Non-Coding RNAs: Regulators of Stress, Ageing, and Developmental Decisions in Yeast?
Abstract
:1. Introduction
2. Transfer RNA—More Than an Adapter
2.1. Abundance of tRNA—Synthesis and Degradation
2.2. Charge Status of tRNAs
2.3. Modifications of tRNA
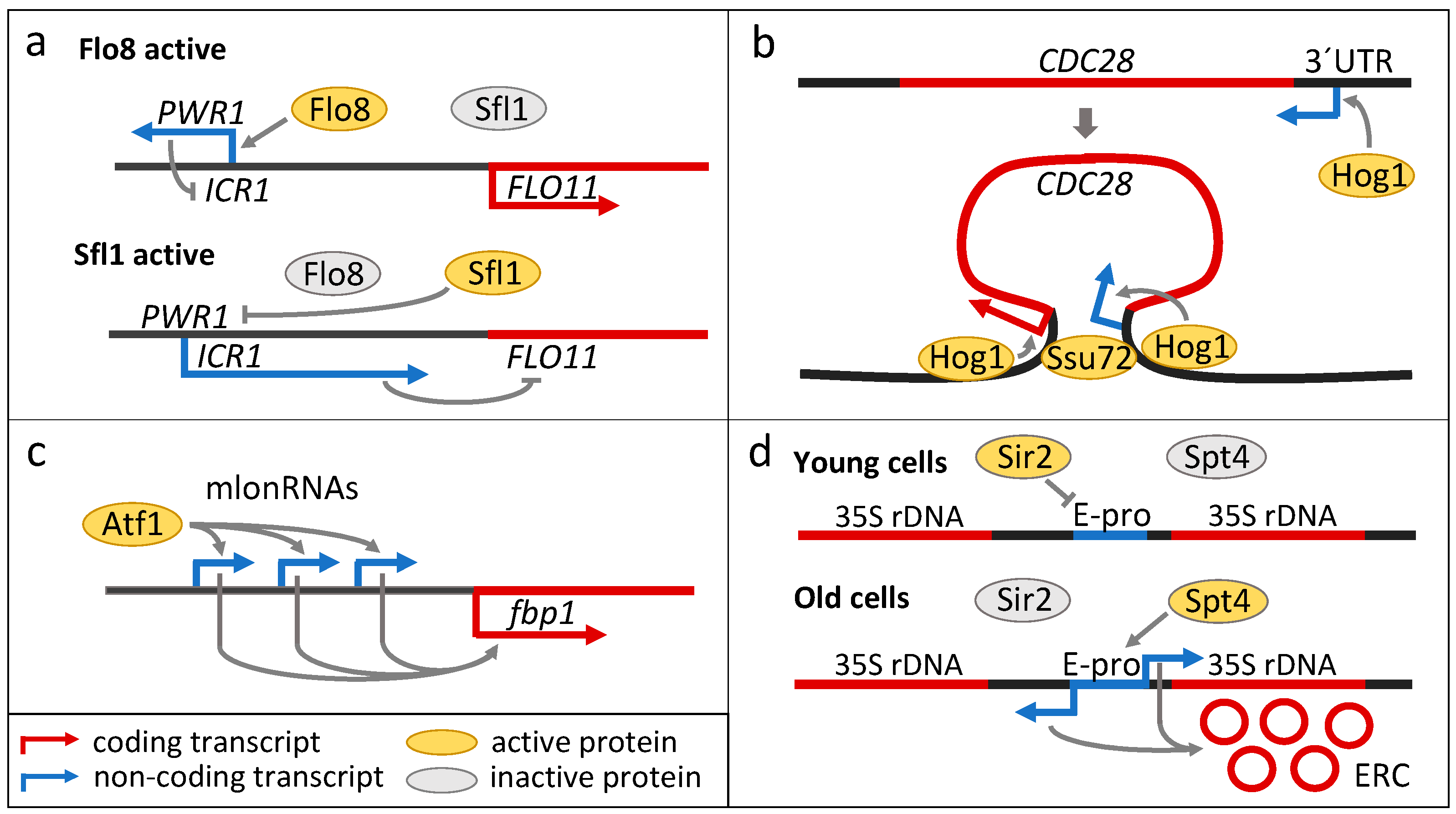
3. Long Non-Coding RNAs
4. Other Types of RNA
4.1. Ribosomal RNAs
4.2. RNAs from Introns
4.3. Small Nuclear RNAs
4.4. RNAs in Extracellular Vesicles
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fatica, A.; Bozzoni, I. Long Non-Coding RNAs: New Players in Cell Differentiation and Development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Grewal, S.S. Why Should Cancer Biologists Care about tRNAs? tRNA Synthesis, mRNA Translation and the Control of Growth. Biochim. Biophys. Acta Gene Regul. Mech. 2015, 1849, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Chery, M.; Drouard, L. Plant tRNA Functions beyond Their Major Role in Translation. J. Exp. Bot. 2023, 74, 2352–2363. [Google Scholar] [CrossRef] [PubMed]
- Chekanova, J.A. Long Non-Coding RNAs and Their Functions in Plants. Curr. Opin. Plant Biol. 2015, 27, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Chauvier, A.; Walter, N.G. Regulation of Bacterial Gene Expression by Non-Coding RNA: It Is All about Time! Cell Chem. Biol. 2024, 31, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Katz, A.; Elgamal, S.; Rajkovic, A.; Ibba, M. Non-Canonical Roles of tRNAs and tRNA Mimics in Bacterial Cell Biology. Mol. Microbiol. 2016, 101, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Marchand, V. Analysis of RNA Modifications by Second- and Third-Generation Deep Sequencing: 2020 Update. Genes 2021, 12, 278. [Google Scholar] [CrossRef] [PubMed]
- Leger, A.; Amaral, P.P.; Pandolfini, L.; Capitanchik, C.; Capraro, F.; Miano, V.; Migliori, V.; Toolan-Kerr, P.; Sideri, T.; Enright, A.J.; et al. RNA Modifications Detection by Comparative Nanopore Direct RNA Sequencing. Nat. Commun. 2021, 12, 7198. [Google Scholar] [CrossRef] [PubMed]
- Lakhotia, S.C. Long Non-Coding RNAs Coordinate Cellular Responses to Stress. WIREs RNA 2012, 3, 779–796. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Begley, T.J.; Dedon, P.C. tRNA Modifications Regulate Translation during Cellular Stress. FEBS Lett. 2014, 588, 4287–4296. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Shichino, Y.; Yamamoto, M. The Long Non-Coding RNA World in Yeasts. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Parker, R. Stressing Out over tRNA Cleavage. Cell 2009, 138, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Eleutherio, E.; de Araujo Brasil, A.; França, M.B.; de Almeida, D.S.G.; Rona, G.B.; Magalhães, R.S.S. Oxidative Stress and Aging: Learning from Yeast Lessons. Fungal Biol. 2018, 122, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Dawes, I.W.; Perrone, G.G. Stress and Ageing in Yeast. FEMS Yeast Res. 2020, 20, 85. [Google Scholar] [CrossRef]
- Kourtis, N.; Tavernarakis, N. Cellular Stress Response Pathways and Ageing: Intricate Molecular Relationships. EMBO J. 2011, 30, 2520–2531. [Google Scholar] [CrossRef]
- de Magalhães, J.P.; Passos, J.F. Stress, Cell Senescence and Organismal Ageing. Mech. Ageing Dev. 2018, 170, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Váchová, L.; Čáp, M.; Palková, Z. Yeast Colonies: A Model for Studies of Aging, Environmental Adaptation, and Longevity. Oxid. Med. Cell. Longev. 2012, 2012, 601836. [Google Scholar] [CrossRef] [PubMed]
- Váchová, L.; Palková, Z. How Structured Yeast Multicellular Communities Live, Age and Die? FEMS Yeast Res. 2018, 18, foy033. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Zhou, G.; Munyon, R.; Ghannoum, M.A. Candida Biofilm: A Well-Designed Protected Environment. Med. Mycol. 2005, 43, 191–208. [Google Scholar] [CrossRef] [PubMed]
- Waldron, C.; Lacroute, F. Effect of Growth Rate on the Amounts of Ribosomal and Transfer Ribonucleic Acids in Yeast. J. Bacteriol. 1975, 122, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, Q.; Zhao, F. Synonymous but Not Silent: The Codon Usage Code for Gene Expression and Protein Folding. Annu. Rev. Biochem. 2021, 90, 375–401. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. A Code within the Genetic Code: Codon Usage Regulates Co-Translational Protein Folding. Cell Commun. Signal. 2020, 18, 145. [Google Scholar] [CrossRef]
- Hanson, G.; Coller, J. Codon Optimality, Bias and Usage in Translation and mRNA Decay. Nat. Rev. Mol. Cell Biol. 2018, 19, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Arias, L.; Martínez, F.; González, D.; Flores-Ríos, R.; Katz, A.; Tello, M.; Moreira, S.; Orellana, O. Modification of Transfer RNA Levels Affects Cyclin Aggregation and the Correct Duplication of Yeast Cells. Front. Microbiol. 2021, 11, 607693. [Google Scholar] [CrossRef]
- Hia, F.; Takeuchi, O. The Effects of Codon Bias and Optimality on mRNA and Protein Regulation. Cell. Mol. Life Sci. 2021, 78, 1909–1928. [Google Scholar] [CrossRef]
- Iben, J.R.; Maraia, R.J. tRNAomics: tRNA Gene Copy Number Variation and Codon Use Provide Bioinformatic Evidence of a New Anticodon:Codon Wobble Pair in a Eukaryote. RNA 2012, 18, 1358–1372. [Google Scholar] [CrossRef]
- Jackman, J.E.; Alfonzo, J.D. Transfer RNA Modifications: Nature’s Combinatorial Chemistry Playground. WIREs RNA 2013, 4, 35–48. [Google Scholar] [CrossRef]
- Nedialkova, D.D.; Leidel, S.A. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015, 161, 1606–1618. [Google Scholar] [CrossRef] [PubMed]
- Raabe, C.A.; Tang, T.H.; Brosius, J.; Rozhdestvensky, T.S. Biases in Small RNA Deep Sequencing Data. Nucleic Acids Res. 2014, 42, 1414–1426. [Google Scholar] [CrossRef]
- Nagai, A.; Mori, K.; Shiomi, Y.; Yoshihisa, T. OTTER, a New Method for Quantifying Absolute Amounts of tRNAs. RNA 2021, 27, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Walsh, I.M.; Bowman, M.A.; Soto Santarriaga, I.F.; Rodriguez, A.; Clark, P.L. Synonymous Codon Substitutions Perturb Cotranslational Protein Folding In Vivo and Impair Cell Fitness. Proc. Natl. Acad. Sci. USA 2020, 117, 3528–3534. [Google Scholar] [CrossRef] [PubMed]
- Rapino, F.; Zhou, Z.; Roncero Sanchez, A.M.; Joiret, M.; Seca, C.; El Hachem, N.; Valenti, G.; Latini, S.; Shostak, K.; Geris, L.; et al. Wobble tRNA Modification and Hydrophilic Amino Acid Patterns Dictate Protein Fate. Nat. Commun. 2021, 12, 2170. [Google Scholar] [CrossRef] [PubMed]
- Presnyak, V.; Alhusaini, N.; Chen, Y.-H.; Martin, S.; Morris, N.; Kline, N.; Olson, S.; Weinberg, D.; Baker, K.E.; Graveley, B.R.; et al. Codon Optimality Is a Major Determinant of mRNA Stability. Cell 2015, 160, 1111–1124. [Google Scholar] [CrossRef] [PubMed]
- Hanson, G.; Alhusaini, N.; Morris, N.; Sweet, T.; Coller, J. Translation Elongation and mRNA Stability Are Coupled through the Ribosomal A-Site. RNA 2018, 24, 1377–1389. [Google Scholar] [CrossRef] [PubMed]
- Harigaya, Y.; Parker, R. Analysis of the Association between Codon Optimality and mRNA Stability in Schizosaccharomyces pombe. BMC Genom. 2016, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Maier, K.C.; Avsec, Ž.; Petra, R.U.S.; Gagneur, J. Cis-Regulatory Elements Explain Most of the mRNA Stability Variation across Genes in Yeast. RNA 2017, 23, 1648–1659. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Medina, S.G.; Kushawah, G.; Devore, M.L.; Castellano, L.A.; Hand, J.M.; Wright, M.; Bazzini, A.A. Translation Affects mRNA Stability in a Codon-Dependent Manner in Human Cells. eLife 2019, 8, e45396. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.; Coller, J. Codon Optimality-Mediated mRNA Degradation: Linking Translational Elongation to mRNA Stability. Mol. Cell 2022, 82, 1467–1476. [Google Scholar] [CrossRef]
- Rahaman, S.; Faravelli, S.; Voegeli, S.; Becskei, A. Polysome Propensity and Tunable Thresholds in Coding Sequence Length Enable Differential mRNA Stability. Sci. Adv. 2023, 9, eadh9545. [Google Scholar] [CrossRef] [PubMed]
- Heyer, E.E.; Moore, M.J. Redefining the Translational Status of 80S Monosomes. Cell 2016, 164, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.; Güell, M.; Serrano, L. Correlation of mRNA and Protein in Complex Biological Samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Marcotte, E.M. Insights into the Regulation of Protein Abundance from Proteomic and Transcriptomic Analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Payne, S.H. The Utility of Protein and mRNA Correlation. Trends Biochem. Sci. 2015, 40, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef] [PubMed]
- Moir, R.D.; Willis, I.M. Regulation of Pol III Transcription by Nutrient and Stress Signaling Pathways. Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Whitney, M.L.; Hurto, R.L.; Shaheen, H.H.; Hopper, A.K. Rapid and Reversible Nuclear Accumulation of CytoplasmictRNA in Response to Nutrient Availability. Mol. Biol. Cell 2007, 18, 2678–2686. [Google Scholar] [CrossRef] [PubMed]
- Bloom-Ackermann, Z.; Navon, S.; Gingold, H.; Towers, R.; Pilpel, Y.; Dahan, O. A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool. PLoS Genet. 2014, 10, e1004084. [Google Scholar] [CrossRef] [PubMed]
- Cieśla, M.; Towpik, J.; Graczyk, D.; Oficjalska-Pham, D.; Harismendy, O.; Suleau, A.; Balicki, K.; Conesa, C.; Lefebvre, O.; Boguta, M. Maf1 Is Involved in Coupling Carbon Metabolism to RNA Polymerase III Transcription. Mol. Cell. Biol. 2007, 27, 7693–7702. [Google Scholar] [CrossRef]
- Shukla, A.; Bhalla, P.; Potdar, P.K.; Jampala, P.; Bhargava, P. Transcription-Dependent Enrichment of the Yeast FACT Complex Influences Nucleosome Dynamics on the RNA Polymerase III-Transcribed Genes. RNA 2021, 27, 273–290. [Google Scholar] [CrossRef] [PubMed]
- Gerber, A.; Ito, K.; Chu, C.S.; Roeder, R.G. Gene-Specific Control of tRNA Expression by RNA Polymerase II. Mol. Cell 2020, 78, 765–778. [Google Scholar] [CrossRef]
- Yague-Sanz, C.; Migeot, V.; Larochelle, M.; Bachand, F.; Wéry, M.; Morillon, A.; Hermand, D. Chromatin Remodeling by Pol II Primes Efficient Pol III Transcription. Nat. Commun. 2023, 14, 3587. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Smith, D.K.; Ni, H.; Wu, K.; Huang, D.; Pan, S.; Sathe, A.A.; Tang, Y.; Liu, M.-L.; Xing, C.; et al. SOX4-Mediated Repression of Specific tRNAs Inhibits Proliferation of Human Glioblastoma Cells. Proc. Natl. Acad. Sci. USA 2020, 117, 5782–5790. [Google Scholar] [CrossRef] [PubMed]
- van Breugel, M.E.; van Kruijsbergen, I.; Mittal, C.; Lieftink, C.; Brouwer, I.; van den Brand, T.; Kluin, R.J.C.; Hoekman, L.; Menezes, R.X.; van Welsem, T.; et al. Locus-Specific Proteome Decoding Reveals Fpt1 as a Chromatin-Associated Negative Regulator of RNA Polymerase III Assembly. Mol. Cell 2023, 83, 4205–4221.e9. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.L.J.; Abo, R.; Levine, S.S.; Dedon, P.C. Diverse Cell Stresses Induce Unique Patterns of tRNA Up- and down-Regulation: tRNA-Seq for Quantifying Changes in tRNA Copy Number. Nucleic Acids Res. 2014, 42, e170. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Chalancon, G.; de Groot, N.S.; Wuster, A.; Madan Babu, M. Cells Alter Their tRNA Abundance to Selectively Regulate Protein Synthesis during Stress Conditions. Sci. Signal. 2018, 11, eaat6409. [Google Scholar] [CrossRef] [PubMed]
- Fisher, D.L.; Nurse, P. A Single Fission Yeast Mitotic Cyclin B P34cdc2 Kinase Promotes Both S-Phase and Mitosis in the Absence of G1 Cyclins. EMBO J. 1996, 15, 850–860. [Google Scholar] [CrossRef]
- Thompson, D.M.; Parker, R. The RNase Rny1p Cleaves tRNAs and Promotes Cell Death during Oxidative Stress in Saccharomyces cerevisiae. J. Cell Biol. 2009, 185, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Luhtala, N.; Parker, R. Structure-Function Analysis of Rny1 in tRNA Cleavage and Growth Inhibition. PLoS ONE 2012, 7, e41111. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V.; Wei, W.; Steinmetz, L.M. Widespread Co-Translational RNA Decay Reveals Ribosome Dynamics. Cell 2015, 161, 1400–1412. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Lu, C.; Green, P.J.; Parker, R. tRNA Cleavage Is a Conserved Response to Oxidative Stress in Eukaryotes. RNA 2008, 14, 2095–2103. [Google Scholar] [CrossRef]
- Tyczewska, A.; Grzywacz, K. tRNA-Derived Fragments as New Players in Regulatory Processes in Yeast. Yeast 2023, 40, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.S.; Nogueira, F.T.S. Plant Small RNA World Growing Bigger: tRNA-Derived Fragments, Longstanding Players in Regulatory Processes. Front. Mol. Biosci. 2021, 8, 638911. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Yao, L.; Yu, X.; Ruan, Y.; Li, Z.; Guo, J. Action Mechanisms and Research Methods of tRNA-Derived Small RNAs. Signal Transduct. Target. Ther. 2020, 5, 109. [Google Scholar] [CrossRef] [PubMed]
- Bąkowska-Żywicka, K.; Mleczko, A.M.; Kasprzyk, M.; Machtel, P.; Żywicki, M.; Twardowski, T. The Widespread Occurrence of tRNA-Derived Fragments in Saccharomyces cerevisiae. FEBS Open Bio 2016, 6, 1186–1200. [Google Scholar] [CrossRef] [PubMed]
- Zywicki, M.; Bakowska-Zywicka, K.; Polacek, N. Revealing Stable Processing Products from Ribosome-Associated Small RNAs by Deep-Sequencing Data Analysis. Nucleic Acids Res. 2012, 40, 4013–4024. [Google Scholar] [CrossRef] [PubMed]
- Mleczko, A.M.; Celichowski, P.; Bąkowska-Żywicka, K. Transfer RNA-Derived Fragments Target and Regulate Ribosome-Associated Aminoacyl-Transfer RNA Synthetases. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Bakowska-Zywicka, K.; Kasprzyk, M.; Twardowski, T. tRNA-Derived Short RNAs Bind to Saccharomyces cerevisiae Ribosomes in a Stress-Dependent Manner and Inhibit Protein Synthesis In Vitro. FEMS Yeast Res. 2016, 16, fow077. [Google Scholar] [CrossRef]
- Streit, R.S.A.; Ferrareze, P.A.G.; Vainstein, M.H.; Staats, C.C. Analysis of tRNA-Derived RNA Fragments (TRFs) in Cryptococcus spp.: RNAi-Independent Generation and Possible Compensatory Effects in a RNAi-Deficient Genotype. Fungal Biol. 2021, 125, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-Analysis of tRNA Derived RNA Fragments Reveals That They Are Evolutionarily Conserved and Associate with AGO Proteins to Recognize Specific RNA Targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef]
- Hu, Y.; Wu, L.; Zhang, P.; Wang, Z.; Shang, J.; Huang, Y. Global View of Dynamic Expression and Precise Mapping of Mitochondrial tRNAs-Derived Fragments during Stressed Conditions in S. pombe. Mitochondrion 2021, 60, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, K.; Meyer, M.R.; Jackson, B.M.; Slade, D.; Roberts, C.; Hinnebusch, A.G.; Marton, M.J. Transcriptional Profiling Shows That Gcn4p Is a Master Regulator of Gene Expression during Amino Acid Starvation in Yeast. Mol. Cell. Biol. 2001, 21, 4347. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G. Translational Regulation of GCN4 and the General Amino Acid Control of Yeast. Annu. Rev. Microbiol. 2005, 59, 407–450. [Google Scholar] [CrossRef] [PubMed]
- Kamada, Y. Novel tRNA Function in Amino Acid Sensing of Yeast TOR Complex1. Genes Cells 2017, 22, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Otsubo, Y.; Matsuo, T.; Nishimura, A.; Yamamoto, M.; Yamashita, A. tRNA Production Links Nutrient Conditions to the Onset of Sexual Differentiation through the TORC 1 Pathway. EMBO Rep. 2018, 19, e44867. [Google Scholar] [CrossRef] [PubMed]
- Hueso, G.; Aparicio-Sanchis, R.; Montesinos, C.; Lorenz, S.; Murguía, J.R.; Serrano, R. A Novel Role for Protein Kinase Gcn2 in Yeast Tolerance to Intracellular Acid Stress. Biochem. J. 2012, 441, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-J.; Wu, Y.-H.; Huang, H.-Y.; Wang, C.-C. Saccharomyces cerevisiae Possesses a Stress-Inducible Glycyl-tRNA Synthetase Gene. PLoS ONE 2012, 7, e33363. [Google Scholar] [CrossRef] [PubMed]
- Simos, G.; Sauer, A.; Fasiolo, F.; Hurt, E.C. A Conserved Domain within Arc1p Delivers tRNA to Aminoacyl-tRNA Synthetases. Mol. Cell 1998, 1, 235–242. [Google Scholar] [CrossRef]
- Godinic, V.; Mocibob, M.; Rocak, S.; Ibba, M.; Weygand-Durasevic, I. Peroxin Pex21p Interacts with the C-Terminal Noncatalytic Domain of Yeast Seryl-tRNA Synthetase and Forms a Specific Ternary Complex with tRNA Ser. FEBS J. 2007, 274, 2788–2799. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-Y.; Chang, C.-P.; Chakraborty, S.; Wang, S.-W.; Tseng, Y.-K.; Wang, C.-C. Modulating the Structure and Function of an Aminoacyl-tRNA Synthetase Cofactor by Biotinylation. J. Biol. Chem. 2016, 291, 17102–17111. [Google Scholar] [CrossRef]
- Frechin, M.; Enkler, L.; Tetaud, E.; Laporte, D.; Senger, B.; Blancard, C.; Hammann, P.; Bader, G.; Clauder-Münster, S.; Steinmetz, L.M.; et al. Expression of Nuclear and Mitochondrial Genes Encoding ATP Synthase Is Synchronized by Disassembly of a Multisynthetase Complex. Mol. Cell 2014, 56, 763–776. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, C.; Lünse, C.E.; Mörl, M. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules 2017, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Gustilo, E.M.; Vendeix, F.A.; Agris, P.F. tRNA’s Modifications Bring Order to Gene Expression. Curr. Opin. Microbiol. 2008, 11, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Tavares, J.F.; Davis, N.K.; Poim, A.; Reis, A.; Kellner, S.; Sousa, I.; Soares, A.R.; Moura, G.M.R.; Dedon, P.C.; Santos, M. tRNA-Modifying Enzyme Mutations Induce Codon-Specific Mistranslation and Protein Aggregation in Yeast. RNA Biol. 2021, 18, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Gieg, R.; Eriani, G. The tRNA Identity Landscape for Aminoacylation and Beyond. Nucleic Acids Res. 2023, 51, 1528–1570. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Waldor, M.K. The RNA Degradosome Promotes tRNA Quality Control through Clearance of Hypomodified tRNA. Proc. Natl. Acad. Sci. USA 2019, 116, 1394–1403. [Google Scholar] [CrossRef]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA Decay Can Result from Lack of Nonessential Modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.Y.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef]
- Yoluç, Y.; van de Logt, E.; Kellner-Kaiser, S. The Stress-Dependent Dynamics of Saccharomyces cerevisiae tRNA and rRNA Modification Profiles. Genes 2021, 12, 1344. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.Y.; Deng, W.; Li, F.; Demott, M.S.; Babu, I.R.; Begley, T.J.; Dedon, P.C. Highly Predictive Reprogramming of tRNA Modifications Is Linked to Selective Expression of Codon-Biased Genes. Chem. Res. Toxicol. 2015, 28, 978–988. [Google Scholar] [CrossRef]
- Chan, C.T.Y.; Pang, Y.L.J.; Deng, W.; Babu, I.R.; Dyavaiah, M.; Begley, T.J.; Dedon, P.C. Reprogramming of tRNA Modifications Controls the Oxidative Stress Response by Codon-Biased Translation of Proteins. Nat. Commun. 2012, 3, 937. [Google Scholar] [CrossRef] [PubMed]
- Nakai, Y.; Nakai, M.; Yano, T. Sulfur Modifications of the Wobble U34 in tRNAs and Their Intracellular Localization in Eukaryotic Cells. Biomolecules 2017, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Damon, J.R.; Pincus, D.; Ploegh, H.L. tRNA Thiolation Links Translation to Stress Responses in Saccharomyces cerevisiae. Mol. Biol. Cell 2015, 26, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Laxman, S.; Sutter, B.M.; Wu, X.; Kumar, S.; Guo, X.; Trudgian, D.C.; Mirzaei, H.; Tu, B.P. Sulfur Amino Acids Regulate Translational Capacity and Metabolic Homeostasis through Modulation of tRNA Thiolation. Cell 2013, 154, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Kaduhr, L.; Brachmann, C.; Ravichandran, K.E.; West, J.D.; Glatt, S.; Schaffrath, R. Urm1, Not Quite a Ubiquitin-like Modifier? Microb. Cell 2021, 8, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Goehring, A.S.; Rivers, D.M.; Sprague, G.F. Attachment of the Ubiquitin-Related Protein Urm1p to the Antioxidant Protein Ahp1p. Eukaryot. Cell 2003, 2, 930–936. [Google Scholar] [CrossRef]
- Joshi, K.; Bhatt, M.J.; Farabaugh, P.J. Codon-Specific Effects of tRNA Anticodon Loop Modifications on Translational Misreading Errors in the Yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2018, 46, 10331–10339. [Google Scholar] [CrossRef] [PubMed]
- Eaglestone, S.S.; Cox, B.S.; Tuite, M.F. Translation Termination Efficiency Can Be Regulated in Saccharomyces cerevisiae by Environmental Stress through a Prion-Mediated Mechanism. EMBO J. 1999, 18, 1974–1981. [Google Scholar] [CrossRef] [PubMed]
- True, H.L.; Lindquist, S.L. A Yeast Prion Provides a Mechanism for Genetic Variation and Phenotypic Diversity. Nature 2000, 407, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jia, H.; Jankowsky, E.; Anderson, J.T. Degradation of Hypomodified tRNAiMet In Vivo Involves RNA-Dependent ATPase Activity of the DExH Helicase Mtr4p. RNA 2008, 14, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Vanácová, S.; Wolf, J.; Martin, G.; Blank, D.; Dettwiler, S.; Friedlein, A.; Langen, H.; Keith, G.; Keller, W. A New Yeast Poly(A) Polymerase Complex Involved in RNA Quality Control. PLoS Biol. 2005, 3, e189. [Google Scholar] [CrossRef]
- Chernyakov, I.; Whipple, J.M.; Kotelawala, L.; Grayhack, E.J.; Phizicky, E.M. Degradation of Several Hypomodified Mature tRNA Species in Saccharomyces cerevisiae Is Mediated by Met22 and the 5′–3′ Exonucleases Rat1 and Xrn1. Genes Dev. 2008, 22, 1369–1380. [Google Scholar] [CrossRef] [PubMed]
- Tasak, M.; Phizicky, E.M. Initiator tRNA Lacking 1-Methyladenosine Is Targeted by the Rapid tRNA Decay Pathway in Evolutionarily Distant Yeast Species. PLOS Genet. 2022, 18, e1010215. [Google Scholar] [CrossRef] [PubMed]
- Kramer, E.B.; Hopper, A.K. Retrograde Transfer RNA Nuclear Import Provides a New Level of tRNA Quality Control in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2013, 110, 21042–21047. [Google Scholar] [CrossRef]
- Liu, F.; Clark, W.; Luo, G.; Wang, X.; Fu, Y.; Wei, J.; Wang, X.; Hao, Z.; Dai, Q.; Zheng, G.; et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 2016, 167, 816–828.e16. [Google Scholar] [CrossRef] [PubMed]
- You, X.-J.; Zhang, S.; Chen, J.-J.; Tang, F.; He, J.; Wang, J.; Qi, C.-B.; Feng, Y.-Q.; Yuan, B.-F. Formation and Removal of 1, N 6-Dimethyladenosine in Mammalian Transfer RNA. Nucleic Acids Res. 2022, 50, 9858–9872. [Google Scholar] [CrossRef] [PubMed]
- Nawrot, B.; Sochacka, E.; Düchler, M. tRNA Structural and Functional Changes Induced by Oxidative Stress. Cell. Mol. Life Sci. 2011, 68, 4023–4032. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; McCabe, V.M.; Norris, D.P.; Cooper, P.J.; Swift, S.; Rastan, S. The Product of the Mouse Xist Gene Is a 15 Kb Inactive X-Specific Transcript Containing No Conserved ORF and Located in the Nucleus. Cell 1992, 71, 515–526. [Google Scholar] [CrossRef] [PubMed]
- David, L.; Huber, W.; Granovskaia, M.; Toedling, J.; Palm, C.J.; Bofkin, L.; Jones, T.; Davis, R.W.; Steinmetz, L.M. A High-Resolution Map of Transcription in the Yeast Genome. Proc. Natl. Acad. Sci. USA 2006, 103, 5320–5325. [Google Scholar] [CrossRef] [PubMed]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D.; Gerstein, M.; Snyder, M. The Transcriptional Landscape of the Yeast Genome Defined by RNA Sequencing. Science 2008, 320, 1344–1349. [Google Scholar] [CrossRef] [PubMed]
- Hovhannisyan, H.; Gabaldón, T. The Long Non-Coding RNA Landscape of Candida Yeast Pathogens. Nat. Commun. 2021, 12, 7317. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, S.R.; Marguerat, S.; Bitton, D.A.; Rodríguez-López, M.; Rallis, C.; Lemay, J.F.; Cotobal, C.; Malecki, M.; Smialowski, P.; Mata, J.; et al. Long Noncoding RNA Repertoire and Targeting by Nuclear Exosome, Cytoplasmic Exonuclease, and RNAi in Fission Yeast. RNA 2018, 24, 1195–1213. [Google Scholar] [CrossRef] [PubMed]
- Kalem, M.C.; Panepinto, J.C. Long Non-Coding RNAs in Cryptococcus neoformans: Insights Into Fungal Pathogenesis. Front. Cell. Infect. Microbiol. 2022, 12, 261. [Google Scholar] [CrossRef] [PubMed]
- Jenjaroenpun, P.; Wongsurawat, T.; Pereira, R.; Patumcharoenpol, P.; Ussery, D.W.; Nielsen, J.; Nookaew, I. Complete Genomic and Transcriptional Landscape Analysis Using Third-Generation Sequencing: A Case Study of Saccharomyces cerevisiae CEN.PK113-7D. Nucleic Acids Res. 2018, 46, e38. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wei, W.; Gagneur, J.; Clauder-Münster, S.; Smolik, M.; Huber, W.; Steinmetz, L.M. Antisense Expression Increases Gene Expression Variability and Locus Interdependency. Mol. Syst. Biol. 2011, 7, 468. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wei, W.; Gagneur, J.; Perocchi, F.; Clauder-Münster, S.; Camblong, J.; Guffanti, E.; Stutz, F.; Huber, W.; Steinmetz, L.M. Bidirectional Promoters Generate Pervasive Transcription in Yeast. Nature 2009, 457, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Quintales, L.; Sánchez, M.; Antequera, F. Analysis of DNA Strand-Specific Differential Expression with High Density Tiling Microarrays. BMC Bioinform. 2010, 11, 136. [Google Scholar] [CrossRef] [PubMed]
- Leong, H.S.; Dawson, K.; Wirth, C.; Li, Y.; Connolly, Y.; Smith, D.L.; Wilkinson, C.R.M.; Miller, C.J. A Global Non-Coding RNA System Modulates Fission Yeast Protein Levels in Response to Stress. Nat. Commun. 2014, 5, 3947. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.; Váchová, L.; Hlaváček, O.; Maršíková, J.; Gilfillan, G.D.; Palková, Z. Long Noncoding RNAs in Yeast Cells and Differentiated Subpopulations of Yeast Colonies and Biofilms. Oxid. Med. Cell. Longev. 2018, 2018, 4950591. [Google Scholar] [CrossRef] [PubMed]
- Parker, S.; Fraczek, M.G.; Wu, J.; Shamsah, S.; Manousaki, A.; Dungrattanalert, K.; de Almeida, R.A.; Invernizzi, E.; Burgis, T.; Omara, W.; et al. Large-Scale Profiling of Noncoding RNA Function in Yeast. PLoS Genet. 2018, 14, e1007253. [Google Scholar] [CrossRef] [PubMed]
- Camblong, J.; Beyrouthy, N.; Guffanti, E.; Schlaepfer, G.; Steinmetz, L.M.; Stutz, F. Trans-Acting Antisense RNAs Mediate Transcriptional Gene Cosuppression in S. cerevisiae. Genes Dev. 2009, 23, 1534–1545. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, X.; Yin, Z.; Hu, Z.; Zhang, K.-Q. An Overview on Identification and Regulatory Mechanisms of Long Non-Coding RNAs in Fungi. Front. Microbiol. 2021, 12, 995. [Google Scholar] [CrossRef] [PubMed]
- Niederer, R.O.; Hass, E.P.; Zappulla, D.C. Long Noncoding RNAs in the Yeast S. cerevisiae. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2017; Volume 1008, pp. 119–132. [Google Scholar] [CrossRef]
- Smith, J.E.; Alvarez-Dominguez, J.R.; Kline, N.; Huynh, N.J.; Geisler, S.; Hu, W.; Coller, J.; Baker, K.E. Translation of Small Open Reading Frames within Unannotated RNA Transcripts in Saccharomyces cerevisiae. Cell Rep. 2014, 7, 1858–1866. [Google Scholar] [CrossRef]
- Andjus, S.; Szachnowski, U.; Vogt, N.; Gioftsidi, S.; Hatin, I.; Cornu, D.; Papadopoulos, C.; Lopes, A.; Namy, O.; Wery, M.; et al. Pervasive Translation of Xrn1-Sensitive Unstable Long Non-Coding RNAs in Yeast. RNA 2024, rna.079903.123. [Google Scholar] [CrossRef] [PubMed]
- Erpf, P.E.; Fraser, J.A. The Long History of the Diverse Roles of Short ORFs: sPEPs in Fungi. Proteomics 2018, 18, 1700219. [Google Scholar] [CrossRef] [PubMed]
- Bouyx, C.; Schiavone, M.; François, J.M. Flo11, a Developmental Gene Conferring Impressive Adaptive Plasticity to the Yeast Saccharomyces cerevisiae. Pathogens 2021, 10, 1509. [Google Scholar] [CrossRef]
- Váchová, L.; Štovíček, V.; Hlaváček, O.; Chernyavskiy, O.; Štěpánek, L.; Kubínová, L.; Palková, Z. Flo11p, Drug Efflux Pumps, and the Extracellular Matrix Cooperate to Form Biofilm Yeast Colonies. J. Cell Biol. 2011, 194, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Voordeckers, K.; De Maeyer, D.; van der Zande, E.; Vinces, M.D.; Meert, W.; Cloots, L.; Ryan, O.; Marchal, K.; Verstrepen, K.J. Identification of a Complex Genetic Network Underlying Saccharomyces cerevisiae Colony Morphology. Mol. Microbiol. 2012, 86, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Van Nguyen, P.; Plocek, V.; Váchová, L.; Palková, Z. Glucose, Cyc8p and Tup1p Regulate Biofilm Formation and Dispersal in Wild Saccharomyces cerevisiae. NPJ Biofilms Microbiomes 2020, 6, 7. [Google Scholar] [CrossRef]
- Correia-Melo, C.; Kamrad, S.; Tengölics, R.; Messner, C.B.; Trebulle, P.; Townsend, S.J.; Jayasree Varma, S.; Freiwald, A.; Heineike, B.M.; Campbell, K.; et al. Cell-Cell Metabolite Exchange Creates a pro-Survival Metabolic Environment That Extends Lifespan. Cell 2023, 186, 63–79.e21. [Google Scholar] [CrossRef] [PubMed]
- Palková, Z.; Váchová, L. Life within a Community: Benefit to Yeast Long-Term Survival. FEMS Microbiol. Rev. 2006, 30, 806–824. [Google Scholar] [CrossRef] [PubMed]
- Brückner, S.; Mösch, H.U. Choosing the Right Lifestyle: Adhesion and Development in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2012, 36, 25–58. [Google Scholar] [CrossRef] [PubMed]
- Bumgarner, S.L.; Dowell, R.D.; Grisafi, P.; Gifford, D.K.; Fink, G.R. Toggle Involving Cis-Interfering Noncoding RNAs Controls Variegated Gene Expression in Yeast. Proc. Natl. Acad. Sci. USA 2009, 106, 18321–18326. [Google Scholar] [CrossRef] [PubMed]
- Chacko, N.; Zhao, Y.; Yang, E.; Wang, L.; Cai, J.J.; Lin, X. The lncRNA RZE1 Controls Cryptococcal Morphological Transition. PLoS Genet. 2015, 11, e1005692. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Lin, X. Matricellular Protein Cfl1 Regulates Cell Differentiation. Commun. Integr. Biol. 2013, 6, e26444. [Google Scholar] [CrossRef] [PubMed]
- Solé, C.; Nadal-Ribelles, M.; de Nadal, E.; Posas, F. A Novel Role for lncRNAs in Cell Cycle Control during Stress Adaptation. Curr. Genet. 2015, 61, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Nadal-Ribelles, M.; Solé, C.; Xu, Z.; Steinmetz, L.M.; de Nadal, E.; Posas, F. Control of Cdc28 CDK1 by a Stress-Induced lncRNA. Mol. Cell 2014, 53, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Neil, H.; Malabat, C.; D’Aubenton-Carafa, Y.; Xu, Z.; Steinmetz, L.M.; Jacquier, A. Widespread Bidirectional Promoters Are the Major Source of Cryptic Transcripts in Yeast. Nature 2009, 457, 1038–1042. [Google Scholar] [CrossRef] [PubMed]
- Balarezo-Cisneros, L.N.; Parker, S.; Fraczek, M.G.; Timouma, S.; Wang, P.; O’Keefe, R.T.; Millar, C.B.; Delneri, D. Functional and Transcriptional Profiling of Non-Coding RNAs in Yeast Reveal Context-Dependent Phenotypes and in Trans Effects on the Protein Regulatory Network. PLoS Genet. 2021, 17, e1008761. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Miyoshi, T.; Kugou, K.; Hoffman, C.S.; Shibata, T.; Ohta, K. Stepwise Chromatin Remodelling by a Cascade of Transcription Initiation of Non-Coding RNAs. Nature 2008, 456, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Oda, A.; Takemata, N.; Hirata, Y.; Miyoshi, T.; Suzuki, Y.; Sugano, S.; Ohta, K. Dynamic Transition of Transcription and Chromatin Landscape during Fission Yeast Adaptation to Glucose Starvation. Genes Cells 2015, 20, 392–407. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Lopez, M.; Anver, S.; Cotobal, C.; Kamrad, S.; Malecki, M.; Correia-Melo, C.; Hoti, M.; Townsend, S.; Marguerat, S.; Pong, S.K.; et al. Functional Profiling of Long Intergenic Non-Coding RNAs in Fission Yeast. eLife 2022, 11, e76000. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zhou, C.; Kennedy, B.K. The Yeast Replicative Aging Model. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2690–2696. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Ganley, A.R.D. Recombination Regulation by Transcription-Induced Cohesin Dissociation in rDNA Repeats. Science 2005, 309, 1581–1584. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, M.; Sasaki, M.; Kobayashi, T. Spt4 Promotes Cellular Senescence by Activating Non-Coding RNA Transcription in Ribosomal RNA Gene Clusters. Cell Rep. 2023, 42, 111944. [Google Scholar] [CrossRef] [PubMed]
- Traven, A.; Jänicke, A.; Harrison, P.; Swaminathan, A.; Seemann, T.; Beilharz, T.H. Transcriptional Profiling of a Yeast Colony Provides New Insight into the Heterogeneity of Multicellular Fungal Communities. PLoS ONE 2012, 7, e46243. [Google Scholar] [CrossRef] [PubMed]
- Čáp, M.; Štěpánek, L.; Harant, K.; Váchová, L.; Palková, Z. Cell Differentiation within a Yeast Colony: Metabolic and Regulatory Parallels with a Tumor-Affected Organism. Mol. Cell 2012, 46, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Čáp, M.; Váchová, L.; Palková, Z. Reactive Oxygen Species in the Signaling and Adaptation of Multicellular Microbial Communities. Oxid. Med. Cell. Longev. 2012, 2012, 976753. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.; Maršíková, J.; Hlaváček, O.; Gilfillan, G.D.; Ježková, E.; Aaløkken, R.; Váchová, L.; Palková, Z. Transcriptome Remodeling of Differentiated Cells during Chronological Ageing of Yeast Colonies: New Insights into Metabolic Differentiation. Oxid. Med. Cell. Longev. 2018, 2018, 4932905. [Google Scholar] [CrossRef]
- Gao, J.; Chow, E.W.L.; Wang, H.; Xu, X.; Cai, C.; Song, Y.; Wang, J.; Wang, Y. lncRNA DINOR Is a Virulence Factor and Global Regulator of Stress Responses in Candida auris. Nat. Microbiol. 2021, 6, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Willi, J.; Küpfer, P.; Eviquoz, D.; Fernandez, G.; Katz, A.; Leumann, C.; Polacek, N. Oxidative Stress Damages rRNA inside the Ribosome and Differentially Affects the Catalytic Center. Nucleic Acids Res. 2018, 46, 1945–1957. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, H.; Regnier, F. Protein−RNA Cross-Linking in the Ribosomes of Yeast under Oxidative Stress. J. Proteome Res. 2006, 5, 3249–3259. [Google Scholar] [CrossRef]
- Shedlovskiy, D.; Zinskie, J.A.; Gardner, E.; Pestov, D.G.; Shcherbik, N. Endonucleolytic Cleavage in the Expansion Segment 7 of 25S rRNA Is an Early Marker of Low-Level Oxidative Stress in Yeast. J. Biol. Chem. 2017, 292, 18469–18485. [Google Scholar] [CrossRef] [PubMed]
- Shankar, V.; Rauscher, R.; Reuther, J.; Gharib, W.H.; Koch, M.; Polacek, N. rRNA Expansion Segment 27Lb Modulates the Factor Recruitment Capacity of the Yeast Ribosome and Shapes the Proteome. Nucleic Acids Res. 2020, 48, 3244–3256. [Google Scholar] [CrossRef] [PubMed]
- Knorr, A.G.; Schmidt, C.; Tesina, P.; Berninghausen, O.; Becker, T.; Beatrix, B.; Beckmann, R. Ribosome–NatA Architecture Reveals That rRNA Expansion Segments Coordinate N-Terminal Acetylation. Nat. Struct. Mol. Biol. 2019, 26, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Fujii, K.; Susanto, T.T.; Saurabh, S.; Barna, M. Decoding the Function of Expansion Segments in Ribosomes. Mol. Cell 2018, 72, 1013–1020.e6. [Google Scholar] [CrossRef] [PubMed]
- Zinskie, J.A.; Ghosh, A.; Trainor, B.M.; Shedlovskiy, D.; Pestov, D.G.; Shcherbik, N. Iron-Dependent Cleavage of Ribosomal RNA during Oxidative Stress in the Yeast Saccharomyces cerevisiae. J. Biol. Chem. 2018, 293, 14237–14248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Smith, A.D.; Renfrow, M.B.; Schneider, D.A. The RNA Polymerase-Associated Factor 1 Complex (Paf1C) Directly Increases the Elongation Rate of RNA Polymerase I and Is Required for Efficient Regulation of rRNA Synthesis. J. Biol. Chem. 2010, 285, 14152–14159. [Google Scholar] [CrossRef] [PubMed]
- Koper, M.; Mroczek, S. Analysis of rRNA Synthesis Using Quantitative Transcription Run-on (qTRO) in Yeast. Biotechniques 2018, 65, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Mroczek, S.; Kufel, J. Apoptotic Signals Induce Specific Degradation of Ribosomal RNA in Yeast. Nucleic Acids Res. 2008, 36, 2874–2888. [Google Scholar] [CrossRef] [PubMed]
- Najmi, S.M.; Schneider, D.A. Quorum Sensing Regulates rRNA Synthesis in Saccharomyces cerevisiae. Gene 2021, 776, 145442. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, J.; Rocha, M.A. Nutrient Depletion and TOR Inhibition Induce 18S and 25S Ribosomal RNAs Resistant to a 5′-Phosphate-Dependent Exonuclease in Candida albicans and Other Yeasts. BMC Mol. Biol. 2018, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Rocha, M.A.; Gowda, B.S.; Fleischmann, J. RNAP II Produces Capped 18S and 25S Ribosomal RNAs Resistant to 5′-Monophosphate Dependent Processive 5′ to 3′ Exonuclease in Polymerase Switched Saccharomyces cerevisiae. BMC Mol. Cell Biol. 2022, 23, 17. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.D.; Talkish, J.; Ding, H.; Igel, H.; Duran, A.; Mantripragada, S.; Paten, B.; Ares, M. Concerted Modification of Nucleotides at Functional Centers of the Ribosome Revealed by Single-Molecule RNA Modification Profiling. eLife 2022, 11, e76562. [Google Scholar] [CrossRef]
- Liu, K.; Santos, D.A.; Hussmann, J.A.; Wang, Y.; Sutter, B.M.; Weissman, J.S.; Tu, B.P. Regulation of Translation by Methylation Multiplicity of 18S rRNA. Cell Rep. 2021, 34, 108825. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.T.; Fink, G.R.; Bartel, D.P. Excised Linear Introns Regulate Growth in Yeast. Nature 2019, 565, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Parenteau, J.; Maignon, L.; Berthoumieux, M.; Catala, M.; Gagnon, V.; Abou Elela, S. Introns Are Mediators of Cell Response to Starvation. Nature 2019, 565, 612–617. [Google Scholar] [CrossRef] [PubMed]
- Juneau, K.; Miranda, M.; Hillenmeyer, M.E.; Nislow, C.; Davis, R.W. Introns Regulate RNA and Protein Abundance in Yeast. Genetics 2006, 174, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Xiao, M.; Yang, C.; Yu, Y.-T. U2 snRNA Is Inducibly Pseudouridylated at Novel Sites by Pus7p and snR81 RNP. EMBO J. 2011, 30, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Radwan, M.K.; Xiao, M.; Adachi, H.; Fan, J.; Yu, Y.T. The TOR Signaling Pathway Regulates Starvation-Induced Pseudouridylation of Yeast U2 snRNA. RNA 2016, 22, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Basak, A.; Query, C.C. A Pseudouridine Residue in the Spliceosome Core Is Part of the Filamentous Growth Program in Yeast. Cell Rep. 2014, 8, 966–973. [Google Scholar] [CrossRef] [PubMed]
- Cha, S.; Hong, C.P.; Kang, H.A.; Hahn, J.S. Differential Activation Mechanisms of Two Isoforms of Gcr1 Transcription Factor Generated from Spliced and Un-Spliced Transcripts in Saccharomyces cerevisiae. Nucleic Acids Res. 2021, 49, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wang, Z.; Liu, X.; Tyler, B.M. Biogenesis and Biological Functions of Extracellular Vesicles in Cellular and Organismal Communication With Microbes. Front. Microbiol. 2022, 13, 817844. [Google Scholar] [CrossRef] [PubMed]
- Liebana-Jordan, M.; Brotons, B.; Falcon-Perez, J.M.; Gonzalez, E. Extracellular Vesicles in the Fungi Kingdom. Int. J. Mol. Sci. 2021, 22, 7221. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, R.P.; Puccia, R.; Rodrigues, M.L.; Oliveira, D.L.; Joffe, L.S.; César, G.V.; Nimrichter, L.; Goldenberg, S.; Alves, L.R. Extracellular Vesicle-Mediated Export of Fungal RNA. Sci. Rep. 2015, 5, 7763. [Google Scholar] [CrossRef] [PubMed]
- Smolarz, M.; Zawrotniak, M.; Satala, D.; Rapala-Kozik, M. Extracellular Nucleic Acids Present in the Candida albicans Biofilm Trigger the Release of Neutrophil Extracellular Traps. Front. Cell. Infect. Microbiol. 2021, 11, 466. [Google Scholar] [CrossRef] [PubMed]
- Bielska, E.; Sisquella, M.A.; Aldeieg, M.; Birch, C.; O’Donoghue, E.J.; May, R.C. Pathogen-Derived Extracellular Vesicles Mediate Virulence in the Fatal Human Pathogen Cryptococcus gattii. Nat. Commun. 2018, 9, 1556. [Google Scholar] [CrossRef] [PubMed]
- Halder, L.D.; Babych, S.; Palme, D.I.; Mansouri-Ghahnavieh, E.; Ivanov, L.; Ashonibare, V.; Langenhorst, D.; Prusty, B.; Rambach, G.; Wich, M.; et al. Candida albicans Induces Cross-Kingdom miRNA Trafficking in Human Monocytes To Promote Fungal Growth. mBio 2022, 13, e03563-21. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.M.; McKenna, M.M.; Walsh, C.P.; McKenna, D.J. miR-24 Regulates CDKN1B/P27 Expression in Prostate Cancer. Prostate 2016, 76, 637–648. [Google Scholar] [CrossRef] [PubMed]


| RNA Type | Regulation | Possible Function |
|---|---|---|
| tRNA | modifications | translation rate |
| stability | codon-dependent translation | |
| aminoacylation | ||
| tRF * | production by | inhibition of aa-tRNA synthetases |
| tRNA cleavage | inhibition of ribosomes | |
| lncRNA * | expression | regulation of transcription |
| snRNA * | modifications | regulation of splicing |
| intronic RNA | linearisation and stabilisation | regulation of splicing |
| rRNA | cleavage modifications 5‘capping | regulation of ribosomal functions |
| miRNA-like * | expression | regulation of expression |
| evRNA * | secretion | various/unknown |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Čáp, M.; Palková, Z. Non-Coding RNAs: Regulators of Stress, Ageing, and Developmental Decisions in Yeast? Cells 2024, 13, 599. https://doi.org/10.3390/cells13070599
Čáp M, Palková Z. Non-Coding RNAs: Regulators of Stress, Ageing, and Developmental Decisions in Yeast? Cells. 2024; 13(7):599. https://doi.org/10.3390/cells13070599
Chicago/Turabian StyleČáp, Michal, and Zdena Palková. 2024. "Non-Coding RNAs: Regulators of Stress, Ageing, and Developmental Decisions in Yeast?" Cells 13, no. 7: 599. https://doi.org/10.3390/cells13070599
APA StyleČáp, M., & Palková, Z. (2024). Non-Coding RNAs: Regulators of Stress, Ageing, and Developmental Decisions in Yeast? Cells, 13(7), 599. https://doi.org/10.3390/cells13070599






