dermatomyositis versus Mendelian autoinflammatory interferonopathies. Arthritis Res. Ther. 2020, 22, 69. [Google Scholar] [CrossRef]
- Vlachogiannis, N.I.; Pappa, M.; Ntouros, P.A.; Nezos, A.; Mavragani, C.P.; Souliotis, V.L.; Sfikakis, P.P. Association Between DNA Damage Response, Fibrosis and Type I Interferon Signature in Systemic Sclerosis. Front. Immunol. 2020, 11, 582401. [Google Scholar] [CrossRef]
- Thessen Hedreul, M.; Möller, S.; Stridh, P.; Gupta, Y.; Gillett, A.; Daniel Beyeen, A.; Öckinger, J.; Flytzani, S.; Diez, M.; Olsson, T.; et al. Combining genetic mapping with genome-wide expression in experimental autoimmune encephalomyelitis highlights a gene network enriched for T cell functions and candidate genes regulating autoimmunity. Hum. Mol. Genet. 2013, 22, 4952–4966. [Google Scholar] [CrossRef]
- Quah, H.S.; Miranda-Hernandez, S.; Khoo, A.; Harding, A.; Fynch, S.; Elkerbout, L.; Brodnicki, T.C.; Baxter, A.G.; Kay, T.W.; Thomas, H.E.; et al. Deficiency in type I interferon signaling prevents the early interferon-induced gene signature in pancreatic islets but not type 1 diabetes in NOD mice. Diabetes. 2014, 63, 1032–1040. [Google Scholar] [CrossRef]
- Qian, J.; Li, R.; Chen, Z.; Cao, Z.; Lu, L.; Fu, Q. Type I interferon score is associated with the severity and poor prognosis in anti-MDA5 antibody-positive dermatomyositis patients. Front. Immunol. 2023, 14, 1151695. [Google Scholar] [CrossRef]
- AlFadhli, S.; Al-Mutairi, M.; Al Tameemi, B.; Nizam, R. Influence of MX1 promoter rs2071430 G/T polymorphism on susceptibility to systemic lupus erythematosus. Clin. Rheumatol. 2016, 35, 623–629. [Google Scholar] [CrossRef]
- Maria, N.I.; Brkic, Z.; Waris, M.; van Helden-Meeuwsen, C.G.; Heezen, K.; van de Merwe, J.P.; van Daele, P.L.; Dalm, V.A.; Drexhage, H.A.; Versnel, M.A. MxA as a clinically applicable biomarker for identifying systemic interferon type I in primary Sjogren’s syndrome. Ann. Rheum. Dis. 2014, 73, 1052–1059. [Google Scholar] [CrossRef] [PubMed]
- Nikopensius, T.; Niibo, P.; Haller, T.; Jagomägi, T.; Voog-Oras, Ü.; Tõnisson, N.; Metspalu, A.; Saag, M.; Pruunsild, C. Association analysis of juvenile idiopathic arthritis genetic susceptibility factors in Estonian patients. Clin. Rheumatol. 2021, 40, 4157–4165. [Google Scholar] [CrossRef]
- Pedersen, K.; Haupt-Jorgensen, M.; Krogvold, L.; Kaur, S.; Gerling, I.C.; Pociot, F.; Dahl-Jørgensen, K.; Buschard, K. Genetic predisposition in the 2′-5′A pathway in the development of type 1 diabetes: Potential contribution to dysregulation of innate antiviral immunity. Diabetologia 2021, 64, 1805–1815. [Google Scholar] [CrossRef] [PubMed]
- Carnero-Montoro, E.; Barturen, G.; Povedano, E.; Kerick, M.; Martinez-Bueno, M.; PRECISESADS Clinical Consortium; Ballestar, E.; Martin, J.; Teruel, M.; Alarcón-Riquelme, M.E. Epigenome-Wide Comparative Study Reveals Key Differences Between Mixed Connective Tissue Disease and Related Systemic Autoimmune Diseases. Front. Immunol. 2019, 10, 1880. [Google Scholar] [CrossRef] [PubMed]
- Palli, E.; Kravvariti, E.; Tektonidou, M.G. Type I Interferon Signature in Primary Antiphospholipid Syndrome: Clinical and Laboratory Associations. Front. Immunol. 2019, 10, 487. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Tang, L.; Zhang, L.; Ren, Y.; Peng, H.; Xiao, Y.; Xu, J.; Mao, D.; Liu, L.; Liu, L. Identification of Biomarkers Associated with CD4+ T-Cell Infiltration with Gene Coexpression Network in Dermatomyositis. Front. Immunol. 2022, 13, 854848. [Google Scholar] [CrossRef]
- Xiao, L.; Zhan, F.; Lin, S. Clinical Values of the Identified Hub Genes in Systemic Lupus Erythematosus. Front. Immunol. 2022, 13, 844025. [Google Scholar] [CrossRef]
- Subhi, O.; Schulten, H.J.; Bagatian, N.; Al-Dayini, R.; Karim, S.; Bakhashab, S.; Alotibi, R.; Al-Ahmadi, A.; Ata, M.; Elaimi, A.; et al. Genetic relationship between Hashimoto‘s thyroiditis and papillary thyroid carcinoma with coexisting Hashimoto’s thyroiditis. PLoS ONE 2020, 15, e0234566. [Google Scholar] [CrossRef]
- Castañeda-Delgado, J.E.; Bastián-Hernandez, Y.; Macias-Segura, N.; Santiago-Algarra, D.; Castillo-Ortiz, J.D.; Alemán-Navarro, A.L.; Martínez-Tejada, P.; Enciso-Moreno, L.; Garcia-De Lira, Y.; Olguín-Calderón, D.; et al. Type I Interferon Gene Response Is Increased in Early and Established Rheumatoid Arthritis and Correlates with Autoantibody Production. Front. Immunol. 2017, 8, 285. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jia, W.; Ma, Q.; Li, F.; Ma, Z.; Yang, M.; Pu, J.; Huo, Z.; Dang, J. Identification of IFI44L as a new candidate molecular marker for systemic lupus erythematosus. Clin. Exp. Rheumatol. 2023, 41, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Cooles, F.A.H.; Tarn, J.; Lendrem, D.W.; Naamane, N.; Lin, C.M.; Millar, B.; Maney, N.J.; Anderson, A.E.; Thalayasingam, N.; Diboll, J.; et al. Interferon-α-mediated therapeutic resistance in early rheumatoid arthritis implicates epigenetic reprogramming. Ann. Rheum. Dis. 2022, 81, 1214–1223. [Google Scholar] [CrossRef]
- Mei, X.; Zhang, B.; Zhao, M.; Lu, Q. An update on epigenetic regulation in autoimmune diseases. J. Transl. Autoimmun. 2022, 5, 100176. [Google Scholar] [CrossRef]
- Bronson, P.G.; Chang, D.; Bhangale, T.; Seldin, M.F.; Ortmann, W.; Ferreira, R.C.; Urcelay, E.; Pereira, L.F.; Martin, J.; Plebani, A.; et al. Common variants at PVT1, ATG13-AMBRA1, AHI1 and CLEC16A are associated with selective IgA deficiency. Nat. Genet. 2016, 48, 1425–1429. [Google Scholar] [CrossRef]
- Chen, W.I.; Cao, Y.; Zhong, Y.; Sun, J.; Dong, J. The Mechanisms of Effector Th Cell Responses Contribute to Treg Cell Function: New Insights into Pathogenesis and Therapy of Asthma. Front. Immunol. 2022, 13, 862866. [Google Scholar] [CrossRef]
- Schreiber, T.H.; Wolf, D.; Tsai, M.S.; Chirinos, J.; Deyev, V.V.; Gonzalez, L.; Malek, T.R.; Levy, R.B.; Podack, E.R. Therapeutic Treg expansion in mice by TNFRSF25 prevents allergic lung inflammation. J. Clin. Investig. 2010, 120, 3629–3640. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef]
- Ameratunga, R.; Leung, E.; Woon, S.T.; Lea, E.; Allan, C.; Chan, L.; Steele, R.; Lehnert, K.; Longhurst, H. Selective IgA Deficiency May Be an Underrecognized Risk Factor for Severe COVID-19. J. Allergy Clin. Immunol. Pract. 2023, 11, 181–186. [Google Scholar] [CrossRef]
- Truedsson, L.; Baskin, B.; Pan, Q.; Rabbani, H.; Vorĕchovský, I.; Smith, C.I.; Hammarström, L. Genetics of IgA deficiency. APMIS 1995, 103, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Aghamohammadi, A.; Pouladi, N.; Parvaneh, N.; Yeganeh, M.; Movahedi, M.; Gharagolou, M.; Pourpak, Z.; Rezaei, N.; Salavati, A.; Abdollahzade, S.; et al. Mortality and morbidity in common variable immunodeficiency. J. Trop. Pediatr. 2007, 53, 32–38. [Google Scholar] [CrossRef]
- Aghamohammadi, A.; Abolhassani, H.; Latif, A.; Tabassomi, F.; Shokuhfar, T.; Torabi Sagvand, B.; Shahinpour, S.; Mirminachi, B.; Parvaneh, N.; Movahedi, M.; et al. Long-term evaluation of a historical cohort of Iranian common variable immunodeficiency patients. Expert. Rev. Clin. Immunol. 2014, 10, 1405–1417. [Google Scholar] [CrossRef]
- Taga, T. The signal transducer gp130 is shared by interleukin-6 family of haematopoietic and neurotrophic cytokines. Ann. Med. 1997, 29, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Bin Dhuban, K.; Bartolucci, S.; d’Hennezel, E.; Piccirillo, C.A. Signaling Through gp130 Compromises Suppressive Function in Human FOXP3+ Regulatory T Cells. Front. Immunol. 2019, 10, 1532. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.; Means, A.R.; Liu, J.O. Calmodulin-dependent protein kinase IV regulates nuclear export of Cabin1 during T-cell activation. EMBO J. 2005, 24, 2104–2113. [Google Scholar] [CrossRef]
- Koga, T.; Sumiyoshi, R.; Tsuji, S.; Furukawa, K.; Kawakami, A. CaMK4 expression on effector memory T cells is associated with organ damage in systemic lupus erythematosus: A case report. Clin. Immunol. 2023, 247, 109222. [Google Scholar] [CrossRef] [PubMed]
- Crispin, J.C.; Hedrich, C.M.; Suárez-Fueyo, A.; Comte, D.; Tsokos, G.C. SLE-Associated Defects Promote Altered T Cell Function. Crit. Rev. Immunol. 2017, 37, 39–58. [Google Scholar] [CrossRef]
- Koga, T.; Hedrich, C.M.; Mizui, M.; Yoshida, N.; Otomo, K.; Lieberman, L.A.; Rauen, T.; Crispín, J.C.; Tsokos, G.C. CaMK4-dependent activation of AKT/mTOR and CREM-α underlies autoimmunity-associated Th17 imbalance. J. Clin. Investig. 2014, 124, 2234–2245. [Google Scholar] [CrossRef]
- Kono, M.; Maeda, K.; Stocton-Gavanescu, I.; Pan, W.; Umeda, M.; Katsuyama, E.; Burbano, C.; Orite, S.Y.K.; Vukelic, M.; Tsokos, M.G.; et al. Pyruvate kinase M2 is requisite for Th1 and Th17 differentiation. JCI Insight. 2019, 4, e127395. [Google Scholar] [CrossRef]
- Psarras, A.; Emery, P.; Vital, E.M. Type I interferon-mediated autoimmune diseases: Pathogenesis, diagnosis and targeted therapy. Rheumatology 2017, 56, 1662–1675. [Google Scholar] [CrossRef]
- Gangaplara, A.; Martens, C.; Dahlstrom, E.; Metidji, A.; Gokhale, A.S.; Glass, D.D.; Lopez-Ocasio, M.; Baur, R.; Kanakabandi, K.; Porcella, S.F.; et al. Type I interferon signaling attenuates regulatory T cell function in viral infection and in the tumor microenvironment. PLoS Pathog. 2018, 14, e1006985. [Google Scholar] [CrossRef]
- Banik, S.; Rakshit, S.; Sarkar, K. The Role of STAT1 in T Helper Cell Differentiation during Breast Cancer Progression. J. Breast Cancer 2021, 24, 253–265. [Google Scholar] [CrossRef]
- Shi, D.; Li, Y.; Shi, X.; Yao, M.; Wu, D.; Zheng, Y.; Lin, Q.; Yang, Y. Transcriptional expression of CXCL10 and STAT1 in lupus nephritis and the intervention effect of triptolide. Clin. Rheumatol. 2023, 42, 539–548. [Google Scholar] [CrossRef]
- Brandt, M.; Kim-Hellmuth, S.; Ziosi, M.; Gokden, A.; Wolman, A.; Lam, N.; Recinos, Y.; Daniloski, Z.; Morris, J.A.; Hornung, V.; et al. An autoimmune disease risk variant: A trans master regulatory effect mediated by IRF1 under immune stimulation? PLoS Genet. 2021, 17, e1009684. [Google Scholar] [CrossRef]
- Bonilla, F.A.; Barlan, I.; Chapel, H.; Costa-Carvalho, B.T.; Cunningham-Rundles, C.; de la Morena, M.T.; Espinosa-Rosales, F.J.; Hammarström, L.; Nonoyama, S.; Quinti, I.; et al. International Consensus Document (ICON): Common Variable Immunodeficiency Disorders. J. Allergy Clin. Immunol. Pract. 2016, 4, 38–59. [Google Scholar] [CrossRef]
- Janssen, L.M.A.; van der Flier, M.; de Vries, E. Lessons Learned from the Clinical Presentation of Common Variable Immunodeficiency Disorders: A Systematic Review and Meta-Analysis. Front. Immunol. 2021, 12, 620709. [Google Scholar] [CrossRef]
| CVID | SIgAD | Control Group | |
|---|---|---|---|
| Number of children | 13 | 13 | 11 |
| Age (years ± SD) | 10.9 ± 3.4 | 8.36 ± 3.1 | 6.25 ± 4.35 |
| Sex (F/M) | 5/8 | 7/6 | 6/5 |
| Familial history of immunodeficiencies | 1/13 | 2/13 | 0/11 |
| Accompanying diseases | 2 patients with thrombocytopenia, 1 patient with ulcerative colitis | 1 patient with celiac disease, 1 patient with juvenile arthritis, 1 patient with Sjögren disease | 0 |
| Gene Symbol (Gene Name) | Fold Change in CVID When Compared to Control (p Value) | Fold Change in SIgAD When Compared to Control (p Value) | Functional Annotation |
|---|---|---|---|
| CAMK4 (calcium/calmodulin-dependent protein kinase IV) | −2.65 (0.0043) | −2.31 (0.0253) | Calcium/calmodulin-dependent protein kinase involved in the calcium-triggered CaMKK-CaMK4 signaling cascade, regulating, mainly by phosphorylation, the activity of several transcription activators, such as CREB1, MEF2D, JUN, and RORA, which play a pivotal role in immune response, inflammation, and memory consolidation. |
| IL6ST (interleukin 6 signal transducer) | −2.35 (0.0018) | −2.09 (0.0466) | Gp130 is involved in the receptor complex formation in the case of several cytokines, including IL-6, IL-27, and IL-11. Gp130 functional impairment in hematopoietic cells results in defective lymphocyte development. |
| OGFRL1 (opioid growth factor receptor-like 1) | 5.18 (0.0438) | 5.62 (0.0038) | No UniProt annotations available. |
| ATP6V1B2 (ATPase, H+ transporting, lysosomal 56/58 kDa, V1 subunit B2) | 2.33 (0.0454) | 2.71 (0.0184) | Non-catalytic subunit of the V1 complex of vacuolar(H+)-ATPase (V-ATPase). V-ATPase is associated with acidifying and maintaining the pH of intracellular compartments, and in some cell types is responsible for acidifying the extracellular environment. |
| TNFAIP2 (tumor necrosis factor, alpha-induced protein 2) | 3.16 (0.0354) | 2.03 (0.0481) | May play a role as a mediator of inflammation and angiogenesis. |
| Gene Symbol | Group | Fold Change (p Value) | Description | Selected Association with Autoimmune Diseases |
|---|---|---|---|---|
| IRF1 | Multiple Complex | 4.63 (0.0455) | interferon regulatory factor 1 | pemphigus [31], systemic sclerosis [32], rheumatoid arthritis [33], localized scleroderma [34] |
| STAT1 | Multiple Complex | 3.61 (0.0219) | signal transducer and activator of transcription 1 | systemic lupus erythematosus [35], Sjögren’s syndrome [36], rheumatoid arthritis [37] |
| Gene Symbol | Fold Change (p Value) | Description | Selected Association with Autoimmune Disease |
|---|---|---|---|
| IFIT1 | −5.68 (0.0383) | interferon-induced protein with tetratricopeptide repeats 1 | systemic lupus erythematosus [38], primary Sjögren’s syndrome [39], autoimmune demyelinating CNS disorders [40], ankylosing spondylitis [41], Anti-neutrophil cytoplasmic antibody (ANCA)-Associated Vasculitis (AAV) [42], juvenile dermatomyositis (JDM) [43], systemic sclerosis [44], experimental autoimmune encephalomyelitis [45], type 1 diabetes [46] |
| MX1 | −5.23 (0.0176) | MX dynamin-like GTPase 1 | dermatomyositis [47], systemic lupus erythematosus [48], Sjögren syndrome [49], juvenile idiopathic arthritis [50], type 1 diabetes [51], mixed connective tissue disease [52], primary antiphospholipid syndrome [53] |
| IFI6 | −5.06 (0.0184) | interferon, alpha-inducible protein 6 | dermatomyositis [54], systemic lupus erythematosus [55], Hashimoto’s thyroiditis [56], rheumatoid arthritis [57] |
| IFI44L | −4.42 (0.0294) | interferon-induced protein 44-like | primary Sjogren’s syndrome [39], systemic lupus erythematosus [58], rheumatoid arthritis [59], (ANCA)-Associated Vasculitis (AAV) [42], mixed connective tissue disease [52], systemic sclerosis [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rutkowska-Zapała, M.; Grabowska-Gurgul, A.; Lenart, M.; Szaflarska, A.; Kluczewska, A.; Mach-Tomalska, M.; Baj-Krzyworzeka, M.; Siedlar, M. Gene Signature of Regulatory T Cells Isolated from Children with Selective IgA Deficiency and Common Variable Immunodeficiency. Cells 2024, 13, 417. https://doi.org/10.3390/cells13050417
Rutkowska-Zapała M, Grabowska-Gurgul A, Lenart M, Szaflarska A, Kluczewska A, Mach-Tomalska M, Baj-Krzyworzeka M, Siedlar M. Gene Signature of Regulatory T Cells Isolated from Children with Selective IgA Deficiency and Common Variable Immunodeficiency. Cells. 2024; 13(5):417. https://doi.org/10.3390/cells13050417
Chicago/Turabian StyleRutkowska-Zapała, Magdalena, Agnieszka Grabowska-Gurgul, Marzena Lenart, Anna Szaflarska, Anna Kluczewska, Monika Mach-Tomalska, Monika Baj-Krzyworzeka, and Maciej Siedlar. 2024. "Gene Signature of Regulatory T Cells Isolated from Children with Selective IgA Deficiency and Common Variable Immunodeficiency" Cells 13, no. 5: 417. https://doi.org/10.3390/cells13050417





