Abstract
Despite a long history of research, neurodegenerative diseases and malignant brain tumor gliomas are both considered incurable, facing challenges in the development of treatments. Recent evidence suggests that RNA modifications, previously considered as static components of intracellular RNAs, are in fact dynamically regulated across various RNA species in cells and play a critical role in major biological processes in the nervous system. Innovations in next-generation sequencing have enabled the accurate detection of modifications on bases and sugars within various RNA molecules. These RNA modifications influence the stability and transportation of RNA, and crucially affect its translation. This review delves into existing knowledge on RNA modifications to offer a comprehensive inventory of these modifications across different RNA species. The detailed regulatory functions and roles of RNA modifications within the nervous system are discussed with a focus on neurodegenerative diseases and gliomas. This article presents a comprehensive overview of the fundamental mechanisms and emerging roles of RNA modifications in these diseases, which can facilitate the creation of innovative diagnostics and therapeutics for these conditions.
1. Introduction
Epigenetic modifications play crucial roles in the development, function, and plasticity of the nervous system. These modifications involve DNA methylation, histone modifications, chromatin remodeling, and RNA modifications. Epigenetic dysregulation has been linked to neurodegenerative diseases, brain tumors, and other various neurological disorders. Understanding the roles of epigenetic modifications in the nervous system is essential for gaining insights into normal brain function, neurological disorders, and potential therapeutic interventions.
Among all epigenetic modifications, RNA modifications are generally considered distinct from classical modifications involving DNA and histones [1]. Recently, more studies have been reported on “epitranscriptomics”, which refers to a molecular biology field that focuses on the biochemical modifications of RNA molecules and their effect on gene expression and various cellular functions [2]. Epitranscriptomics research aims to understand the distribution patterns, biogenesis mechanisms, and regulatory functions of modified RNA molecules, as well as the interactome, evolutional conservation, and novel reader proteins. It has become evident that the information carried by RNA molecules is not static and can be dynamically modified by adding, removing, or modifying chemical groups [3]. The link between atypical RNA modifications and a range of neurological disorders highlights the significance of these chemical modifications in understanding the underlying mechanisms of these diseases. Regulating RNA modifications can provide new opportunities for therapies, as they offer a new area of biology to explore. In this review, we will outline the RNA modifications, including non-coding RNAs, and uncover their distinct functions and the consequential effects on cellular and physiological processes in the pathogenesis of neurologic disorders (Figure 1).
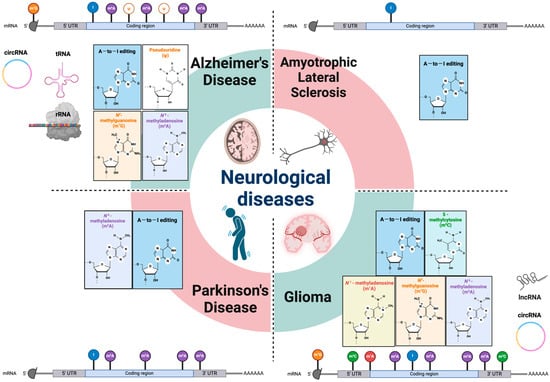
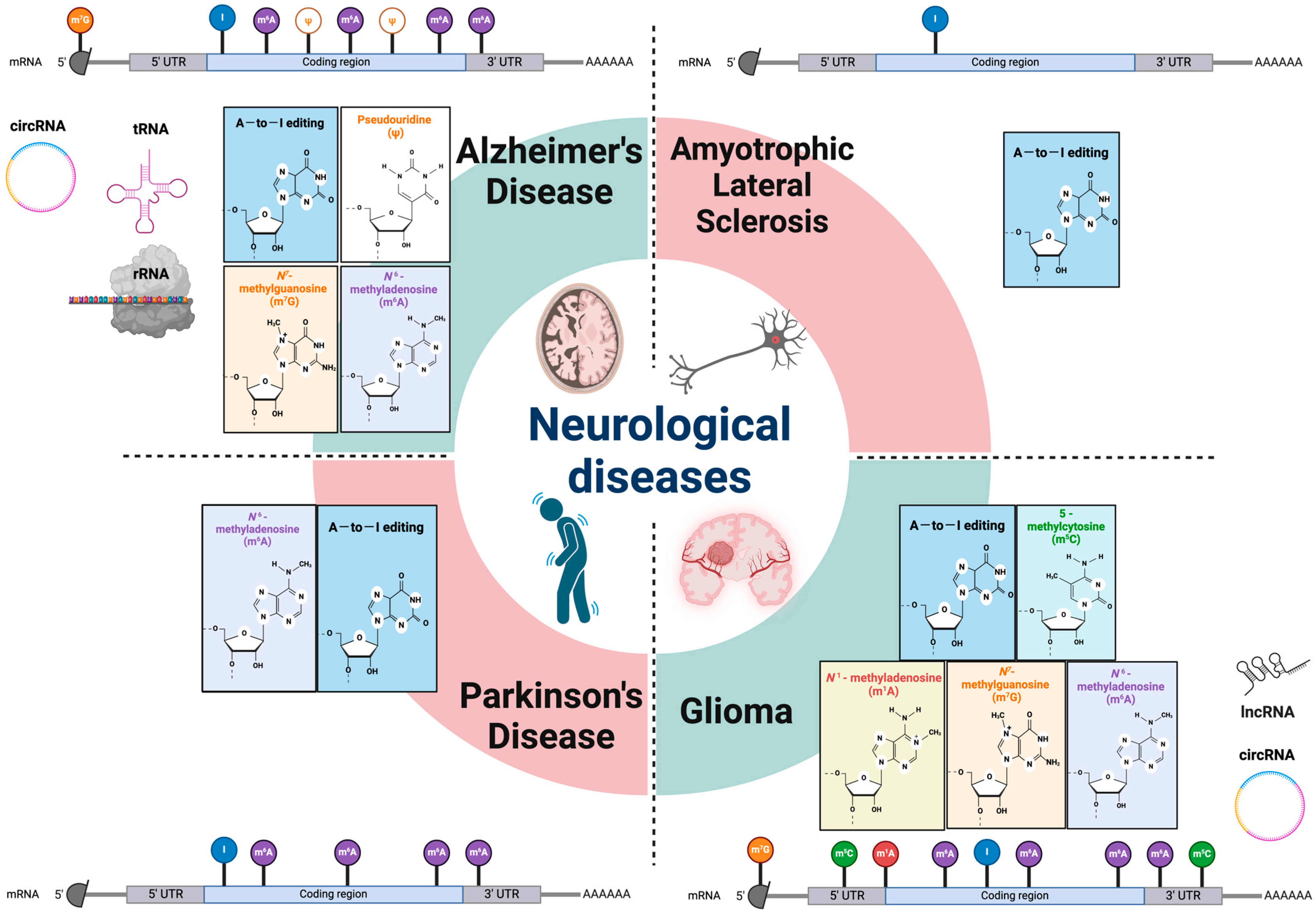
Figure 1.
RNA modifications in various neurological diseases. Epitranscriptomic RNA modifications in both mRNA and different non-coding RNAs are involved in multiple types of neurologic disorders. Genetic analysis and animal model studies have revealed that various RNA modifications and their machineries have critical roles in the etiology of neurodegenerative diseases and gliomas. mRNA: messenger RNA; rRNA: ribosomal RNA; lncRNA: long non-coding RNA; tRNA: transfer RNA; circRNA: circular RNA; UTR: untranslated region; A-to-I editing: adenosine to inosine RNA editing.
2. Modifications on mRNAs, rRNAs, and Non-Coding RNAs
Although numerous RNA modifications have been identified including messenger RNAs (mRNAs), ribosomal RNAs (rRNAs), and non-coding RNAs, only a handful of them have undergone thorough investigation within the nervous system. Some of the well-established RNA modifications are N6-methyladenosine (the methylation of adenosine at position 6, m6A), N1-methyladenosine (m1A), 5-methylcytosine (m5C), pseudouridine, and RNA editing. These modifications have been linked to various processes in the brain, including neurodevelopment, neurogenesis, neuroplasticity, learning and memory, neural regeneration, neurodegeneration, and brain tumorigenesis.
2.1. N6-Methyladenosine (m6A)
Since the 1970s when m6A modification was first identified [4], m6A became the most extensively researched and prevalent RNA modification in human cells [5]. With the advancement of identifying and detecting transcriptome-wide m6A distribution at single-base resolution [6,7], we now know that m6A modifications are crucial in various biological processes, including gene expression regulation, RNA processing, and RNA stability. These modifications are installed by methyltransferase complexes and can be reversed by demethylases, allowing them to have a dynamic and reversible nature. m6A marks have been discovered across different RNA repertoires: mRNAs, rRNAs, transfer RNAs (tRNAs), circular RNAs (circRNAs), and micro RNAs (miRNAs). Specifically for mRNAs, most m6A marks are situated at the start of the final exons, in the 3′-untranslated region (UTR), or near to the stop codons [8,9]. For long non-coding RNAs (lncRNAs), high-throughput methods revealed that human m6A marks were mapped to certain well-characterized lncRNAs, including X-inactive specific transcript (XIST) and metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) [10,11,12]. Writer complexes like Methyltransferase-like 3 and 14 (METTL3 and METTL14), Wilms tumor 1-associated protein (WTAP), Zinc finger CCCH domain-containing protein 13 (ZC3H13), RNA-binding motif protein 15/15B (RBM15/15B), and Vir-like m6A methyltransferase-associated protein (VIRMA) mediate the methylation of adenosine residues at position 6 [13,14,15,16,17]. METTL3 and METTL14 are heterodimers that form the core complex responsible for methyltransferase activity, stabilized by the association of another complex formed by WTAP, RBM15, ZC3H13, and VIRMA called the methylation-associated complex. METTL16 which is also one of the components of the m6A writer complexes, along with WTAP, is known to engage with specific lncRNAs, such as MALAT-1 [10]. YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2, IGF2BP1, IGF2BP2, and IGF2BP3 have been identified as reader proteins that detect the m6A methylation marks. For instance, YTHDC1 facilitates the nuclear export of methylated mRNAs to the cytoplasm by specifically binding with m6A. Suppressing YTHDC1 leads to the nuclear buildup of m6A-modified mRNA, causing these transcripts to accumulate within the nucleus and become scarce in the cytoplasm [18]. In addition, it has been reported that m6A modifications in circRNAs promote efficient protein translation in a cap-independent fashion through YTHDF3 and the translation initiation factors eIF4G2 and eIF3A [19]. This m6A-driven translation by circRNAs can be enhanced by METTL3 and METTL14. Meanwhile, enzymes such as FTO and ALKBH5 act as erasers for removing m6A marks. m6A modifications can either increase or decrease mRNA stability, depending on the specific reader proteins that bind them. Functionally, m6A participates in nearly every stage of the mRNA life cycle, affecting splicing, other nuclear processing, and degradation within the cytoplasm [20].
m6A modifications are known to impact a range of brain functions and development, including neurogenesis, spinogenesis, learning and memory, dendritic structure, axon regeneration, and brain development. Some of the neurologic disease phenotypes are a consequence of disrupted m6A pathways, attributed to either disease-specific mutations or alterations in the levels of various m6A modulators. The m6A marks located in the 3′-UTR regions of mRNAs are believed to play significant mechanistic roles in the onset of age-related disorders such as Alzheimer’s disease (AD), Parkinson’s disease (PD), and Amyotrophic lateral sclerosis (ALS), as they regulate the translation of associated transcripts. In Table 1, we provide an overview of various RNA modifications, including m6A marks, alongside detection methods, and their connections to diverse RNA metabolic activities associated with neurological diseases.

Table 1.
The role of RNA modifications on diverse RNA metabolic activities and their association with neurological diseases.
2.2. N1-Methyladenosine (m1A)
Methylation occurring at the N1 position of adenosine represents another example of dynamic modifications of RNA within mammalian systems [21]. This modification is abundant, prevalent, and conserved across both prokaryotic and eukaryotic RNAs as an internal post-transcriptional alteration, particularly prominent in the RNAs of higher eukaryotic cells [22,23]. This modification halts translation elongation, which is the process of translating the genetic code into a polypeptide chain. It also hinders the incorrect incorporation of nucleotides during reverse transcription (RT) by impeding the formation of Watson–Crick base pairs [24]. While m1A methylations have been reported in the 5′-UTR regions and coding sequences of mRNAs, they have been primarily identified in rRNAs and tRNAs, altering their structure and influencing their capacity for protein binding, stability, and functionality. The most abundant and representative m1A writer in the cytosol is the heterotetrameric tRNA methyltransferase TRMT6/61A, which incorporates a GUUCRA tRNA-like motif with a t-loop structure into specific mRNAs [25]. On the other hand, ALKBH3 and ALKBH1 serve as erasers for m1A, with ALKBH3 responsible for demethylating the m1A mark on mRNAs [26,27]. In addition, reports indicate that YTHDF2 not only serves as the reader for m6A but can also specifically detect m1A-modified sequences and bind to endogenous m1A-modified transcripts with a low affinity [28]. These findings suggest that YTHDF2 could play a more expansive role in RNA modifications beyond its established function as an m6A reader. Modifications of m1A in mRNA are primarily linked to the regulation of mRNA translation and decay, suggesting crucial roles in ensuring proper brain development and function. However, studies on the correlation between m1A marks and neurological disorders are still limited.
2.3. 5-Methylcytosine (m5C)
m5C modifications have been identified across a spectrum of RNA species, including rRNAs, tRNAs, and mRNAs, as well as small nuclear RNAs (snRNAs), vault RNAs, enhancer RNAs, lncRNAs, and miRNAs. These modifications are crucial in regulating RNA stability, transcription, transportation, and translation. They have been associated with gene expression and metabolism. For example, early studies have demonstrated that m5C modifications affect the interactions of lncRNAs, such as MALAT1, antisense non-coding RNA in the INK4 locus (ANRIL), nuclear paraspeckle assembly transcript 1 (NEAT1), growth arrest-specific transcript 5 (GAS5), ribonuclease P RNA component H1 (RPPH1), Pvt1 oncogene non-protein coding (PVT1), SNHG12, Telomerase RNA component (TERC), and XIST with chromatin-associated protein complexes [29,30]. In human cells, the NOL1/NOP2/SUN domain-containing protein family, specifically NOP2/SUN RNA methyltransferase (NSUN) 1 through NSUN7, along with the DNA methyltransferase (DNMT) homolog DNMT2, are responsible for generating m5C marks on RNA molecules [31]. NSUN1, NSUN2, and NSUN5 especially exhibit conservation across eukaryotes, while the remaining family members are exclusive to higher eukaryotes. In terms of functionality, NSUN1 and NSUN5 are involved in methylating cytosine C5 of rRNAs in the cytosol, while NSUN2, NSUN6, and DNMT2 are responsible for methylating cytosolic tRNAs. Additionally, NSUN2, NSUN3, and NSUN4 perform methylation at cytosine C5 within mitochondrial RNAs [32,33]. The mRNA export adaptor protein ALYREF and Y-box binding protein1 (YBX1) bind and recognize the m5C modifications and regulate the nucleo-cytoplasmic transport and stability of mRNA molecules [34,35]. The modifications of RNAs through m5C can be reversed by α-ketoglutarate (α-KG)-dependent dioxygenases, like ten-eleven translocation (TET)1 and TET2, which actively demethylate these marks [36].
A major significant role of m5C RNA methylation is that m5C RNA modifications affect the stability of eukaryotic rRNA by impacting the folding of crucial ribosomal regions, consequently controlling translation [37]. In addition, the modification at the m5C position can influence the aminoacylation step in translation, thereby impacting the overall accuracy of the translation process [38,39]. Notably, the impact of m5C modification on mRNA translation differs based on m5C placement. It hampers translation efficiency if it occurs within the 5′-UTR or coding sequence (CDS). However, m5C modification in the 3′-UTR, when mediated by NSUN2, has been found to increase translation efficiency [40,41,42,43]. The essential functional roles fulfilled by m5C modifications suggest that the disrupted expression of these genes could contribute to diverse neuronal conditions. For instance, a study that involved conditionally knocking out NSUN2 in the prefrontal cortex of mice resulted in bidirectional behavioral changes associated with depression [44]. The deficiency of NSUN2 led to modifications in almost 1500 proteins within the prefrontal cortex, accompanied by reduced translation efficiency linked to a glycine-codon defect. This, in turn, caused a disruption in synaptic communication among pyramidal neurons in the prefrontal cortex. Another example is the interruption of NSUN2’s tRNA methylation activity which can cause an accumulation of tRNA fragments at the 5′ ends, interfering with the formation of upper layer neurons and negatively impacting brain development in mice [45].
2.4. Pseudouridine
Pseudouridine, the isomer of uridine known as 5-ribosyl uracil or Ψ, is less abundant in mRNA; instead, it is more frequently detected in non-coding RNAs such as rRNAs, snRNAs, and tRNAs. Regardless of the abundance among mRNAs, pseudouridine can still influence the secondary structure of these molecules. For example, pseudouridine residues were found in MALAT1 at the positions of U5160, U5590, and U3374 [46,47]. However, the functions of these chemical alterations and their influence on MALAT1 activity remain unclear, requiring more comprehensive research to understand them. Two primary mechanisms for RNA pseudouridylation have been identified. First is the pseudouridylation that is independent of guide RNA and relies on pseudouridine synthase (PUS) enzymes directly catalyzing the transformation of uridine into Ψ within their target sequences (Table 1). On the other hand, pseudouridylation that is dependent on guide RNA involves H/ACA-box small nucleolar RNAs (snoRNAs) binding to target RNAs through sequence-specific interactions [48,49,50]. Codons containing Ψ have been demonstrated to exert a modest influence on ribosomes by incorporating specific amino acids. Additionally, stop codons containing pseudouridine (Ψ) have been found to guide the suppression of translation termination [51,52,53]. Furthermore, the existence of Ψ in numerous RNAs alters their interactions with RNA-binding proteins (RBPs) that participate in nuclear RNA processing, as well as the localization or stability of cytosolic RNA [48,49].
2.5. Adenosine to Inosine RNA Editing (A-to-I Editing)
RNA editing, a post-transcriptional modification, involves converting specific nucleotides into RNA molecules. This includes adenosine-to-inosine (A-to-I) editing, prevalent in vertebrates, with numerous such sites discovered in mice and humans. This editing process is orchestrated by the adenosine deaminase acting on RNA (ADAR) protein family. The adenosine deaminase that acts on tRNA (ADAT) conducts this conversion for tRNAs. The change from adenosine to inosine is due to the deamination of the amino group at adenosine’s C6 position, altering the RNA’s informational content and potentially its secondary structure [54,55]. In addition, A-to-I RNA editing is known to contribute significantly to the diversity of the epitranscriptome and the proteome in various cancers [56,57]. RNA editing plays a crucial role in modulating gene expression, particularly by altering miRNA expression, maturation, and stability [58,59,60]. This regulatory mechanism involves the activity of specific enzymes from the adenosine/cytidine deaminase family, which trigger single nucleotide transformation in primary miRNAs. Such modifications can substantially affect the miRNA’s stability and its ability to mature and target specific mRNAs, ultimately influencing miRNA-guided gene expression regulation. Additionally, RNA editing modifies the primary structure of wild-type mature miRNAs, thereby altering their gene-regulatory functions.
RNA editing, particularly A-to-I editing, is an extensive process in humans, with estimates suggesting that up to 85% of pre-mRNAs are subject to this modification [61,62,63]. This editing process predominantly occurs within introns and untranslated regions (UTRs) of genes encoding proteins and is reportedly involved with neural development and various neurological disorders. For example, RNA editing at the I/V site of the potassium channel Kv1.1, mediated by ADAR2, has been associated with epilepsy [64]. Episodic ataxia type 1, a condition characterized by seizures, ataxia, and myokymia, has been reported to be caused by mutations in the gene KCNA1, which codes for Kv1.1. In addition, ADAR2’s role in editing the RNA of glutamate receptors is also associated with psychiatric diseases, including schizophrenia [65] (Table 1).
2.6. Other Modifications
An extensive array of RNA modifications has been discovered, particularly in tRNAs, which includes m5C, 3-methylcytidine (m3C), 1-methyl-guanosine (m1G), N2-methylguanosine (m2G), N7-methylguanosine (m7G), N2,N2-dimethylguanosine (m22G), N1-methylpseudouridine (m1J), 2-methyladenosine (2 mA), 5-formyl-20-O-methylcytidine (f5Cm), 3-(3-amino-3-carboxypropyl) uridine (acp3U), 5-methyluridine (5 mU), 5-methoxycarbonylmethyluridine (mcm5U), 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U), dihydrouridine, queuosine (Q), galactosylqueuosine (gal Q), mannosyl-queuosine (manQ), N6-threonylcarbamoyladenosine (t6A), N6-methyl-N6-threonylcarbamoyladenosine (m6t6A), 2-methylthio-N6-threonylcarbamoyladenosine (ms2t6A), N4-acetylcytidine (ac4C), N6-isopentenyladenosine(i6A), peroxywybutosine (o2yW), and wybutosine (yW) [66,67]. These modifications are categorized by their position into two principal types: some that are located in the anticodon loop and others found elsewhere. The modifications occurring within the anticodon loop can influence translation by altering the base pairing between the codons in mRNAs and the tRNA that carries amino acids [68]. Modifications situated outside of the anticodon loop primarily influence the secondary structure of tRNAs.
Modifications in rRNA are relatively sparse, with just 2% of nucleotides undergoing changes. Among these, 2′-O-methylation (2′-OMe), where a methyl group is added to the 2′ hydroxyl of ribose, is the most common [69]. These 2′-OMe modifications can occur at the first nucleotide transcribed (denoted as m7GpppNmN-) or the second (m7GpppNmNm-). It has been shown that 2′-OMe modifications enhance the stability of RNA–RNA hybrid duplexes [70].
2.7. Detection and Quantification of RNA Modifications
To elucidate the dynamics of a broad spectrum of RNA modifications, including m6A, m1A, m5C, A-to-I editing, and modifications in tRNA, various sophisticated techniques are employed. Liquid chromatography–tandem mass spectrometry (LC-MS/MS) is pivotal for identifying and quantifying these modifications by leveraging the distinct mass changes in modified nucleotides [8,71]. Methylated RNA immunoprecipitation sequencing (MeRIP-Seq) specifically maps the genome-wide distribution of m6A, capturing m6A-modified RNA fragments [9]. RNA bisulfite sequencing (RNA-BisSeq) differentiates m5C from unmodified cytosine [29], while Ψ-seq specifically labels pseudouridine for precise localization [47]. Additionally, A-to-I RNA editing, mediated by ADAR enzymes, is identified through sequencing technologies that detect inosine as guanosine, due to its base-pairing properties [72]. The complexity of tRNA modifications, critical for the stability, structure, and function of tRNA, is revealed through methods such as LC-MS/MS and specialized sequencing techniques [73].
The regulatory roles of m6A methylation enzymes, including writers, readers, and erasers, are clarified by unveiling the regulation of gene expressions through quantitative Real-Time PCR (qRT-PCR) measuring mRNA levels. This is further investigated through immunoprecipitation coupled with mass spectrometry (IP-MS) and gene editing using CRISPR/Cas9 for gene knockout or knockdown, providing insights into the composition and interactions of these protein complexes. IP-MS elucidates the composition and interactions of these protein complexes [14]. Moreover, next-generation sequencing (NGS)-based detection techniques, such as MeRIP-Seq and Cross-linking and immunoprecipitation sequencing (CLIP-seq), alongside nanopore direct RNA sequencing, offer comprehensive methods for analyzing the regulation of RNA by m6A modifications, A-to-I editing patterns, and tRNA modifications, enriching our understanding of RNA biology in disease [71].
3. RNA Modifications in Neurodegenerative Diseases
3.1. RNA Modifications in Alzheimer’s Disease (AD)
One of the most abundant RNA modifications in the brain is m6A, and most of the recent studies on RNA modifications indicate that the loss of m6A methylation of RNA could promote AD development. Utilizing m6A-sequencing in combination with high-throughput LC–MS/MS, Shafik et al. revealed a substantial reduction in the expression level of METTL3, along with decreased m6A levels, in 5xFAD mice compared to control groups [74]. This result was consistent with other groups’ studies where notable reductions in both neuronal m6A methylation and METTL3 expression have been observed in human AD brains compared to those without the condition, as demonstrated by immunoblot analysis [75]. For circRNA, high-throughput sequencing has revealed significant changes in circRNA m6A methylation in APP/PS1 AD mice compared to control groups [76]. This result was consistent with another group’s study where METTL3-dependent m6A-modified circular RNA, circRIMS2, was significantly upregulated in APP/PS1 AD mice, which mediated synaptic and memory impairments by activating the ubiquitination of the GluN2B subunit of the NMDA receptor [77]. Silencing METTL3 or hindering the GluN2B ubiquitination by a short membrane-permeable peptide significantly rescued synaptic impairment in APP/PS1 AD mice. On the other hand, METTL3 knockdown in the mouse hippocampus has been associated with several negative outcomes, including memory loss, neurodegeneration, spine loss, and gliosis [75]. In terms of mechanism, the lack of METTL3 slows down the mRNA degradation of m6A-modified cell cycle genes like Cyclin D1 and Cyclin D2. This effect was observed in cultivated primary neurons, which resulted in impaired cell cycle control [75]. Furthermore, a recent investigation into the gene expression patterns regulated by m6A in post-mortem brains from AD patients revealed the abnormal expression of METTL3 and the RNA binding motif protein 15B in the hippocampus of individuals with AD. The study indicated that the accumulation of METTL3 in insoluble fractions exhibited a positive correlation with Tau levels in hippocampal lysates. This suggests that disruptions in m6A signaling might contribute to neuronal dysfunction in AD [78]. It is also found that the demethylase FTO triggers mTOR signaling and eventually activates Tau phosphorylation in AD [79]. Notably, the neuron-specific knockout of the gene FTO has been demonstrated to alleviate cognitive impairments in 3xTg AD mice [79]. Moreover, a genetic variant within the FTO locus was significantly linked to Alzheimer’s in the NIA-LOAD study [80,81]. The decreased expression of FTO in the cortex and amygdala of Alzheimer’s patients compared to healthy individuals indicates the functional significance of FTO in AD [80,81]. This is further substantiated by a prospective study indicating that individuals with the AA genotype in the FTO gene face an elevated risk of developing AD and other types of dementia [82].
Other reports demonstrate alterations in small RNA modifications in AD patients compared to healthy individuals. LC–MS/MS analysis showed an increase in m7G, 2′-O-methylcytidine (Cm), and 2′-O-methylguanosine, while m22G and N2,N2,7-trimethylguanosine (m2,2,7G) exhibited significant decreases in the miRNA fraction from the cortex of AD brains [83]. Interestingly, within small RNA fractions of rRNA-derived small RNAs, tRNA-derived small RNAs, Y RNA-derived small RNAs, and other unannotated RNAs, elevated levels of modifications such as Cm, 2′-O-methyluridine (Um), and m7G were observed in comparison to controls. Conversely, modifications such as pseudouridine, m1G, and m2,2,7G were diminished [83]. Additionally, microfluidic high-throughput PCR-based next-generation sequencing has discovered a significant reduction in A-to-I RNA editing levels in AD patients as compared to control samples [84]. In the pre-frontal cortex of individuals with AD, RNA editing of the GluA2 subunit of the AMPA receptor leads to alterations in intracellular Ca2+ levels, which are associated with neuronal dysfunction and neurodegeneration stemming from increased Ca2+ permeability [85]. In healthy individuals, less than 0.1% of all GluA2 RNA molecules are unedited in the pre-frontal cortex, contrasting with 1.0% in AD patients. Another study observed reduced RNA editing at the glutamine/arginine (Q/R) site of the Glu2 subunit in the caudate nucleus and hippocampus of sporadic AD patients who carry the Apo E4 allele [86]. The presence of the ApoE E4 allele, a known genetic risk factor for AD, is suggested to affect the AMPA receptor dynamics and glutamate regulation in the hippocampus [87,88,89]. Comprehensive analysis from the ROSMAP, MayoRNAseq, and MSBB studies identified millions of RNA editing sites across nine brain regions, with 108,010 edits to facilitate and 26,168 edits to impede the progression of AD [90].
3.2. RNA Modifications in Parkinson’s Disease (PD)
Individuals with PD display incapacitating motor impairments characterized by resting tremors, bradykinesia (reduced movement speed), muscle stiffness, and postural instability, commonly recognized as PD’s four cardinal manifestations. The emergence of these symptoms is attributed to the degeneration of dopaminergic neurons in the substantia nigra region of the brain. Some researchers have performed comprehensive genetic analyses, including genome-wide association, differential gene expression, and expression quantitative trait locus analyses, which have identified five m6A SNPs associated with altered gene expressions related to PD [91]. In one study, m6A methylation was reduced and FTO was highly expressed in PC12 cells, a cellular model of PD [92]. The overexpression of FTO in dopaminergic neurons decreases mRNA m6A modification and upregulates ionotropic glutamate receptor 1 (NMDAR1). This escalation contributes to oxidative stress, an increase in calcium influx, and ultimately, it accelerates the degeneration or cell death of these dopaminergic neurons [92]. In another study, METTL3, METTL14, and YTHDF2 were significantly downregulated in PD patient blood mononuclear cells with METTL14 being the main factor involved in the abnormal m6A modification of α-synuclein mRNA [93]. Mettl14 targets and regulates the expression of the α-synuclein gene by binding an m6A motif in the coding region which eventually modifies α-synuclein mRNA and weakens its stability. Spearman correlation analysis showed METTL14 levels inversely correlated with plasma α-synuclein concentrations and motor function of PD patients [93]. Furthermore, the advancement in RNA sequencing capabilities has facilitated the detailed exploration of RNA editing occurrences across the whole transcriptome, offering deeper insights into A-to-I editing in various diseases. A comprehensive transcriptome study linked PD to alterations in Alu insertions, which are the main substrates for the ADAR protein family [94]. A compelling and innovative concept has recently been suggested for leveraging the inherent editing activity of endogenous ADAR2 to correct a disease-causing mutation in PINK1 associated with PD [95]. This mutation, a G-to-A substitution, leads to a premature stop codon that truncates the PINK1 protein, resulting in the truncation of the protein’s C-terminus [95]. By designing guide RNAs that direct ADAR2 to target and edit the specific mRNA, researchers were able to restore PINK1/Parkin-mediated mitophagy in cell models [95].
3.3. RNA Modifications in Amyotrophic Lateral Sclerosis (ALS)
ALS, one of the prevalent neurodegenerative diseases, is marked by a progressive decline in muscle strength and atrophy, leading to both upper and lower motor neuron dysfunction. Many studies exploring the pathogenesis of ALS, where RNA modifications play a role, have demonstrated the involvement of A-to-I RNA editing. ADAR2, an RNA editing enzyme, is mislocated in cases of ALS/FTD caused by C9orf72 repeat expansion [96]. In mice with a conditional knockout of ADAR2, the Q/R site on GluA2, which is crucial for the proper functioning of the AMPA receptors in motor neurons, remains unedited, leading to gradual motor neuron degeneration [97,98]. Interestingly, ADAR2 is uniquely reduced in the motor neurons of ALS patients among all the ADAR family [99]. This led to abnormal Ca2+ influx through AMPA receptors in neurons, resulting in motor neuron degeneration, which is a characteristic feature of ALS [100,101,102]. Furthermore, TAR DNA-binding protein 43 (TDP-43), a nuclear RNA-binding protein implicated in both familial and sporadic forms of ALS, is specifically expressed in motor neurons that lack ADAR2. This suggests that unedited GluA2 at the Q/R locus is pathogenic in ALS [103]. Some reports suggest that the entry of Ca2+ through AMPA receptors that include unedited GluA2 results in calpain activation, which initiates TDP-43 pathology and deficits in nucleocytoplasmic transport, alongside causing excitotoxicity [104,105]. In other studies, the presence of Q/R site-unedited GluA2 followed by the downregulation of ADAR2 has been observed in ALS patients with mutation in fused in sarcoma (FUSP525L mutation) with alterations of several circRNA expression levels [106,107].
4. RNA Modifications in Glioma
Gliomas account for nearly 80% of malignant brain tumors in adults [108]. They are broadly defined but generally are known to develop from the support cells of the brain called glia. The glial cells of the brain that most commonly undergo gliomagenesis are astrocytes, oligodendrocytes, and ependymal cells. Even with the standard of care, which includes a combination of surgery, radiation, and chemotherapy, gliomas recur at a high rate, which leads to a low overall survival (OS) for patients. The 2021 WHO classification implemented a new grading system for gliomas that integrates histopathological features and molecular diagnostic criteria to classify CNS tumors into more distinct subtypes further, improving approaches to patient care [109]. Although this improves diagnosis and allows for more targeted therapeutic approaches, patient outcomes remain poor, necessitating further molecular characterizations of these tumors to inspire novel treatments.
Epi-transcriptomics has been shown to be involved in the progression and malignancy of various cancer types, including gliomas [110]. m6A RNA methylation is the most heavily studied form of RNA modification. Various studies have shown that m6A RNA modification could inhibit or promote tumor progression depending on factors such as whether the target gene is an oncogene or a tumor suppressor and which components of the m6A methylation regulators are at play. In this section of the review, we explore the various ways that m6A methylation and its regulators, known as “Writers”, “Erasers”, and “Readers”, play roles in the emergence and progression of gliomas and the potential therapeutic approaches that target m6A methylation and its regulators.
4.1. m6A Methylation Regulators “Writers” in Glioma Pathogenesis and Treatment Resistance
METTL3 is a key component of the m6A methyltransferase complex, also comprising METTL14, and is crucial in the m6A methylation of nuclear RNA. It binds specifically to S-adenosyl methionine (SAM), facilitating m6A methylation, while METTL14 plays a supportive role despite its lack of a catalytic site [14,111]. Research on METTL3’s function in glioma has produced varied and sometimes conflicting results. Cui et al.’s initial studies showed that METTL3 knockdown results in increased growth, self-renewal, and tumorigenesis in glioma stem cells (GSCs), which are critical for the growth, invasion, and drug resistance in glioblastoma [112]. Conversely, other research indicates that silencing METTL3 or enhancing its mutated version suppresses GSC growth and proliferation. METTL3 downregulation reduced m6A methylation of serine- and arginine-rich splicing factors (SASFs), leading to YTHDC1-dependent nonsense-mediated decay of SASFs mRNA and lowered protein levels. Additionally, METTL3 influences the splicing of BCL-X and NCOR2, which play roles in cancer cell death and motility [113]. In contrast, Visvanathan et al. observed that overexpressing METTL3 correlates with enhanced GSC stemness and reduced differentiation, with the knockout of METTL3 or METTL14 increasing GSC sensitivity to gamma-irradiation [114]. These discrepancies may be due to different m6A methylation target genes and the specific “readers” recognizing these modifications.
METTL3 is involved in the methylation of non-coding RNAs, including lncRNAs and circRNAs, which are critical in glioma initiation and progression. One study demonstrated that METTL3-mediated methylation of LINC00839, regulated by YTHDF2, enhances its expression in GSC lines. This stabilization activates the Wnt/β-Catenin signaling pathway, increasing radioresistance and GSC proliferation [115]. Additionally, METTL3-induced m6A methylation of LINC01003 was found to regulate the focal adhesion kinase (FAK) pathway, promoting glioma cell proliferation and migration [116]. For circRNAs, Wu and colleagues found that METTL3′s m6A modification process increases the stability and expression of circDLC1 [117]. This elevation aids circDLC1 in its competitive binding with miR-671-5p, which in turn supports the transcription of Catenin Beta Interacting Protein 1 (CTNNBIP1) and ultimately inhibits the excessive growth of glioma cells.
METTL3-mediated m6A methylation is associated with treatment resistance in gliomas. Shi et al. reported that METTL3 methylation is elevated in glioblastoma tissues resistant to temozolomide (TMZ), with METTL3 overexpression increasing the stability of DNA repair enzymes, suggesting a mechanism for TMZ resistance [118]. Additionally, it has been shown that METTL3-mediated methylation indirectly fosters TMZ resistance by stabilizing Oxidized Low-Density Lipoprotein Receptor 1 (OLR1), a receptor involved in lipid metabolism and cellular signaling, and by activating the Wnt/β-Catenin pathway [119].
miR-1208 targets METTL3’s 3′UTR region, diminishing NUP214 levels, and inhibiting glioma cell proliferation [120]. Knockdown of the histone methyltransferase SETD2, which influences m6A modification, results in decreased levels of METTL3/14 and WTAP, reducing glioma cell proliferation and migration [121]. The resistance of glioma cells to mTOR inhibitors is attributed to m6A methylation at IRES sites, enhancing the translation of oncogenes, a process disrupted by METTL3/14 knockout [122]. Under fear stress, increased METTL3 expression stabilizes FSP1 through m6A methylation and inhibits ferroptosis in glioma cells, indicating a role in stress responses [123].
Another central component of the m6A methyltransferase complex is WTAP that has been implicated in various aspects of glioma progression and treatment response. One study found that Flotilin-1 (FLOT1), upregulated in gliomas and associated with advanced progression and poor prognosis, is stabilized by m6A methylation, with WTAP acting as the writer. The silencing of FLOT1 led to reduced glioma cell proliferation, highlighting the significance of WTAP in glioma biology [124]. Additionally, WTAP has been strongly linked to microsatellite instability, indicative of a compromised DNA mismatch repair system. Together with other genes like TRMT6, DNMT1, and DNMT3B, WTAP can predict overall survival in glioma patients and is correlated with poor post-operative outcomes [125]. Increased WTAP expression in glioblastoma compared to normal tissue further underscores its role in glioma pathogenesis [126].
WTAP also plays a role in the regulation of cell proliferation, migration, and apoptosis. A study demonstrated that miR-29a, which is typically underexpressed in GSCs, can inhibit the Quaking gene isoform 6 (QKI-6) and subsequently reduce WTAP expression. This inhibition leads to decreased activity in key pathways like phosphoinositide 3-kinase/AKT and extracellular signal-related kinase, reducing cell proliferation and invasion while promoting apoptosis [127]. Furthermore, WTAP expression levels have been closely correlated with tumor grading levels [128].
The modulation of WTAP expression can directly impact glioma tumorigenicity. Knockdown and overexpression studies have shown that WTAP can regulate epidermal growth factor receptor, influencing tumorigenicity in glioma. This was further confirmed in xenograft models [129].
Other m6A writers in glioma involve KIAA1429, RBM15, and ZC3H13. KIAA1429, also known as VIRMA, is consistently upregulated in glioblastoma and negatively correlated with the response to anti-cancer drugs, suggesting its role in drug resistance and tumor progression [130]. RBM15 is implicated in the proneural to mesenchymal transition (PMT) in GSCs. Studies have shown that neuronal activation can lead to changes in GSC behavior via miR-184-3p mediated inhibition of RBM15 expression, thus impacting radioresistance and progression. RBM15 knockdown led to decreased m6A modification and DLG3 mRNA levels, which in turn increased p-STAT3, a key signaling molecule in PMT [131]. Additionally, RBM15 expression has been shown to have prognostic value in glioma, particularly in predicting overall survival in patients with low-grade glioma (LGG) [132,133]. ZC3H13 plays a role in glioblastoma progression through its interaction with the tumor microenvironment. Under hypoxic conditions, ZC3H13 expression in microglia is influenced by neuron-derived exosomes, leading to changes in microglial polarization and subsequently affecting glioblastoma progression [134]. A prognostic model has indicated that ZC3H13 levels are positively associated with glioblastoma prognosis, suggesting its potential as a tumor suppressor [126]. Furthermore, the knockdown of ZC3H13 has been linked to increased TMZ resistance in Rb1 mutant glioblastoma cells [135]. In advancing glioma treatment, investigating m6A RNA methylation’s role in GSC biology, especially regarding stem cell maintenance and drug resistance, is a promising avenue. Such research would entail studying the impact of altering m6A writers such as METTL3, WTAP, VIRMA, RBM15, and ZC3H13 on GSCs. Employing CRISPR-Cas9 gene editing or RNA interference, modifications in m6A writers could be analyzed for their effects on GSC characteristics, including proliferation, differentiation, and survival under chemotherapy and radiotherapy. Although small molecule inhibitors of these writers have been designed and found to have promising results in other cancers, such as acute myeloid leukemia, they have not been heavily investigated in gliomas. These investigations are expected to uncover new molecular pathways affected by m6A modifications, leading to novel therapeutic targets that could effectively prevent glioma recurrence and contribute to improved glioma treatments.
4.2. m6A Methylation Regulators “Erasers” in Glioma Pathogenesis and Treatment Resistance
The role of erasers in m6A methylation, particularly in the context of glioma, is vital for understanding their influence on tumor progression and therapeutic resistance. Particularly, the erasers FTO and ALKBH5 exhibit a range of activities impacting various aspects of glioma biology. FTO, known for its role in fat consumption and overall metabolic rate regulation [136], also acts as a demethylase for m6A, affecting pre-mRNAs’ alternative splicing and 3′-end processing [137]. FTO’s influence extends to mitochondrial functions, affecting the expression of genes like SDHA and regulating the STAT3/FTO axis [138]. Prognostic studies suggest that FTO can predict poor outcomes in glioma patients, and inhibitors targeting FTO have shown promise in reducing tumorigenicity and aggressiveness in glioma models [139,140,141,142]. FTO expression is also linked to the decreased apoptosis and increased proliferation of glioma cells [143], and its inhibition suppresses GSC growth and self-renewal [112]. Intriguingly, FTO knockdown affects the nuclear localization of FOXO3a, influencing the expression of target genes like BIM, BNIP3, and BCL-6 [144].
FOXM1 is a transcription factor essential for tumorigenicity and invasion [145]. Silencing ALKBH5 suppresses FOXM1 and GSC proliferation, and its inhibitors have shown effectiveness in decreasing glioma cell proliferation [146,147]. ALKBH5 overexpression in glioblastoma stem cells contributes to increased resistance to radiation and enhanced invasive capabilities, and its activity is regulated by EGFR signaling, impacting ferroptosis through m6A modulation [148,149]. ALKBH5 also promotes PYCR2 expression, influencing glioma cell proliferation, migration, and PMT [150]. The regulation of ALKBH5 by USP36 underscores its role in glioblastoma progression and sensitivity to TMZ [151]. Its involvement in the proliferation, migration, and invasion of glioma cells has been well-established [152,153], and inhibitors targeting ALKBH5 reduce glioma cell migration and invasiveness [154]. Additionally, ALKBH5 influences immune responses in glioma, affecting cytokine expression and Programmed Death-Ligand 1 (PD-L1) protein levels [155]. The hypoxia-induced activity of ALKBH5 stabilizes transcripts like NEAT1, facilitating tumor-associated macrophage recruitment and immunosuppression, and enhances glioma cell growth by activating pathways like PPP [156,157]. In the context of TMZ resistance, ALKBH5 demethylates transcripts like NANOG, contributing to the development of resistance, and regulates TMZ sensitivity by interacting with transcripts like SOX2 [158,159].
Given the roles of FTO and ALKBH5 in modulating cell death in glioma, focusing on how these m6A erasers influence apoptosis and ferroptosis, especially in GSCs, could be valuable. Exploring the interplay between apoptosis inhibition in FTO and ferroptosis resistance in ALKBH5, particularly under radiotherapy, using gene editing techniques, offers promising insights. Such research could pave the way for innovative treatments aimed at overcoming radio-resistance and curbing GSC proliferation, ultimately improving the efficacy of glioma therapies. Moreover, further investigation is needed for inhibitors of these erasers to be fully established as promising treatment options.
4.3. m6A Methylation Regulators “Readers” in Glioma Pathogenesis and Treatment Resistance
In the intricate network of glioma pathogenesis, the YTHDF family, consisting of YTHDF1, YTHDF2, and YTHDF3, and YTHDC1 and YTHDC2, as members of the YTH domain-containing family, play significant roles in the prognosis and molecular mechanisms underlying glioma [158]. The YTHDC1 protein is more nuclear-focused, influencing mRNA splicing and export, whereas the YTHDC2 and YTHDF proteins are primarily involved in controlling the stability, degradation, and translation of m6A-modified mRNAs in the cytoplasm [160,161,162,163].
YTHDF1 is involved in various mechanisms that contribute to glioma resistance and progression. For instance, it stabilizes OLR1 mRNA, influencing the OLR1-mediated Wnt/β-Catenin pathway activation, which is linked to TMZ resistance in glioma [119]. Additionally, YTHDF1 plays a role in RNA editing, as it binds and promotes the translation of m6A-modified ADAR1, a molecule implicated in glioma progression [164]. YTHDF1’s expression, increased by C-myc overexpression, leads to higher levels of FDX1, a protein associated with several cancer signaling pathways [159]. Moreover, the protein Musashi-1, overexpressed in glioblastoma, upregulates YTHDF1, thereby sensitizing glioblastoma cells to TMZ and inhibiting their proliferation [165]. YTHDF2 is particularly significant in maintaining oncogene expression in glioblastoma stem cells. GSCs express YTHDF2 preferentially, and its targeting can inhibit cell growth and viability, suggesting the potential of the YTHDF2-MYC-IGFBP3 axis as a therapeutic target [156]. Furthermore, YTHDF2’s role extends to immune regulation, where its deficiency impairs the stability of ZDHHC3 mRNA, affecting PD-L1 expression and degradation in glioma [148]. YTHDF2 is also involved in key signaling pathways, including the receptor tyrosine kinase MET pathway, which is essential for glioblastoma stem cell renewal and tumorigenicity [166]. Its expression correlates with various immune cells in low-grade gliomas, and it has been shown to enhance TMZ resistance in glioblastoma [167,168]. Additionally, YTHDF2 promotes glioblastoma cell proliferation and tumorigenesis, largely through the downregulation of LXRα and HIVEP2 [169]. YTHDF3 has emerged as a potential target for treating Osimertinib-resistant glioblastoma cells. Research indicates that silencing YTHDF3 increases the sensitivity of these cells to Osimertinib, affecting their migratory and sphere-forming abilities [157]. Moreover, YTHDF3’s role in mTOR inhibitor resistance involves the methylation of IRES-mediated mRNA translation, highlighting its importance in resistance mechanisms [122].
On the other hand, YTHDC1 has been determined to be a prognostic marker for overall survival in patients with low-grade glioma (LGG) [133]. It plays a role in the regulation of key molecules and pathways that influence glioma cell behavior. For instance, YTHDC1 interacts with the circRNAs from the EPHB4 gene, which is implicated in glioma. METTL3-mediated m6A methylation of CircEPHB4 leads to its recognition by YTHDC1, which then localizes the transcript to the cytoplasm. This localization facilitates the stabilization of SOX2 mRNA, promoting the transcription of PHLDB2, a molecule associated with epithelial–mesenchymal transformation in various cancers. This mechanism underscores the role of YTHDC1 in enhancing stemness in glioma spheres, as evidenced by the increased expression of stemness marker proteins and the promotion of cell proliferation, invasion, and migration [170,171,172]. Additionally, YTHDC1 influences glioma cell proliferation through its impact on VPS25, a protein upregulated in gliomas. Knockdown of YTHDC1 leads to decreased VPS25 levels and reduced cell proliferation [173]. Furthermore, YTHDC1 is implicated in the non-sense mediated mRNA decay (NMD) of SASFs, a process dependent on the m6A methylation status of these factors [113]. YTHDC2, similarly, is crucial in the context of LGG. It is overexpressed in these tumors and correlates with patient prognosis, where higher levels of YTHDC2 are associated with poorer outcomes [174]. Moreover, YTHDC2 has been identified as an independent negative prognostic indicator for overall survival in gliomas. This finding, derived from Cox regression multivariate analysis, suggests that YTHDC2 levels could be a significant marker in evaluating the progression and potential outcomes of glioma treatments [175]. YTHDC1 and YTHDC2 are integral components in the molecular landscape of glioma, influencing various aspects of tumor biology. Their roles in RNA processing, interaction with circRNAs, and impact on key signaling pathways underscore their potential as prognostic biomarkers and as targets for therapeutic intervention in glioma.
HNRNPC and HNRNPA2B1 are extensively expressed m6A regulators in the tumor microenvironment, suggesting their importance in a variety of tumor-relevant cell types [176]. HNRNPC shows significant expression in glioblastoma, serving as an essential splicing factor. Higher expression levels in glioblastoma tissues compared to normal tissues are associated with a poorer prognosis in high-risk patients. Additionally, HNRNPC expression positively correlates with PD-L1, highlighting its prognostic significance and role in immune modulation [166]. HNRNPC interacts with long non-coding RNA DDX11 antisense RNA 1 (DDX11-AS1) to promote the Wnt/β-Catenin and AKT pathways, influencing the epithelial–mesenchymal transition (EMT) and glioma cell migration. Knockdown of HNRNPC disrupts these pathways and hinders EMT. Furthermore, HNRNPC regulates microRNA-21 (miR-21) expression, thereby affecting glioblastoma progression via Programmed Cell Death 4 (PDCD4) [177,178]. HNRNPA2B1 is identified as an independent prognostic factor for glioma. HNRNPA2B1 promotes GSC self-renewal and tumorigenesis by modulating cholesterol biosynthesis, notably through the stabilization of SREBP2 mRNA. This stabilization boosts the expression of HMGCR and LDLR mRNA, key components in cholesterol biosynthesis and reuptake. Combining HNRNPA2B1 suppression with cholesterol metabolism drugs yields potent inhibitory effects on glioma cells [179]. HNRNPA2B1 also plays a role where circular RNA from the NEIL3 gene (known as circNEIL3) is packed into exosomes and transferred to tumor-associated macrophages within the tumor microenvironment [180]. This transfer promotes the suppression of the immune response by stabilizing IGF2BP3, thereby facilitating glioma progression. HNRNPA2B1 modulates cell cycle dynamics, apoptosis, and treatment responses, particularly to β-asarone. Knockdown of HNRNPA2B1 reduces cell proliferation and increases apoptosis, affecting signaling pathways such as AKT and STAT3. This modulation alters the expression of proteins like B-cell lymphoma-2 (Bcl-2), CyclinD1, and PCNA, underscoring the role of HNRNPA2B1 in glioma cell proliferation and its therapeutic potential [181,182,183].
The Insulin-like Growth Factor 2 mRNA-Binding Proteins (IGF2BP) family, comprising IGF2BP1, IGF2BP2, and IGF2BP3, plays a crucial role in glioma development and response to treatment. IGF2BP1 is upregulated in mesenchymal glioblastoma compared to proneural glioblastoma, correlating with poor patient outcomes. Its overexpression in proneural glioblastoma increases cell stemness, while its knockdown in mesenchymal glioblastoma leads to decreased proliferation and sphere formation. IGF2BP1 specifically binds m6A on YAP mRNA to stabilize it, increasing protein expression and activating the YAP/TAZ complex involved in the Hippo signaling pathway. This indicates a feed-forward loop enhancing tumorigenicity and stemness, particularly in mesenchymal glioblastoma [184]. Furthermore, IGF2BP1 is targeted by various non-coding RNAs and microRNAs, such as LINC00689, PCAT6, Lnc-THOR, and miR-4500, which regulate its expression and thus impact glioma progression and apoptosis [185,186,187,188,189,190]. IGF2BP2’s involvement in glioma includes interactions with various RNA elements and transcription factors. For instance, it participates in vascular mimicry through its interaction with HOTAIRM1 [191] and engages in the LINC00265/miR-let-7d-5p/IFI30/ZNF384/IGF2BP2 axis to regulate EMT and stemness [192]. The protein complex formed between HOXD-AS2 and IGF2BP2 is associated with poorer prognosis in glioma, with STAT3 playing a role in this feedback loop [193]. Additionally, IGF2BP2 stabilizes transcripts like NUP214 and FLOT1, influencing glioma progression and TMZ resistance [120,124,194,195,196,197,198]. In glioma endothelial cells, IGF2BP3, in combination with METTL3, stabilizes CPEB2, maintaining the blood–tumor barrier and thereby influencing drug delivery [199]. It is also targeted by YTHDF2, influencing oncogene expression in GSCs [164]. Compared to other IGF2BPs, IGF2BP3 shows a higher correlation with stemness markers and immune infiltration in gliomas. Knockdown studies in glioma cells reveal that IGF2BP3 is integral to cell proliferation, invasion, and migration [200].
Another important m6A reader is eukaryotic initiation factor 3 (eIF3), which is crucial in the initiation of mRNA translation and exhibits significant implications in gliomas. eIF3 is unique in its ability to recognize m6A modifications on the 5′UTR of mRNA. This recognition allows for the initiation of mRNA translation independent of the conventional 5′ 7-methylguanosine (m7G) cap recognition by eukaryotic initiation factor 4E (eIF4E). eIF3 recruits the 43S preinitiation complex, which includes the 40S ribosomal subunit and eIF1A, eIF1, eIF2, and eIF3, initiating the translation of m6A-modified mRNA [201]. In U251 glioma cells, the knockdown of eIF3 subunit e led to an increase in mRNAs related to the p53 pathway, such as FAS and GADD45α, and a decrease in mRNAs related to survival and DNA replication/repair, such as UBE2V1, FGF11, CDC45, and JAK3. Furthermore, silencing of the eIF3 subunit e resulted in increased radiosensitivity in LN18 and U251 glioblastoma cell lines, suggesting a potential therapeutic avenue [202]. Targeting the eIF3 subunit c in U87 cells via siRNA led to suppressed cell proliferation, reduced colony formation, arrested cell cycle progression at the G0/G1 phase, promoted apoptosis, and prevented tumorsphere formation in xenograft models. Similar effects were observed in U251 cells, where knockdown of the eIF3 subunit c decreased proliferation and increased apoptosis [203,204]. Studies targeting the eIF3 subunit b in U87 cells and subunit d in both U87 and U251 cells showed that the knockdown of these subunits impeded glioma cell growth and proliferation, indicating their crucial role in glioma cell survival [205,206]. While not directly related to m6A methylation reading, the various subunits of eIF3 have potential prognostic value in gliomas. The expression levels of these subunits correlate with the severity and progression of the disease, suggesting their usefulness as biomarkers for glioma prognosis [207]. eIF3, particularly its various subunits, plays a significant role in the molecular mechanisms of glioma, influencing cell proliferation, survival, and response to treatments such as radiotherapy. The ability of eIF3 to initiate translation of m6A-modified mRNA independent of the m7G cap adds another layer of complexity to its role in glioma biology, presenting potential targets for therapeutic intervention. Targeting m6A reader proteins, like eIF3, to enhance radiotherapy sensitivity and modulate YTHDF, YTHDC, and HNRNP proteins to counteract chemotherapeutic resistance represents future research avenues to improve glioma treatment efficacy. Additionally, investigating the IGF2BP family’s influence on stemness and immune evasion could lead to novel interventions. The development of biomarkers to quantify these proteins’ expression will facilitate personalized treatment approaches.
4.4. Other RNA Modifications in Glioma Pathogenesis and Treatment Resistance
Knowledge of other RNA modifications in glioma pathogenesis is still limited. Some reports have demonstrated the influence of RNA m5C modifications on glioma’s biological characteristics, including proliferation, differentiation, migration, and malignancy [208]. Regulators of m5C, including NOP2, NSUN4, NSUN5, and NSUN7, have been linked to poor prognosis, while NSUN6 is associated with better outcomes [209]. In the glioblastoma cell line U87, NSUN2 has been shown to facilitate cell migration through the enhancement of mRNA cytosine methylation [210]. Compared to healthy tissue, glioma specimens show varied expression of m5C regulators, including DNMTs, NSUNs, TETs, YTHDF2, ALYREF, and YBX1 [211].
A-to-I editing is another significant RNA modification in glioma. It involves the re-editing of the glutamate receptor subunit B at the Q/R site, crucial for receptor functionality in the central nervous system [212]. Low-activity ADAR2, prevalent in gliomas, leads to reduced RNA editing of the GluA2 subunit at the Q/R site [213,214]. Similarly, ADAR3, a brain-specific adenosine deaminase, plays a comparable role in gliomas [215]. Decreased ADAR3 expression correlates with glioma progression, and A-to-I editing has been extensively linked to glioblastoma cell proliferation, migration, and invasion [216,217,218]. In terms of treatment resistance, experimental findings have revealed that increased expression of ADAR3 enhances the resistance to TMZ [219]. This expression alteration affects 641 genes, primarily regulated by NF-κB signaling pathways. Additionally, GSCs are central to A-to-I RNA editing in glioblastoma, with ADARs influencing GSC self-renewal and stem-like traits, potentially affecting the response to TMZ.
The role of other RNA modifications, such as m7G RNA methylation, in glioma, is still being unraveled. Research has indicated that out of 31 m7G methylation regulators studied, 17 exhibited higher expression levels in gliomas [220]. Moreover, an imbalance in m1A regulators is closely related to glioma onset and progression [220,221]. The inhibition of TRMT6, an m1A methyltransferase, impairs glioma cell proliferation, migration, and invasion (Figure 2).
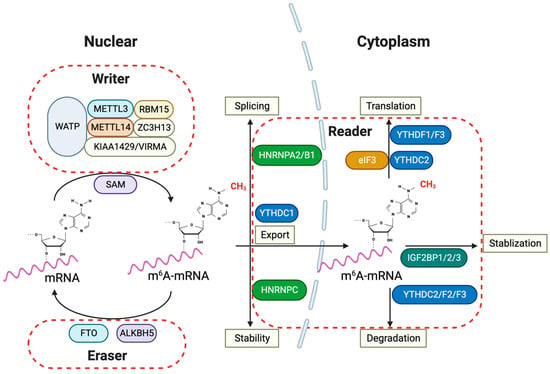
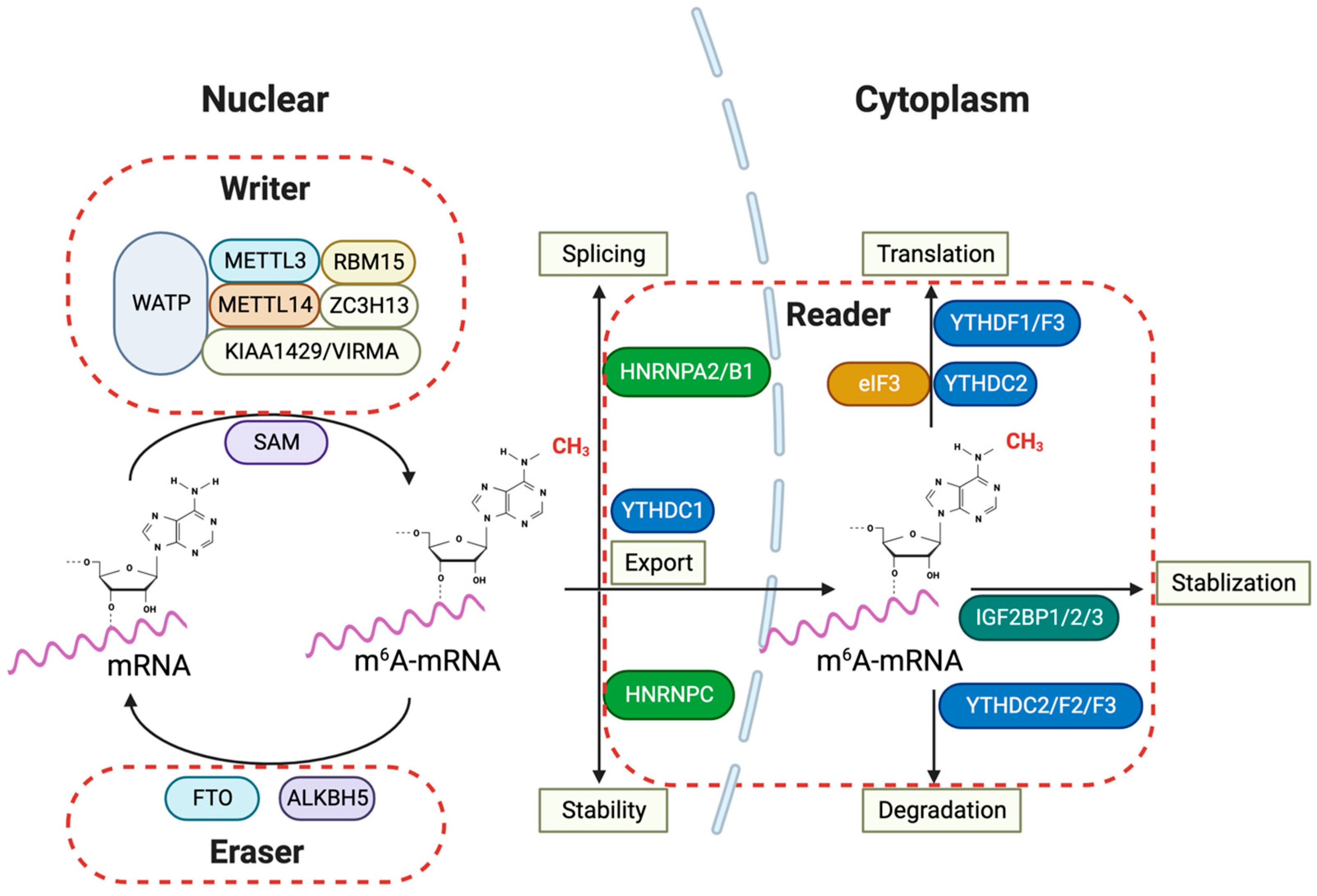
Figure 2.
m6A RNA modification and its regulatory proteins in glioma pathogenesis. The m6A RNA modification pathways implied for glioma pathogenesis involve various nuclear and cytoplasmic processes in the cells. The ‘writers’ of the m6A mark, which include METTL3, METTL14, WTAP, RBM15, ZC3H13, and KIAA1429/VIRMA, collectively form the methyltransferase complex in the nucleus, utilizing SAM as a methyl donor to catalyze mRNA methylation. Conversely, ‘erasers’ such as FTO and ALKBH5 demethylate m6A-mRNA, reversing the modifications introduced by the writers. Within the cytoplasm, ‘readers’ such as YTHDFs, YTHDC2, HNRNPC, and the IGF2BP family recognize and bind to m6A-mRNA, affecting its stability, nuclear export, splicing, and translation. The involvement of IGF2BPs indicates their role in stabilizing m6A-mRNA, which is crucial for regulating gene expression that is vital for glioma progression. Furthermore, proteins like HNRNPs and eIF3 are implicated in splicing and translation, underscoring the complexity and importance of the m6A modification system in glioma pathogenesis. METTL3: Methyltransferase Like 3; METTL14: Methyltransferase Like 14; WTAP: Wilms’ Tumor 1-Associating Protein; RBM15: RNA Binding Motif Protein 15; ZC3H13: Zinc Finger CCCH-Type Containing 13; KIAA1429/VIRMA: Vir Like m6A Methyltransferase Associated; SAM: S-adenosylmethionine; FTO: Fat Mass and Obesity-Associated Protein; ALKBH5: AlkB Homolog 5; YTHDF: YTH Domain Family; YTHDC: YTH Domain Containing; HNRNP: Heterogeneous Nuclear Ribonucleoprotein; IGF2BP: Insulin-like Growth Factor 2 mRNA-Binding Proteins; eIF3: Eukaryotic Translation Initiation Factor 3.
5. Conclusions
In summary, the rapidly advancing field of epitranscriptomics, particularly RNA modifications, has gained significant attention as a novel focus in understanding and potentially targeting neurological diseases. This review has highlighted the essential role of RNA modification in various diseases, emphasizing its impact on mRNA stability, translation, and the control of protein levels in disease-associated pathways. Specifically, m6A modification in glial cells is crucial for the onset and progression of neurological diseases like Alzheimer’s disease and glioma. However, despite the current focus on DNA methylation, histone modifications, and chromatin rearrangement in neurological diseases, the critical biological functions of RNA modification have been relatively underexplored and warrant further investigation. Additionally, while m6A modification is increasingly studied in various fields, a significant number of enzymes responsible for these modifications—known as writers, erasers, and readers—remain unidentified. This holds significant pertinence when considering neurodegenerative diseases like Parkinson’s disease (PD), where m6A-modification genes have been identified, but their association with the disease needs more comprehensive study. Therefore, future efforts should delve deeper into the role of RNA modifications in the nervous system, which could reveal new molecular targets for pharmacological and clinical therapy development for uncurable neurological diseases.
Author Contributions
Conceptualization, A.K. and A.N.; writing-original draft preparation, A.K. and Y.K.; writing-review and editing, A.N., K.Y. and H.W.; supervision, K.Y., H.W. and S.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by funding from the Overseas Assistance Fellowship program from Nakatani Foundation for Advancement of Measuring Technologies in Biomedical Engineering (Y.K.), Health Science Research Grant for Young Investigators from Meiji Yasuda Life Foundation of Health and Welfare (A.K.), Overseas Research Fellowship from Uehara Memorial Foundation (A.K. and Y.K.), and Grant-in-Aid for Challenging Research (Exploratory) from the Japan Society for the Promotion of Science 23K18338 (S.T.).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Li, S.; Mason, C.E. The Pivotal Regulatory Landscape of RNA Modifications. Annu. Rev. Genom. Hum. Genet. 2014, 15, 127–150. [Google Scholar] [CrossRef] [PubMed]
- Cerneckis, J.; Ming, G.-L.; Song, H.; He, C.; Shi, Y. The Rise of Epitranscriptomics: Recent Developments and Future Directions. Trends Pharmacol. Sci. 2023, 45, P24–P28. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Jaffrey, S.R.; Pan, T.; Rechavi, G.; Suzuki, T. RNA Modifications: What Have We Learned and Where Are We Headed? Nat. Rev. Genet. 2016, 17, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Dubin, D.T.; Taylor, R.H. The Methylation State of Poly A-Containing Messenger RNA from Cultured Hamster Cells. Nucleic Acids Res. 1975, 2, 1653–1668. [Google Scholar] [CrossRef]
- Molinie, B.; Wang, J.; Lim, K.S.; Hillebrand, R.; Lu, Z.-X.; Van Wittenberghe, N.; Howard, B.D.; Daneshvar, K.; Mullen, A.C.; Dedon, P.; et al. M(6)A-LAIC-Seq Reveals the Census and Complexity of the m(6)A Epitranscriptome. Nat. Methods 2016, 13, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Liu, S.; Peng, Y.; Ge, R.; Su, R.; Senevirathne, C.; Harada, B.T.; Dai, Q.; Wei, J.; Zhang, L.; et al. M6A RNA Modifications Are Measured at Single-Base Resolution across the Mammalian Transcriptome. Nat. Biotechnol. 2022, 40, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-Nucleotide-Resolution Mapping of M6A and M6Am throughout the Transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the Human and Mouse M6A RNA Methylomes Revealed by M6A-Seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of MRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Brown, J.A.; Kinzig, C.G.; DeGregorio, S.J.; Steitz, J.A. Methyltransferase-like Protein 16 Binds the 3′-Terminal Triple Helix of MALAT1 Long Noncoding RNA. Proc. Natl. Acad. Sci. USA 2016, 113, 14013–14018. [Google Scholar] [CrossRef]
- Amodio, N.; Raimondi, L.; Juli, G.; Stamato, M.A.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. MALAT1: A Druggable Long Non-Coding RNA for Targeted Anti-Cancer Approaches. J. Hematol. Oncol. 2018, 11, 63. [Google Scholar] [CrossRef]
- Patil, D.P.; Chen, C.-K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. M6A RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Knuckles, P.; Bühler, M. Adenosine Methylation as a Molecular Imprint Defining the Fate of RNA. FEBS Lett. 2018, 592, 2845–2859. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Warda, A.S.; Kretschmer, J.; Hackert, P.; Lenz, C.; Urlaub, H.; Höbartner, C.; Sloan, K.E.; Bohnsack, M.T. Human METTL16 Is a N6-methyladenosine (M6A) Methyltransferase That Targets Pre-mRNAs and Various Non-coding RNAs. EMBO Rep. 2017, 18, 2004–2014. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA M6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038.e6. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA Mediates Preferential M6A MRNA Methylation in 3′UTR and near Stop Codon and Associates with Alternative Polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Luo, G.-Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 Mediates Nuclear Export of N6-Methyladenosine Methylated MRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y.; et al. Extensive Translation of Circular RNAs Driven by N6-Methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-Transcriptional Gene Regulation by MRNA Modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Dunn, D.B. The Occurrence of 1-Methyladenine in Ribonucleic Acid. Biochim. Biophys. Acta 1961, 46, 198–200. [Google Scholar] [CrossRef]
- Dominissini, D.; Nachtergaele, S.; Moshitch-Moshkovitz, S.; Peer, E.; Kol, N.; Ben-Haim, M.S.; Dai, Q.; Di Segni, A.; Salmon-Divon, M.; Clark, W.C.; et al. The Dynamic N(1)-Methyladenosine Methylome in Eukaryotic Messenger RNA. Nature 2016, 530, 441–446. [Google Scholar] [CrossRef]
- Li, X.; Xiong, X.; Wang, K.; Wang, L.; Shu, X.; Ma, S.; Yi, C. Transcriptome-Wide Mapping Reveals Reversible and Dynamic N(1)-Methyladenosine Methylome. Nat. Chem. Biol. 2016, 12, 311–316. [Google Scholar] [CrossRef]
- Zhou, H.; Kimsey, I.J.; Nikolova, E.N.; Sathyamoorthy, B.; Grazioli, G.; McSally, J.; Bai, T.; Wunderlich, C.H.; Kreutz, C.; Andricioaei, I.; et al. M(1)A and m(1)G Disrupt A-RNA Structure through the Intrinsic Instability of Hoogsteen Base Pairs. Nat. Struct. Mol. Biol. 2016, 23, 803–810. [Google Scholar] [CrossRef]
- Safra, M.; Sas-Chen, A.; Nir, R.; Winkler, R.; Nachshon, A.; Bar-Yaacov, D.; Erlacher, M.; Rossmanith, W.; Stern-Ginossar, N.; Schwartz, S. The M1A Landscape on Cytosolic and Mitochondrial MRNA at Single-Base Resolution. Nature 2017, 551, 251–255. [Google Scholar] [CrossRef]
- Aas, P.A.; Otterlei, M.; Falnes, P.O.; Vågbø, C.B.; Skorpen, F.; Akbari, M.; Sundheim, O.; Bjørås, M.; Slupphaug, G.; Seeberg, E.; et al. Human and Bacterial Oxidative Demethylases Repair Alkylation Damage in Both RNA and DNA. Nature 2003, 421, 859–863. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Clark, W.; Luo, G.; Wang, X.; Fu, Y.; Wei, J.; Wang, X.; Hao, Z.; Dai, Q.; Zheng, G.; et al. ALKBH1-Mediated TRNA Demethylation Regulates Translation. Cell 2016, 167, 1897. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Wang, T.; Gonzalez, G.; Wang, Y. Identification of YTH Domain-Containing Proteins as the Readers for N1-Methyladenosine in RNA. Anal. Chem. 2018, 90, 6380–6384. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread Occurrence of 5-Methylcytosine in Human Coding and Non-Coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef] [PubMed]
- Amort, T.; Souliere, M.F.; Wille, A.; Jia, X.Y.; Fiegl, H.; Worle, H. Long Noncoding RNAs as Targets for Cytosine Methylation. RNA Biol. 2013, 10, 1002–1008. [Google Scholar] [CrossRef] [PubMed]
- Reid, R.; Greene, P.J.; Santi, D.V. Exposition of a Family of RNA m(5)C Methyltransferases from Searching Genomic and Proteomic Sequences. Nucleic Acids Res. 1999, 27, 3138–3145. [Google Scholar] [CrossRef] [PubMed]
- Van Haute, L.; Lee, S.-Y.; McCann, B.J.; Powell, C.A.; Bansal, D.; Vasiliauskaitė, L.; Garone, C.; Shin, S.; Kim, J.-S.; Frye, M.; et al. NSUN2 Introduces 5-Methylcytosines in Mammalian Mitochondrial TRNAs. Nucleic Acids Res. 2019, 47, 8720–8733. [Google Scholar] [CrossRef]
- Bohnsack, K.E.; Höbartner, C.; Bohnsack, M.T. Eukaryotic 5-Methylcytosine (M5C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes 2019, 10, 102. [Google Scholar] [CrossRef]
- Yang, X.; Yang, Y.; Sun, B.-F.; Chen, Y.-S.; Xu, J.-W.; Lai, W.-Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-Methylcytosine Promotes MRNA Export-NSUN2 as the Methyltransferase and ALYREF as an M5C Reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, L.; Han, X.; Yang, W.-L.; Zhang, M.; Ma, H.-L.; Sun, B.-F.; Li, A.; Xia, J.; Chen, J.; et al. RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal MRNA Decay. Mol. Cell 2019, 75, 1188–1202.e11. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Lan, M.-D.; Qi, C.-B.; Zheng, S.-J.; Wei, S.-Z.; Yuan, B.-F.; Feng, Y.-Q. Formation and Determination of the Oxidation Products of 5-Methylcytosine in RNA. Chem. Sci. 2016, 7, 5495–5502. [Google Scholar] [CrossRef]
- Schosserer, M.; Minois, N.; Angerer, T.B.; Amring, M.; Dellago, H.; Harreither, E.; Calle-Perez, A.; Pircher, A.; Gerstl, M.P.; Pfeifenberger, S.; et al. Methylation of Ribosomal RNA by NSUN5 Is a Conserved Mechanism Modulating Organismal Lifespan. Nat. Commun. 2015, 6, 6158. [Google Scholar] [CrossRef]
- Shanmugam, R.; Fierer, J.; Kaiser, S.; Helm, M.; Jurkowski, T.P.; Jeltsch, A. Cytosine Methylation of TRNA-Asp by DNMT2 Has a Role in Translation of Proteins Containing Poly-Asp Sequences. Cell Discov. 2015, 1, 15010. [Google Scholar] [CrossRef]
- Tuorto, F.; Herbst, F.; Alerasool, N.; Bender, S.; Popp, O.; Federico, G.; Reitter, S.; Liebers, R.; Stoecklin, G.; Gröne, H.-J.; et al. The TRNA Methyltransferase Dnmt2 Is Required for Accurate Polypeptide Synthesis during Haematopoiesis. EMBO J. 2015, 34, 2350–2362. [Google Scholar] [CrossRef]
- Schumann, U.; Zhang, H.-N.; Sibbritt, T.; Pan, A.; Horvath, A.; Gross, S.; Clark, S.J.; Yang, L.; Preiss, T. Multiple Links between 5-Methylcytosine Content of MRNA and Translation. BMC Biol. 2020, 18, 40. [Google Scholar] [CrossRef]
- Cui, X.; Liang, Z.; Shen, L.; Zhang, Q.; Bao, S.; Geng, Y.; Zhang, B.; Leo, V.; Vardy, L.A.; Lu, T.; et al. 5-Methylcytosine RNA Methylation in Arabidopsis Thaliana. Mol. Plant 2017, 10, 1387–1399. [Google Scholar] [CrossRef]
- Huang, T.; Chen, W.; Liu, J.; Gu, N.; Zhang, R. Genome-Wide Identification of MRNA 5-Methylcytosine in Mammals. Nat. Struct. Mol. Biol. 2019, 26, 380–388. [Google Scholar] [CrossRef]
- Li, Q.; Li, X.; Tang, H.; Jiang, B.; Dou, Y.; Gorospe, M.; Wang, W. NSUN2-Mediated M5C Methylation and METTL3/METTL14-Mediated M6A Methylation Cooperatively Enhance P21 Translation. J. Cell. Biochem. 2017, 118, 2587–2598. [Google Scholar] [CrossRef]
- Blaze, J.; Navickas, A.; Phillips, H.L.; Heissel, S.; Plaza-Jennings, A.; Miglani, S.; Asgharian, H.; Foo, M.; Katanski, C.D.; Watkins, C.P.; et al. Neuronal Nsun2 Deficiency Produces TRNA Epitranscriptomic Alterations and Proteomic Shifts Impacting Synaptic Signaling and Behavior. Nat. Commun. 2021, 12, 4913. [Google Scholar]
- Flores, J.V.; Cordero-Espinoza, L.; Oeztuerk-Winder, F.; Andersson-Rolf, A.; Selmi, T.; Blanco, S.; Tailor, J.; Dietmann, S.; Frye, M. Cytosine-5 RNA Methylation Regulates Neural Stem Cell Differentiation and Motility. Stem Cell Rep. 2017, 8, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R.; Zander, S.; Gutschner, T. The Dark Side of the Epitranscriptome: Chemical Modifications in Long Non-Coding RNAs. Int. J. Mol. Sci. 2017, 18, 2387. [Google Scholar] [CrossRef] [PubMed]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine Profiling Reveals Regulated MRNA Pseudouridylation in Yeast and Human Cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Borchardt, E.K.; Martinez, N.M.; Gilbert, W.V. Regulation and Function of RNA Pseudouridylation in Human Cells. Annu. Rev. Genet. 2020, 54, 309–336. [Google Scholar] [CrossRef] [PubMed]
- Angelova, M.T.; Dimitrova, D.G.; Dinges, N.; Lence, T.; Worpenberg, L.; Carré, C.; Roignant, J.-Y. The Emerging Field of Epitranscriptomics in Neurodevelopmental and Neuronal Disorders. Front. Bioeng. Biotechnol. 2018, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Nainar, S.; Marshall, P.R.; Tyler, C.R.; Spitale, R.C.; Bredy, T.W. Evolving Insights into RNA Modifications and Their Functional Diversity in the Brain. Nat. Neurosci. 2016, 19, 1292–1298. [Google Scholar] [CrossRef]
- Eyler, D.E.; Franco, M.K.; Batool, Z.; Wu, M.Z.; Dubuke, M.L.; Dobosz-Bartoszek, M.; Jones, J.D.; Polikanov, Y.S.; Roy, B.; Koutmou, K.S. Pseudouridinylation of MRNA Coding Sequences Alters Translation. Proc. Natl. Acad. Sci. USA 2019, 116, 23068–23074. [Google Scholar] [CrossRef] [PubMed]
- Fernández, I.S.; Ng, C.L.; Kelley, A.C.; Wu, G.; Yu, Y.-T.; Ramakrishnan, V. Unusual Base Pairing during the Decoding of a Stop Codon by the Ribosome. Nature 2013, 500, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Karijolich, J.; Yu, Y.-T. Converting Nonsense Codons into Sense Codons by Targeted Pseudouridylation. Nature 2011, 474, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.X.; So, E.; Devlin, J.L.; Zhao, Y.; Wu, M.; Cheung, V.G. ADAR Regulates RNA Editing, Transcript Stability, and Gene Expression. Cell Rep. 2013, 5, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Solomon, O.; Di Segni, A.; Cesarkas, K.; Porath, H.T.; Marcu-Malina, V.; Mizrahi, O.; Stern-Ginossar, N.; Kol, N.; Farage-Barhom, S.; Glick-Saar, E.; et al. RNA Editing by ADAR1 Leads to Context-Dependent Transcriptome-Wide Changes in RNA Secondary Structure. Nat. Commun. 2017, 8, 1440. [Google Scholar] [CrossRef] [PubMed]
- Gassner, F.J.; Zaborsky, N.; Buchumenski, I.; Levanon, E.Y.; Gatterbauer, M.; Schubert, M.; Rauscher, S.; Hebenstreit, D.; Nadeu, F.; Campo, E.; et al. RNA Editing Contributes to Epitranscriptome Diversity in Chronic Lymphocytic Leukemia. Leukemia 2021, 35, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Xu, X.; Wang, Y.; Hawke, D.H.; Yu, S.; Han, L.; Zhou, Z.; Mojumdar, K.; Jeong, K.J.; Labrie, M.; et al. A-to-I RNA Editing Contributes to Proteomic Diversity in Cancer. Cancer Cell 2018, 33, 817–828.e7. [Google Scholar] [CrossRef] [PubMed]
- Correia de Sousa, M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering MiRNAs’ Action through MiRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef]
- Kawahara, Y.; Zinshteyn, B.; Sethupathy, P.; Iizasa, H.; Hatzigeorgiou, A.G.; Nishikura, K. Redirection of Silencing Targets by Adenosine-to-Inosine Editing of MiRNAs. Science 2007, 315, 1137–1140. [Google Scholar] [CrossRef]
- Yang, W.; Chendrimada, T.P.; Wang, Q.; Higuchi, M.; Seeburg, P.H.; Shiekhattar, R.; Nishikura, K. Modulation of MicroRNA Processing and Expression through RNA Editing by ADAR Deaminases. Nat. Struct. Mol. Biol. 2006, 13, 13–21. [Google Scholar] [CrossRef]
- Athanasiadis, A.; Rich, A.; Maas, S. Widespread A-to-I RNA Editing of Alu-Containing MRNAs in the Human Transcriptome. PLoS Biol. 2004, 2, e391. [Google Scholar] [CrossRef] [PubMed]
- Bazak, L.; Haviv, A.; Barak, M.; Jacob-Hirsch, J.; Deng, P.; Zhang, R.; Isaacs, F.J.; Rechavi, G.; Li, J.B.; Eisenberg, E.; et al. A-to-I RNA Editing Occurs at over a Hundred Million Genomic Sites, Located in a Majority of Human Genes. Genome Res. 2014, 24, 365–376. [Google Scholar] [CrossRef]
- Tan, M.H.; Li, Q.; Shanmugam, R.; Piskol, R.; Kohler, J.; Young, A.N.; Liu, K.I.; Zhang, R.; Ramaswami, G.; Ariyoshi, K.; et al. Dynamic Landscape and Regulation of RNA Editing in Mammals. Nature 2017, 550, 249–254. [Google Scholar] [CrossRef]
- Konen, L.M.; Wright, A.L.; Royle, G.A.; Morris, G.P.; Lau, B.K.; Seow, P.W.; Zinn, R.; Milham, L.T.; Vaughan, C.W.; Vissel, B. A New Mouse Line with Reduced GluA2 Q/R Site RNA Editing Exhibits Loss of Dendritic Spines, Hippocampal CA1-Neuron Loss, Learning and Memory Impairments and NMDA Receptor-Independent Seizure Vulnerability. Mol. Brain 2020, 13, 27. [Google Scholar] [CrossRef]
- Kubota-Sakashita, M.; Iwamoto, K.; Bundo, M.; Kato, T. A Role of ADAR2 and RNA Editing of Glutamate Receptors in Mood Disorders and Schizophrenia. Mol. Brain 2014, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- de Crécy-Lagard, V.; Boccaletto, P.; Mangleburg, C.G.; Sharma, P.; Lowe, T.M.; Leidel, S.A.; Bujnicki, J.M. Matching TRNA Modifications in Humans to Their Known and Predicted Enzymes. Nucleic Acids Res. 2019, 47, 2143–2159. [Google Scholar] [CrossRef]
- Jühling, F.; Mörl, M.; Hartmann, R.K.; Sprinzl, M.; Stadler, P.F.; Pütz, J. TRNAdb 2009: Compilation of TRNA Sequences and TRNA Genes. Nucleic Acids Res. 2009, 37, D159–D162. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T. The Expanding World of TRNA Modifications and Their Disease Relevance. Nat. Rev. Mol. Cell Biol. 2021, 22, 375–392. [Google Scholar] [CrossRef]
- Hoernes, T.P.; Clementi, N.; Faserl, K.; Glasner, H.; Breuker, K.; Lindner, H.; Hüttenhofer, A.; Erlacher, M.D. Nucleotide Modifications within Bacterial Messenger RNAs Regulate Their Translation and Are Able to Rewire the Genetic Code. Nucleic Acids Res. 2016, 44, 852–862. [Google Scholar] [CrossRef]
- Yildirim, I.; Kierzek, E.; Kierzek, R.; Schatz, G.C. Interplay of LNA and 2′-O-Methyl RNA in the Structure and Thermodynamics of RNA Hybrid Systems: A Molecular Dynamics Study Using the Revised AMBER Force Field and Comparison with Experimental Results. J. Phys. Chem. B 2014, 118, 14177–14187. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. The Dynamic Epitranscriptome: N6-Methyladenosine and Gene Expression Control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Jantsch, M.F.; Öhman, M. RNA Editing by Adenosine Deaminases That Act on RNA (ADARs). In Nucleic Acids and Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2008; pp. 51–84. ISBN 9783540737865. [Google Scholar]
- Motorin, Y.; Helm, M. TRNA Stabilization by Modified Nucleotides. Biochemistry 2010, 49, 4934–4944. [Google Scholar] [CrossRef] [PubMed]
- Shafik, A.M.; Zhang, F.; Guo, Z.; Dai, Q.; Pajdzik, K.; Li, Y.; Kang, Y.; Yao, B.; Wu, H.; He, C.; et al. N6-Methyladenosine Dynamics in Neurodevelopment and Aging, and Its Potential Role in Alzheimer’s Disease. Genome Biol. 2021, 22, 17. [Google Scholar] [CrossRef]
- Zhao, F.; Xu, Y.; Gao, S.; Qin, L.; Austria, Q.; Siedlak, S.L.; Pajdzik, K.; Dai, Q.; He, C.; Wang, W.; et al. METTL3-Dependent RNA M6A Dysregulation Contributes to Neurodegeneration in Alzheimer’s Disease through Aberrant Cell Cycle Events. Mol. Neurodegener. 2021, 16, 70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, S.; Han, S.; Sun, Y.; Han, M.; Zheng, X.; Li, F.; Wei, Y.; Wang, Y.; Bi, J. Differential Methylation of CircRNA M6A in an APP/PS1 Alzheimer’s Disease Mouse Model. Mol. Med. Rep. 2023, 27, 55. [Google Scholar] [CrossRef]
- Wang, X.; Xie, J.; Tan, L.; Lu, Y.; Shen, N.; Li, J.; Hu, H.; Li, H.; Li, X.; Cheng, L. N6-Methyladenosine-Modified CircRIMS2 Mediates Synaptic and Memory Impairments by Activating GluN2B Ubiquitination in Alzheimer’s Disease. Transl. Neurodegener. 2023, 12, 53. [Google Scholar] [CrossRef]
- Huang, H.; Camats-Perna, J.; Medeiros, R.; Anggono, V.; Widagdo, J. Altered Expression of the M6A Methyltransferase METTL3 in Alzheimer’s Disease. eNeuro 2020, 7, 1–10. [Google Scholar] [CrossRef]
- Li, H.; Ren, Y.; Mao, K.; Hua, F.; Yang, Y.; Wei, N.; Yue, C.; Li, D.; Zhang, H. FTO Is Involved in Alzheimer’s Disease by Targeting TSC1-MTOR-Tau Signaling. Biochem. Biophys. Res. Commun. 2018, 498, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, S.; Shen, L.; Kim, S.; Inlow, M.; West, J.D.; Faber, K.M.; Foroud, T.; Mayeux, R.; Saykin, A.J.; Alzheimer’s Disease Neuroimaging Initiative; et al. Analysis of Copy Number Variation in Alzheimer’s Disease: The NIALOAD/NCRAD Family Study. Curr. Alzheimer Res. 2012, 9, 801–814. [Google Scholar] [CrossRef]
- Reitz, C.; Tosto, G.; Mayeux, R.; Luchsinger, J.A.; The NIA-LOAD/NCRAD Family Study Group; The Alzheimer’s Disease Neuroimaging Initiative. Genetic Variants in the Fat and Obesity Associated (FTO) Gene and Risk of Alzheimer’s Disease. PLoS ONE 2012, 7, e50354. [Google Scholar] [CrossRef]
- Keller, L.; Xu, W.; Wang, H.-X.; Winblad, B.; Fratiglioni, L.; Graff, C. The Obesity Related Gene, FTO, Interacts with APOE, and Is Associated with Alzheimer’s Disease Risk: A Prospective Cohort Study. J. Alzheimers Dis. 2011, 23, 461–469. [Google Scholar] [CrossRef]
- Zhang, X.; Trebak, F.; Souza, L.A.C.; Shi, J.; Zhou, T.; Kehoe, P.G.; Chen, Q.; Feng Earley, Y. Small RNA Modifications in Alzheimer’s Disease. Neurobiol. Dis. 2020, 145, 105058. [Google Scholar] [CrossRef]
- Khermesh, K.; D’Erchia, A.M.; Barak, M.; Annese, A.; Wachtel, C.; Levanon, E.Y.; Picardi, E.; Eisenberg, E. Reduced Levels of Protein Recoding by A-to-I RNA Editing in Alzheimer’s Disease. RNA 2016, 22, 290–302. [Google Scholar] [CrossRef]
- Akbarian, S.; Smith, M.A.; Jones, E.G. Editing for an AMPA Receptor Subunit RNA in Prefrontal Cortex and Striatum in Alzheimer’s Disease, Huntington’s Disease and Schizophrenia. Brain Res. 1995, 699, 297–304. [Google Scholar] [CrossRef]
- Gaisler-Salomon, I.; Kravitz, E.; Feiler, Y.; Safran, M.; Biegon, A.; Amariglio, N.; Rechavi, G. Hippocampus-Specific Deficiency in RNA Editing of GluA2 in Alzheimer’s Disease. Neurobiol. Aging 2014, 35, 1785–1791. [Google Scholar] [CrossRef] [PubMed]
- Payami, H.; Kaye, J.; Heston, L.L.; Bird, T.D.; Schellenberg, G.D. Apolipoprotein E Genotype and Alzheimer’s Disease. Lancet 1993, 342, 738. [Google Scholar]
- Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.; George-Hyslop, P.H.; Pericak-Vance, M.A.; Joo, S.H.; Rosi, B.L.; Gusella, J.F.; Crapper-MacLachlan, D.R.; Alberts, M.J. Association of Apolipoprotein E Allele Epsilon 4 with Late-Onset Familial and Sporadic Alzheimer’s Disease. Neurology 1993, 43, 1467–1472. [Google Scholar] [CrossRef] [PubMed]
- Valastro, B.; Ghribi, O.; Poirier, J.; Krzywkowski, P.; Massicotte, G. AMPA Receptor Regulation and LTP in the Hippocampus of Young and Aged Apolipoprotein E-Deficient Mice. Neurobiol. Aging 2001, 22, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Yang, M.; Kim, P.; Zhou, X. ADeditome Provides the Genomic Landscape of A-to-I RNA Editing in Alzheimer’s Disease. Brief. Bioinform. 2021, 22, bbaa384. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; He, H.; Huang, Y.; Wang, J.; Xiao, Y. Genome-Wide Identification of M6A-Associated Single-Nucleotide Polymorphisms in Parkinson’s Disease. Neurosci. Lett. 2020, 737, 135315. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yu, C.; Guo, M.; Zheng, X.; Ali, S.; Huang, H.; Zhang, L.; Wang, S.; Huang, Y.; Qie, S.; et al. Down-Regulation of M6A MRNA Methylation Is Involved in Dopaminergic Neuronal Death. ACS Chem. Neurosci. 2019, 10, 2355–2363. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Zhang, Q.; Liao, J.; Lei, J.; Luo, M.; Huang, J.; Chen, M.; Shen, Y.; Wang, J.; Xu, P.; et al. METTL14 Is Decreased and Regulates M6 A Modification of α-Synuclein in Parkinson’s Disease. J. Neurochem. 2023, 166, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Paz-Yaacov, N.; Levanon, E.Y.; Nevo, E.; Kinar, Y.; Harmelin, A.; Jacob-Hirsch, J.; Amariglio, N.; Eisenberg, E.; Rechavi, G. Adenosine-to-Inosine RNA Editing Shapes Transcriptome Diversity in Primates. Proc. Natl. Acad. Sci. USA 2010, 107, 12174–12179. [Google Scholar] [CrossRef] [PubMed]
- Wettengel, J.; Reautschnig, P.; Geisler, S.; Kahle, P.J.; Stafforst, T. Harnessing Human ADAR2 for RNA Repair—Recoding a PINK1 Mutation Rescues Mitophagy. Nucleic Acids Res. 2016, 45, gkw911. [Google Scholar] [CrossRef] [PubMed]
- Sommer, B.; Köhler, M.; Sprengel, R.; Seeburg, P.H. RNA Editing in Brain Controls a Determinant of Ion Flow in Glutamate-Gated Channels. Cell 1991, 67, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Hideyama, T.; Yamashita, T.; Suzuki, T.; Tsuji, S.; Higuchi, M.; Seeburg, P.H.; Takahashi, R.; Misawa, H.; Kwak, S. Induced Loss of ADAR2 Engenders Slow Death of Motor Neurons from Q/R Site-Unedited GluR2. J. Neurosci. 2010, 30, 11917–11925. [Google Scholar] [CrossRef]
- Hideyama, T.; Kwak, S. When Does ALS Start? ADAR2-GluA2 Hypothesis for the Etiology of Sporadic ALS. Front. Mol. Neurosci. 2011, 4, 33. [Google Scholar] [CrossRef]
- Hideyama, T.; Yamashita, T.; Aizawa, H.; Tsuji, S.; Kakita, A.; Takahashi, H.; Kwak, S. Profound Downregulation of the RNA Editing Enzyme ADAR2 in ALS Spinal Motor Neurons. Neurobiol. Dis. 2012, 45, 1121–1128. [Google Scholar] [CrossRef]
- Takuma, H.; Kwak, S.; Yoshizawa, T.; Kanazawa, I. Reduction of GluR2 RNA Editing, a Molecular Change That Increases Calcium Influx through AMPA Receptors, Selective in the Spinal Ventral Gray of Patients with Amyotrophic Lateral Sclerosis. Ann. Neurol. 1999, 46, 806–815. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; Ito, K.; Sun, H.; Aizawa, H.; Kanazawa, I.; Kwak, S. Glutamate Receptors: RNA Editing and Death of Motor Neurons. Nature 2004, 427, 801. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.; Kawahara, Y. Deficient RNA Editing of GluR2 and Neuronal Death in Amyotropic Lateral Sclerosis. J. Mol. Med. 2005, 83, 110–120. [Google Scholar] [CrossRef]
- Aizawa, H.; Sawada, J.; Hideyama, T.; Yamashita, T.; Katayama, T.; Hasebe, N.; Kimura, T.; Yahara, O.; Kwak, S. TDP-43 Pathology in Sporadic ALS Occurs in Motor Neurons Lacking the RNA Editing Enzyme ADAR2. Acta Neuropathol. 2010, 120, 75–84. [Google Scholar] [CrossRef]
- Yamashita, T.; Aizawa, H.; Teramoto, S.; Akamatsu, M.; Kwak, S. Calpain-Dependent Disruption of Nucleo-Cytoplasmic Transport in ALS Motor Neurons. Sci. Rep. 2017, 7, 39994. [Google Scholar] [CrossRef]
- Yamashita, T.; Akamatsu, M.; Kwak, S. Altered Intracellular Milieu of ADAR2-Deficient Motor Neurons in Amyotrophic Lateral Sclerosis. Genes 2017, 8, 60. [Google Scholar] [CrossRef]
- Colantoni, A.; Capauto, D.; Alfano, V.; D’Ambra, E.; D’Uva, S.; Tartaglia, G.G.; Morlando, M. FUS Alters CircRNA Metabolism in Human Motor Neurons Carrying the ALS-Linked P525L Mutation. Int. J. Mol. Sci. 2023, 24, 3181. [Google Scholar] [CrossRef] [PubMed]
- Aizawa, H.; Hideyama, T.; Yamashita, T.; Kimura, T.; Suzuki, N.; Aoki, M.; Kwak, S. Deficient RNA-Editing Enzyme ADAR2 in an Amyotrophic Lateral Sclerosis Patient with a FUS(P525L) Mutation. J. Clin. Neurosci. 2016, 32, 128–129. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro. Oncol. 2021, 23, iii1–iii105. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro. Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Barbieri, I.; Kouzarides, T. Role of RNA Modifications in Cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef]
- Huang, J.; Dong, X.; Gong, Z.; Qin, L.-Y.; Yang, S.; Zhu, Y.-L.; Wang, X.; Zhang, D.; Zou, T.; Yin, P.; et al. Solution Structure of the RNA Recognition Domain of METTL3-METTL14 N6-Methyladenosine Methyltransferase. Protein Cell 2019, 10, 272–284. [Google Scholar] [CrossRef]
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.G.; et al. M6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017, 18, 2622–2634. [Google Scholar] [CrossRef]
- Li, F.; Yi, Y.; Miao, Y.; Long, W.; Long, T.; Chen, S.; Cheng, W.; Zou, C.; Zheng, Y.; Wu, X.; et al. N6-Methyladenosine Modulates Nonsense-Mediated MRNA Decay in Human Glioblastoma. Cancer Res. 2019, 79, 5785–5798. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, A.; Patil, V.; Arora, A.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Somasundaram, K. Essential Role of METTL3-Mediated M6A Modification in Glioma Stem-like Cells Maintenance and Radioresistance. Oncogene 2018, 37, 522–533. [Google Scholar] [CrossRef]
- Yin, J.; Ding, F.; Cheng, Z.; Ge, X.; Li, Y.; Zeng, A.; Zhang, J.; Yan, W.; Shi, Z.; Qian, X.; et al. METTL3-Mediated M6A Modification of LINC00839 Maintains Glioma Stem Cells and Radiation Resistance by Activating Wnt/β-Catenin Signaling. Cell Death Dis. 2023, 14, 417. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wu, X.; Yang, H.; Xu, Q.; Zhang, M.; Liu, X.; Lv, K. M6A-Mediated Upregulation of LINC01003 Regulates Cell Migration by Targeting the CAV1/FAK Signaling Pathway in Glioma. Biol. Direct 2023, 18, 27. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yin, X.; Zhao, W.; Xu, W.; Chen, L. Molecular Mechanism of M6A Methylation of CircDLC1 Mediated by RNA Methyltransferase METTL3 in the Malignant Proliferation of Glioma Cells. Cell Death Discov. 2022, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Chen, G.; Dong, X.; Li, H.; Li, S.; Cheng, S.; Li, Y.; Wang, L.; Yuan, J.; Qian, Z.; et al. METTL3 Promotes the Resistance of Glioma to Temozolomide via Increasing MGMT and ANPG in a M6A Dependent Manner. Front. Oncol. 2021, 11, 702983. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, J.; Huang, S.; Li, J.; Zhou, L.; Sun, J.; Chen, L. CircTTLL13 Promotes TMZ Resistance in Glioma via Modulating OLR1-Mediated Activation of the Wnt/β-Catenin Pathway. Mol. Cell. Biol. 2023, 43, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Song, Y.; Qiao, W.; Sun, L.; Wang, X.; Yi, B.; Yang, X.; Ji, L.; Su, P.; Zhao, W.; et al. Focused Ultrasound Combined with MiR-1208-Equipped Exosomes Inhibits Malignant Progression of Glioma. Br. J. Cancer 2023, 129, 1083–1094. [Google Scholar] [CrossRef]
- Kumari, S.; Singh, M.; Kumar, S.; Muthuswamy, S. SETD2 Controls M6A Modification of Transcriptome and Regulates the Molecular Oncogenesis of Glioma. Med. Oncol. 2023, 40, 249. [Google Scholar] [CrossRef]
- Benavides-Serrato, A.; Saunders, J.T.; Kumar, S.; Holmes, B.; Benavides, K.E.; Bashir, M.T.; Nishimura, R.N.; Gera, J. M6A-Modification of Cyclin D1 and c-Myc IRESs in Glioblastoma Controls ITAF Activity and Resistance to MTOR Inhibition. Cancer Lett. 2023, 562, 216178. [Google Scholar] [CrossRef] [PubMed]
- Bu, C.; Hu, S.; Yu, J.; Li, N.; Gu, J.; Sheng, Z.; Yan, Z.; Bu, X. Fear Stress Promotes Glioma Progression through Inhibition of Ferroptosis by Enhancing FSP1 Stability. Clin. Transl. Oncol. 2023, 25, 1378–1388. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Hu, Z.; Zeng, C.; Luo, H.; Liu, J. FLOT1, Stabilized by WTAP/IGF2BP2 Mediated N6-Methyladenosine Modification, Predicts Poor Prognosis and Promotes Growth and Invasion in Gliomas. Heliyon 2023, 9, e16280. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Li, W.; Yang, Y.; Hu, X.; Dai, Y.; Huang, M.; Luo, J.; Zhang, K.; Zhao, N. Comprehensive Analysis of M6A/M5C/M1A-Related Gene Expression, Immune Infiltration, and Sensitivity of Antineoplastic Drugs in Glioma. Front. Immunol. 2022, 13, 955848. [Google Scholar] [CrossRef]
- Wang, L.-C.; Chen, S.-H.; Shen, X.-L.; Li, D.-C.; Liu, H.-Y.; Ji, Y.-L.; Li, M.; Yu, K.; Yang, H.; Chen, J.-J.; et al. M6A RNA Methylation Regulator HNRNPC Contributes to Tumorigenesis and Predicts Prognosis in Glioblastoma Multiforme. Front. Oncol. 2020, 10, 536875. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Wang, P.; Xue, Y.; Shang, C.; Liu, X.; Ma, J.; Li, Z.; Li, Z.; Bao, M.; Liu, Y. Overexpression of MiR-29a Reduces the Oncogenic Properties of Glioblastoma Stem Cells by Downregulating Quaking Gene Isoform 6. Oncotarget 2017, 8, 24949–24963. [Google Scholar] [CrossRef]
- Xi, Z.; Xue, Y.; Zheng, J.; Liu, X.; Ma, J.; Liu, Y. WTAP Expression Predicts Poor Prognosis in Malignant Glioma Patients. J. Mol. Neurosci. 2016, 60, 131–136. [Google Scholar] [CrossRef]
- Jin, D.-I.; Lee, S.W.; Han, M.-E.; Kim, H.-J.; Seo, S.-A.; Hur, G.-Y.; Jung, S.; Kim, B.-S.; Oh, S.-O. Expression and Roles of Wilms’ Tumor 1-Associating Protein in Glioblastoma. Cancer Sci. 2012, 103, 2102–2109. [Google Scholar] [CrossRef]
- Cong, P.; Wu, T.; Huang, X.; Liang, H.; Gao, X.; Tian, L.; Li, W.; Chen, A.; Wan, H.; He, M.; et al. Identification of the Role and Clinical Prognostic Value of Target Genes of M6A RNA Methylation Regulators in Glioma. Front. Cell Dev. Biol. 2021, 9, 709022. [Google Scholar] [CrossRef]
- Guo, X.; Qiu, W.; Wang, C.; Qi, Y.; Li, B.; Wang, S.; Zhao, R.; Cheng, B.; Han, X.; Du, H.; et al. Neuronal Activity Promotes Glioma Progression by Inducing Proneural-to-Mesenchymal Transition in Glioma Stem Cells. Cancer Res. 2023, 84, 372–387. [Google Scholar] [CrossRef]
- Guan, S.; He, Y.; Su, Y.; Zhou, L. A Risk Signature Consisting of Eight M6A Methylation Regulators Predicts the Prognosis of Glioma. Cell. Mol. Neurobiol. 2022, 42, 2733–2743. [Google Scholar] [CrossRef] [PubMed]
- Maimaiti, A.; Tuersunniyazi, A.; Meng, X.; Pei, Y.; Ji, W.; Feng, Z.; Jiang, L.; Wang, Z.; Kasimu, M.; Wang, Y.; et al. N6-Methyladenosine RNA Methylation Regulator-Related Alternative Splicing Gene Signature as Prognostic Predictor and in Immune Microenvironment Characterization of Patients with Low-Grade Glioma. Front. Genet. 2022, 13, 872186. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Qiu, W.; Li, B.; Qi, Y.; Wang, S.; Zhao, R.; Cheng, B.; Han, X.; Du, H.; Pan, Z.; et al. Hypoxia-Induced Neuronal Activity in Glioma Patients Polarizes Microglia by Potentiating RNA M6A Demethylation. Clin. Cancer Res. 2023, OF1–OF15. [Google Scholar] [CrossRef] [PubMed]
- Chow, R.D.; Guzman, C.D.; Wang, G.; Schmidt, F.; Youngblood, M.W.; Ye, L.; Errami, Y.; Dong, M.B.; Martinez, M.A.; Zhang, S.; et al. AAV-Mediated Direct in Vivo CRISPR Screen Identifies Functional Suppressors in Glioblastoma. Nat. Neurosci. 2017, 20, 1329–1341. [Google Scholar] [CrossRef]
- Gerken, T.; Girard, C.A.; Tung, Y.-C.L.; Webby, C.J.; Saudek, V.; Hewitson, K.S.; Yeo, G.S.H.; McDonough, M.A.; Cunliffe, S.; McNeill, L.A.; et al. The Obesity-Associated FTO Gene Encodes a 2-Oxoglutarate-Dependent Nucleic Acid Demethylase. Science 2007, 318, 1469–1472. [Google Scholar] [CrossRef]
- Bartosovic, M.; Molares, H.C.; Gregorova, P.; Hrossova, D.; Kudla, G.; Vanacova, S. N6-Methyladenosine Demethylase FTO Targets Pre-MRNAs and Regulates Alternative Splicing and 3′-End Processing. Nucleic Acids Res. 2017, 45, 11356–11370. [Google Scholar] [CrossRef]
- Sun, M.; Yan, Q.; Qiao, Y.; Zhao, H.; Wang, Y.; Shan, C.; Zhang, S. PIKE-A Modulates Mitochondrial Metabolism through Increasing SDHA Expression Mediated by STAT3/FTO Axis. Int. J. Mol. Sci. 2022, 23, 11304. [Google Scholar] [CrossRef]
- Liu, P.; Yan, X.; Ma, C.; Gu, J.; Tian, F.; Qu, J. Prognostic Value of M6A Regulators and the Nomogram Construction in Glioma Patients. Medicine 2022, 101, e30643. [Google Scholar] [CrossRef]
- Huff, S.; Kummetha, I.R.; Zhang, L.; Wang, L.; Bray, W.; Yin, J.; Kelley, V.; Wang, Y.; Rana, T.M. Rational Design and Optimization of M6A-RNA Demethylase FTO Inhibitors as Anticancer Agents. J. Med. Chem. 2022, 65, 10920–10937. [Google Scholar] [CrossRef]
- Huff, S.; Tiwari, S.K.; Gonzalez, G.M.; Wang, Y.; Rana, T.M. M6A-RNA Demethylase FTO Inhibitors Impair Self-Renewal in Glioblastoma Stem Cells. ACS Chem. Biol. 2021, 16, 324–333. [Google Scholar] [CrossRef]
- Xiao, L.; Li, X.; Mu, Z.; Zhou, J.; Zhou, P.; Xie, C.; Jiang, S. FTO Inhibition Enhances the Antitumor Effect of Temozolomide by Targeting MYC-MiR-155/23a Cluster-MXI1 Feedback Circuit in Glioma. Cancer Res. 2020, 80, 3945–3958. [Google Scholar] [CrossRef]
- Li, F.; Zhang, C.; Zhang, G. M6A RNA Methylation Controls Proliferation of Human Glioma Cells by Influencing Cell Apoptosis. Cytogenet. Genome Res. 2019, 159, 119–125. [Google Scholar] [CrossRef]
- Tao, B.; Huang, X.; Shi, J.; Liu, J.; Li, S.; Xu, C.; Zhong, J.; Wan, L.; Feng, B.; Li, B. FTO Interacts with FOXO3a to Enhance Its Transcriptional Activity and Inhibits Aggression in Gliomas. Signal Transduct. Target. Ther. 2020, 5, 130. [Google Scholar] [CrossRef]
- Joshi, K.; Banasavadi-Siddegowda, Y.; Mo, X.; Kim, S.-H.; Mao, P.; Kig, C.; Nardini, D.; Sobol, R.W.; Chow, L.M.L.; Kornblum, H.I.; et al. MELK-Dependent FOXM1 Phosphorylation Is Essential for Proliferation of Glioma Stem Cells. Stem Cells 2013, 31, 1051–1063. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bögler, O.; et al. M6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606.e6. [Google Scholar] [CrossRef]
- Takahashi, H.; Hase, H.; Yoshida, T.; Tashiro, J.; Hirade, Y.; Kitae, K.; Tsujikawa, K. Discovery of Two Novel ALKBH5 Selective Inhibitors That Exhibit Uncompetitive or Competitive Type and Suppress the Growth Activity of Glioblastoma Multiforme. Chem. Biol. Drug Des. 2022, 100, 1–12. [Google Scholar] [CrossRef]
- Kowalski-Chauvel, A.; Lacore, M.G.; Arnauduc, F.; Delmas, C.; Toulas, C.; Cohen-Jonathan-Moyal, E.; Seva, C. The M6A RNA Demethylase ALKBH5 Promotes Radioresistance and Invasion Capability of Glioma Stem Cells. Cancers 2020, 13, 40. [Google Scholar] [CrossRef]
- Lv, D.; Zhong, C.; Dixit, D.; Yang, K.; Wu, Q.; Godugu, B.; Prager, B.C.; Zhao, G.; Wang, X.; Xie, Q.; et al. EGFR Promotes ALKBH5 Nuclear Retention to Attenuate N6-Methyladenosine and Protect against Ferroptosis in Glioblastoma. Mol. Cell 2023, 83, P4334–P4351. [Google Scholar] [CrossRef]
- Li, L.; Yang, M.; Pu, X.; Tang, Y.; Fei, F.; Li, Z.; Hou, H.; Chen, Q.; Wang, Q.; Wu, Y.; et al. ALKBH5-PYCR2 Positive Feedback Loop Promotes Proneural-Mesenchymal Transition via Proline Synthesis in GBM. J. Cancer 2023, 14, 1579–1591. [Google Scholar] [CrossRef]
- Chang, G.; Xie, G.S.; Ma, L.; Li, P.; Li, L.; Richard, H.T. USP36 Promotes Tumorigenesis and Drug Sensitivity of Glioblastoma by Deubiquitinating and Stabilizing ALKBH5. Neuro. Oncol. 2023, 25, 841–853. [Google Scholar] [CrossRef]
- Wei, C.; Wang, B.; Peng, D.; Zhang, X.; Li, Z.; Luo, L.; He, Y.; Liang, H.; Du, X.; Li, S.; et al. Pan-Cancer Analysis Shows That ALKBH5 Is a Potential Prognostic and Immunotherapeutic Biomarker for Multiple Cancer Types Including Gliomas. Front. Immunol. 2022, 13, 849592. [Google Scholar]
- Tao, M.; Li, X.; He, L.; Rong, X.; Wang, H.; Pan, J.; Lu, Z.; Zhang, X.; Peng, Y. Decreased RNA M6A Methylation Enhances the Process of the Epithelial Mesenchymal Transition and Vasculogenic Mimicry in Glioblastoma. Am. J. Cancer Res. 2022, 12, 893–906. [Google Scholar]
- Malacrida, A.; Rivara, M.; Di Domizio, A.; Cislaghi, G.; Miloso, M.; Zuliani, V.; Nicolini, G. 3D Proteome-Wide Scale Screening and Activity Evaluation of a New ALKBH5 Inhibitor in U87 Glioblastoma Cell Line. Bioorg. Med. Chem. 2020, 28, 115300. [Google Scholar] [CrossRef]
- Tang, W.; Xu, N.; Zhou, J.; He, Z.; Lenahan, C.; Wang, C.; Ji, H.; Liu, B.; Zou, Y.; Zeng, H.; et al. ALKBH5 Promotes PD-L1-Mediated Immune Escape through M6A Modification of ZDHHC3 in Glioma. Cell Death Discov. 2022, 8, 497. [Google Scholar] [CrossRef]
- Dong, F.; Qin, X.; Wang, B.; Li, Q.; Hu, J.; Cheng, X.; Guo, D.; Cheng, F.; Fang, C.; Tan, Y.; et al. ALKBH5 Facilitates Hypoxia-Induced Paraspeckle Assembly and IL8 Secretion to Generate an Immunosuppressive Tumor Microenvironment. Cancer Res. 2021, 81, 5876–5888. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, Y.; Wang, L.; Ji, S. ALKBH5 Promotes the Proliferation of Glioma Cells via Enhancing the MRNA Stability of G6PD. Neurochem. Res. 2021, 46, 3003–3011. [Google Scholar] [CrossRef]
- Ding, C.; Yi, X.; Chen, X.; Wu, Z.; You, H.; Chen, X.; Zhang, G.; Sun, Y.; Bu, X.; Wu, X.; et al. Warburg Effect-Promoted Exosomal Circ_0072083 Releasing up-Regulates NANGO Expression through Multiple Pathways and Enhances Temozolomide Resistance in Glioma. J. Exp. Clin. Cancer Res. 2021, 40, 164. [Google Scholar] [CrossRef]
- Liu, B.; Zhou, J.; Wang, C.; Chi, Y.; Wei, Q.; Fu, Z.; Lian, C.; Huang, Q.; Liao, C.; Yang, Z.; et al. LncRNA SOX2OT Promotes Temozolomide Resistance by Elevating SOX2 Expression via ALKBH5-Mediated Epigenetic Regulation in Glioblastoma. Cell Death Dis. 2020, 11, 384. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-Methyladenosine-Dependent Regulation of Messenger RNA Stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 Facilitates Translation and Decay of N6-Methyladenosine-Modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.-S.; Hao, Y.-J.; Sun, B.-F.; Sun, H.-Y.; Li, A.; Ping, X.-L.; Lai, W.-Y.; et al. Nuclear M6A Reader YTHDC1 Regulates MRNA Splicing. Mol. Cell 2016, 61, 925. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Dixit, D.; Prager, B.C.; Gimple, R.C.; Poh, H.X.; Wang, Y.; Wu, Q.; Qiu, Z.; Kidwell, R.L.; Kim, L.J.Y.; Xie, Q.; et al. The RNA M6A Reader YTHDF2 Maintains Oncogene Expression and Is a Targetable Dependency in Glioblastoma Stem Cells. Cancer Discov. 2021, 11, 480–499. [Google Scholar] [CrossRef]
- Lee, H.-H.; Hsieh, C.-C.; Chang, C.-C.; Liao, W.-T.; Chi, H.-C. YTHDF3 Modulates EGFR/ATK/ERK/P21 Signaling Axis to Promote Cancer Progression and Osimertinib Resistance of Glioblastoma Cells. Anticancer Res. 2023, 43, 5485–5498. [Google Scholar] [CrossRef]
- Deng, X.; Sun, X.; Hu, Z.; Wu, Y.; Zhou, C.; Sun, J.; Gao, X.; Huang, Y. Exploring the Role of M6A Methylation Regulators in Glioblastoma Multiforme and Their Impact on the Tumor Immune Microenvironment. FASEB J. 2023, 37, e23155. [Google Scholar] [CrossRef] [PubMed]
- Guowei, L.; Xiufang, L.; Qianqian, X.; Yanping, J. The FDX1 Methylation Regulatory Mechanism in the Malignant Phenotype of Glioma. Genomics 2023, 115, 110601. [Google Scholar] [CrossRef] [PubMed]
- Tassinari, V.; Cesarini, V.; Tomaselli, S.; Ianniello, Z.; Silvestris, D.A.; Ginistrelli, L.C.; Martini, M.; De Angelis, B.; De Luca, G.; Vitiani, L.R.; et al. ADAR1 Is a New Target of METTL3 and Plays a Pro-Oncogenic Role in Glioblastoma by an Editing-Independent Mechanism. Genome Biol. 2021, 22, 51. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Wu, X.; Gao, Y.; Chen, J.; Zhang, M.; Zhou, H.; Yang, J.; Xiao, F.; Yang, X.; Huang, N.; et al. Circular RNA Encoded MET Variant Promotes Glioblastoma Tumorigenesis. Nat. Commun. 2023, 14, 4467. [Google Scholar] [CrossRef]
- Jin, C.; Zhao, J.; Zhang, Z.-P.; Wu, M.; Li, J.; Liu, B.; Bin Liao, X.-; Liao, Y.-X.; Liu, J.-P. CircRNA EPHB4 Modulates Stem Properties and Proliferation of Gliomas via Sponging MiR-637 and up-Regulating SOX10. Mol. Oncol. 2021, 15, 596–622. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhang, S.; Liu, Y.; Li, C.; Guo, T.; Zhang, S.; Li, Z.; Jiao, Z.; Sun, H.; Zhang, Y.; et al. NR2F1-AS1/MiR-190a/PHLDB2 Induces the Epithelial-Mesenchymal Transformation Process in Gastric Cancer by Promoting Phosphorylation of AKT3. Front. Cell Dev. Biol. 2021, 9, 688949. [Google Scholar] [CrossRef]
- Liao, Y.; Qiu, X.; Liu, J.; Zhang, Z.; Liu, B.; Jin, C. The Role of M6A-Modified CircEPHB4 in Glioma Pathogenesis: Insights into Cancer Stemness Metastasis. Ann. Clin. Transl. Neurol. 2023, 10, 1749–1767. [Google Scholar] [CrossRef]
- Zhu, X.; Yang, H.; Zhang, M.; Wu, X.; Jiang, L.; Liu, X.; Lv, K. YTHDC1-Mediated VPS25 Regulates Cell Cycle by Targeting JAK-STAT Signaling in Human Glioma Cells. Cancer Cell Int. 2021, 21, 645. [Google Scholar] [CrossRef]
- Zhang, C.; Guo, C.; Li, Y.; Ouyang, L.; Zhao, Q.; Liu, K. The Role of YTH Domain Containing 2 in Epigenetic Modification and Immune Infiltration of Pan-Cancer. J. Cell. Mol. Med. 2021, 25, 8615–8627. [Google Scholar] [CrossRef]
- Sun, C.; Zheng, X.; Sun, Y.; Yu, J.; Sheng, M.; Yan, S.; Zhu, Q.; Lan, Q. Identification of IGF2BP3 as an Adverse Prognostic Biomarker of Gliomas. Front. Genet. 2021, 12, 743738. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Cai, X.; Cong, Z.; Wang, Y.; Geng, Y.; Aili, Y.; Du, C.; Zhu, J.; Yang, J.; Tang, C.; et al. Roles of the M6A Modification of RNA in the Glioblastoma Microenvironment as Revealed by Single-Cell Analyses. Front. Immunol. 2022, 13, 798583. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.; Lv, Q.; Zhang, Y.; Chen, X.; Guo, R.; Liu, S.; Peng, X. Long Non-Coding RNA DDX11-AS1 Promotes the Proliferation and Migration of Glioma Cells by Combining with HNRNPC. Mol. Ther. Nucleic Acids 2022, 28, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.M.; Hwang, S.J.; Masuda, K.; Choi, K.-M.; Jeong, M.-R.; Nam, D.-H.; Gorospe, M.; Kim, H.H. Heterogeneous Nuclear Ribonucleoprotein C1/C2 Controls the Metastatic Potential of Glioblastoma by Regulating PDCD4. Mol. Cell. Biol. 2012, 32, 4237–4244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, B.; Xu, C.; Ji, C.; Yin, A.; Liu, Y.; Yao, Y.; Li, B.; Chen, T.; Shen, L.; et al. Cholesterol Homeostasis Confers Glioma Malignancy Triggered by HnRNPA2B1-Dependent Regulation of SREBP2 and LDLR. Neuro. Oncol. 2023, noad233. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Zhao, R.; Li, B.; Qi, Y.; Qiu, W.; Guo, Q.; Zhang, S.; Zhao, S.; Xu, H.; Li, M.; et al. EWSR1-Induced CircNEIL3 Promotes Glioma Progression and Exosome-Mediated Macrophage Immunosuppressive Polarization via Stabilizing IGF2BP3. Mol. Cancer 2022, 21, 16. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Kong, C.; Chen, M. Effect of HnRNPA2/B1 on the Proliferation and Apoptosis of Glioma U251 Cells via the Regulation of AKT and STAT3 Pathways. Biosci. Rep. 2020, 40, BSR20190318. [Google Scholar] [CrossRef]
- Li, L.; Yang, Y.; Wu, M.; Yu, Z.; Wang, C.; Dou, G.; He, H.; Wang, H.; Yang, N.; Qi, H.; et al. Β-Asarone Induces Apoptosis and Cell Cycle Arrest of Human Glioma U251 Cells via Suppression of HnRNP A2/B1-Mediated Pathway in Vitro and in Vivo. Molecules 2018, 23, 1072. [Google Scholar] [CrossRef]
- Li, L.; Wu, M.; Wang, C.; Yu, Z.; Wang, H.; Qi, H.; Xu, X. Β-Asarone Inhibits Invasion and EMT in Human Glioma U251 Cells by Suppressing Splicing Factor HnRNP A2/B1. Molecules 2018, 23, 671. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wu, X.; Wang, J.; Guo, X.; Chen, J.; Yang, X.; Zhong, J.; Li, X.; Deng, Z. Feedforward Loop between IMP1 and YAP/TAZ Promotes Tumorigenesis and Malignant Progression in Glioblastoma. Cancer Sci. 2023, 114, 2053–2062. [Google Scholar] [CrossRef] [PubMed]
- Zhan, W.-L.; Gao, N.; Tu, G.-L.; Tang, H.; Gao, L.; Xia, Y. LncRNA LINC00689 Promotes the Tumorigenesis of Glioma via Mediation of MiR-526b-3p/IGF2BP1 Axis. Neuromol. Med. 2021, 23, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhao, P.; Li, B.; Xu, D.; Wang, K. LncRNA PCAT6 Regulated by YY1 Accelerates the Progression of Glioblastoma via MiR-513/IGF2BP1. Neurochem. Res. 2020, 45, 2894–2902. [Google Scholar] [CrossRef]
- Xue, J.; Zhong, S.; Sun, B.-M.; Sun, Q.-F.; Hu, L.-Y.; Pan, S.-J. Lnc-THOR Silencing Inhibits Human Glioma Cell Survival by Activating MAGEA6-AMPK Signaling. Cell Death Dis. 2019, 10, 866. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-W.; Xue, M.; Zhu, B.-X.; Yue, C.-L.; Chen, M.; Qin, H.-H. MicroRNA-4500 Inhibits Human Glioma Cell Progression by Targeting IGF2BP1. Biochem. Biophys. Res. Commun. 2019, 513, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Sun, R.; Zhang, J.; Sun, T.; Liu, X.; Yang, B. MiR-506 Inhibits the Proliferation and Invasion by Targeting IGF2BP1 in Glioblastoma. Am. J. Transl. Res. 2015, 7, 2007–2014. [Google Scholar] [PubMed]
- Wang, R.-J.; Li, J.-W.; Bao, B.-H.; Wu, H.-C.; Du, Z.-H.; Su, J.-L.; Zhang, M.-H.; Liang, H.-Q. MicroRNA-873 (MiRNA-873) Inhibits Glioblastoma Tumorigenesis and Metastasis by Suppressing the Expression of IGF2BP1. J. Biol. Chem. 2015, 290, 8938–8948. [Google Scholar] [CrossRef]
- Wu, Z.; Wei, N. METTL3-Mediated HOTAIRM1 Promotes Vasculogenic Mimicry Icontributionsn Glioma via Regulating IGFBP2 Expression. J. Transl. Med. 2023, 21, 855. [Google Scholar] [CrossRef]
- Yang, J.; Yang, S.; Cai, J.; Chen, H.; Sun, L.; Wang, J.; Hou, G.; Gu, S.; Ma, J.; Ge, J. A Transcription Factor ZNF384, Regulated by LINC00265, Activates the Expression of IFI30 to Stimulate Malignant Progression in Glioma. ACS Chem. Neurosci. 2023, 15, 290–299. [Google Scholar] [CrossRef]
- Zhang, Z.-X.; Ren, P.; Cao, Y.-Y.; Wang, T.-T.; Huang, G.-H.; Li, Y.; Zhou, S.; Yang, W.; Yang, L.; Liu, G.-L.; et al. HOXD-AS2-STAT3 Feedback Loop Attenuates Sensitivity to Temozolomide in Glioblastoma. CNS Neurosci. Ther. 2023, 29, 3430–3445. [Google Scholar] [CrossRef]
- Tan, L.-M.; Chen, P.; Nie, Z.-Y.; Liu, X.-F.; Wang, B. Circular RNA XRCC5 Aggravates Glioma Progression by Activating CLC3/SGK1 Axis via Recruiting IGF2BP2. Neurochem. Int. 2023, 166, 105534. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, Y.; Yuan, Y.; Jin, Z.; Zhai, H.; Liu, B.; Li, Y.; Zhang, C.; Chen, M.; Shi, Y.; et al. LncRNA GSCAR Promotes Glioma Stem Cell Maintenance via Stabilizing SOX2 Expression. Int. J. Biol. Sci. 2023, 19, 1681–1697. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, X.; Zhou, Y.; Huang, X.; Jiang, X. Long Non-Coding RNA OIP5-AS1 Inhibition Upregulates MicroRNA-129-5p to Repress Resistance to Temozolomide in Glioblastoma Cells via Downregulating IGF2BP2. Cell Biol. Toxicol. 2022, 38, 963–977. [Google Scholar] [CrossRef]
- Li, H.; Wang, D.; Yi, B.; Cai, H.; Wang, Y.; Lou, X.; Xi, Z.; Li, Z. SUMOylation of IGF2BP2 Promotes Vasculogenic Mimicry of Glioma via Regulating OIP5-AS1/MiR-495-3p Axis. Int. J. Biol. Sci. 2021, 17, 2912–2930. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, P.; Su, R.; Xue, Y.; Yang, C.; Wang, D.; Ruan, X.; Zheng, J.; Yang, Y.; Li, Z.; et al. IGF2BP2 Stabilized FBXL19-AS1 Regulates the Blood-Tumour Barrier Permeability by Negatively Regulating ZNF765 by STAU1-Mediated MRNA Decay. RNA Biol. 2020, 17, 1777–1788. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yang, C.; Ruan, X.; Liu, X.; Wang, D.; Liu, L.; Shao, L.; Wang, P.; Dong, W.; Xue, Y. CPEB2 M6A Methylation Regulates Blood-Tumor Barrier Permeability by Regulating Splicing Factor SRSF5 Stability. Commun. Biol. 2022, 5, 908. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Zhao, H.; Zhang, S.; Ding, Q.; Guo, Y.; Hou, K.; Kan, Y.; Deng, F.; Xu, Q. A Pan-Cancer Landscape of IGF2BPs and Their Association with Prognosis, Stemness and Tumor Immune Microenvironment. Front. Oncol. 2022, 12, 1049183. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.-B.; Jaffrey, S.R. 5′ UTR M6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Bertorello, J.; Sesen, J.; Gilhodes, J.; Evrard, S.; Courtade-Saïdi, M.; Augustus, M.; Uro-Coste, E.; Toulas, C.; Moyal, E.C.-J.; Seva, C.; et al. Translation Reprogramming by EIF3 Linked to Glioblastoma Resistance. NAR Cancer 2020, 2, zcaa020. [Google Scholar] [CrossRef]
- Hao, J.; Liang, C.; Jiao, B. Eukaryotic Translation Initiation Factor 3, Subunit C Is Overexpressed and Promotes Cell Proliferation in Human Glioma U-87 MG Cells. Oncol. Lett. 2015, 9, 2525–2533. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Wang, Z.; Wang, Y.; Liang, Z.; Zhang, X.; Zhao, Z.; Jiao, B. Eukaryotic Initiation Factor 3C Silencing Inhibits Cell Proliferation and Promotes Apoptosis in Human Glioma. Oncol. Rep. 2015, 33, 2954–2962. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Zhou, C.; Liang, H.; Wang, X.; Xu, L. RNAi-Mediated Silencing of EIF3D Alleviates Proliferation and Migration of Glioma U251 and U87MG Cells. Chem. Biol. Drug Des. 2015, 86, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Ding, X.; Zhou, C.; Zhang, Y.; Xu, M.; Zhang, C.; Xu, L. Knockdown of Eukaryotic Translation Initiation Factors 3B (EIF3B) Inhibits Proliferation and Promotes Apoptosis in Glioblastoma Cells. Neurol. Sci. 2012, 33, 1057–1062. [Google Scholar] [CrossRef]
- Chai, R.-C.; Wang, N.; Chang, Y.-Z.; Zhang, K.-N.; Li, J.-J.; Niu, J.-J.; Wu, F.; Liu, Y.-Q.; Wang, Y.-Z. Systematically Profiling the Expression of EIF3 Subunits in Glioma Reveals the Expression of EIF3i Has Prognostic Value in IDH-Mutant Lower Grade Glioma. Cancer Cell Int. 2019, 19, 155. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wu, M.; Tu, Z.; Tao, C.; Hu, Q.; Li, K.; Zhu, X.; Huang, K. Identification of RNA: 5-Methylcytosine Methyltransferases-Related Signature for Predicting Prognosis in Glioma. Front. Oncol. 2020, 10, 1119. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Yang, L.; Sun, X.; He, L.; Du, T. Physical Performance Is Associated with Visual Acuity in University Students: Results of a School-Based Study. Rev. Assoc. Med. Bras. 2020, 66, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, Y.; Zhang, J.; Zhang, X. NSun2 Promotes Cell Migration through Methylating Autotaxin MRNA. J. Biol. Chem. 2020, 295, 18134–18147. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, N.; Wu, J.; Gao, X.; Zhao, H.; Liu, Z.; Yan, X.; Dong, J.; Wang, F.; Ba, Y.; et al. 5-Methylcytosine Related LncRNAs Reveal Immune Characteristics, Predict Prognosis and Oncology Treatment Outcome in Lower-Grade Gliomas. Front. Immunol. 2022, 13, 844778. [Google Scholar] [CrossRef]
- Maas, S.; Patt, S.; Schrey, M.; Rich, A. Underediting of Glutamate Receptor GluR-B MRNA in Malignant Gliomas. Proc. Natl. Acad. Sci. USA 2001, 98, 14687–14692. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, Z.; Du, C.; Qi, B.; Zhao, X.; Wang, L.; Bi, L.; Wang, G.; Zhang, X.; Su, X.; et al. Abnormal Expression of an ADAR2 Alternative Splicing Variant in Gliomas Downregulates Adenosine-to-Inosine RNA Editing. Acta Neurochir. 2014, 156, 1135–1142. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tian, Y.; Tian, N.; Zhao, X.; Du, C.; Han, L.; Zhang, H. Aberrant Alternative Splicing Pattern of ADAR2 Downregulates Adenosine-to-Inosine Editing in Glioma. Oncol. Rep. 2015, 33, 2845–2852. [Google Scholar] [CrossRef]
- Oakes, E.; Anderson, A.; Cohen-Gadol, A.; Hundley, H.A. Adenosine Deaminase That Acts on RNA 3 (ADAR3) Binding to Glutamate Receptor Subunit B Pre-MRNA Inhibits RNA Editing in Glioblastoma. J. Biol. Chem. 2017, 292, 4326–4335. [Google Scholar] [CrossRef]
- Cesarini, V.; Silvestris, D.A.; Tassinari, V.; Tomaselli, S.; Alon, S.; Eisenberg, E.; Locatelli, F.; Gallo, A. ADAR2/MiR-589-3p Axis Controls Glioblastoma Cell Migration/Invasion. Nucleic Acids Res. 2018, 46, 2045–2059. [Google Scholar] [CrossRef]
- Patil, V.; Pal, J.; Mahalingam, K.; Somasundaram, K. Global RNA Editome Landscape Discovers Reduced RNA Editing in Glioma: Loss of Editing of Gamma-Amino Butyric Acid Receptor Alpha Subunit 3 (GABRA3) Favors Glioma Migration and Invasion. PeerJ 2020, 8, e9755. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, K.; Zhao, Z.; Sun, S.; Zhang, K.; Huang, R.; Zeng, F.; Hu, H. ADAR3 Expression Is an Independent Prognostic Factor in Lower-Grade Diffuse Gliomas and Positively Correlated with the Editing Level of GRIA2Q607R. Cancer Cell Int. 2018, 18, 196. [Google Scholar] [CrossRef]
- Raghava Kurup, R.; Oakes, E.K.; Vadlamani, P.; Nwosu, O.; Danthi, P.; Hundley, H.A. ADAR3 Activates NF-ΚB Signaling and Promotes Glioblastoma Cell Resistance to Temozolomide. Sci. Rep. 2022, 12, 13362. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Z.; Ding, W.; Zhang, J.-H.; Tan, Z.-L.; Mei, Y.-R.; He, W.; Wang, X.-J. Expression and Potential Biomarkers of Regulators for M7G RNA Modification in Gliomas. Front. Neurol. 2022, 13, 886246. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Niu, L.; Wang, Z.; Zhao, Z. RNA M1A Methyltransferase TRMT6 Predicts Poorer Prognosis and Promotes Malignant Behavior in Glioma. Front. Mol. Biosci. 2021, 8, 692130. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).