Unraveling Desmin’s Head Domain Structure and Function
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Two-Hybrid Library Screening
2.2. Generation and Purification of Glutathione S-Transferase (GST) Fusion Proteins
2.3. Preparation of Desmin-Enriched Cardiac Extracts
2.4. Preparation of Total Protein Cardiac Extracts
2.5. Desmin Head Deletion Protein Preparation
Plasmid Vector Construction
2.6. Cell Culture Transfection and Total Protein Extracts Isolation
2.7. Glutathione S-Transferase Pulldown Assay
2.8. Immunofluorescent Staining
2.9. Homology Modeling and Model Evaluation
2.10. Molecular Electrostatic Potential (MEP)
2.11. Docking Studies and Protein–Protein Interactions
2.12. Energy Minimizations before Dynamics
2.13. Molecular Dynamics
2.14. Post Molecular Dynamics Analysis
3. Results
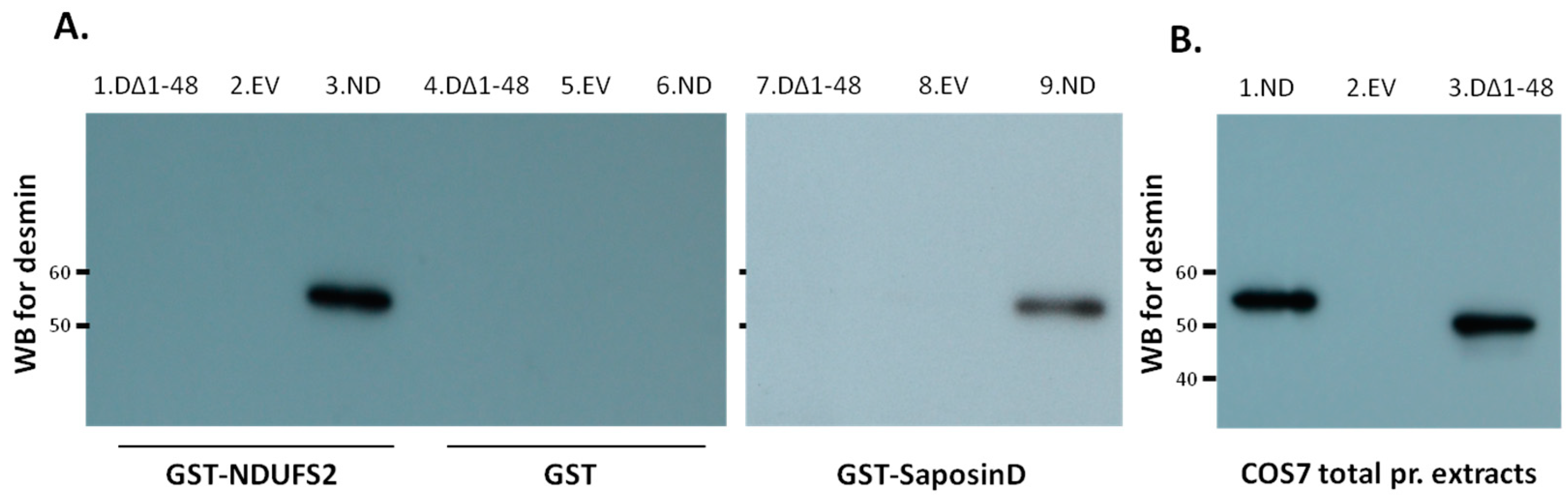
3.1. Desmin Binds to Mitochondrial and Lysosomal Proteins NDUFS2 and Saposin D, Respectively
3.2. In Silico Analysis of Interactions
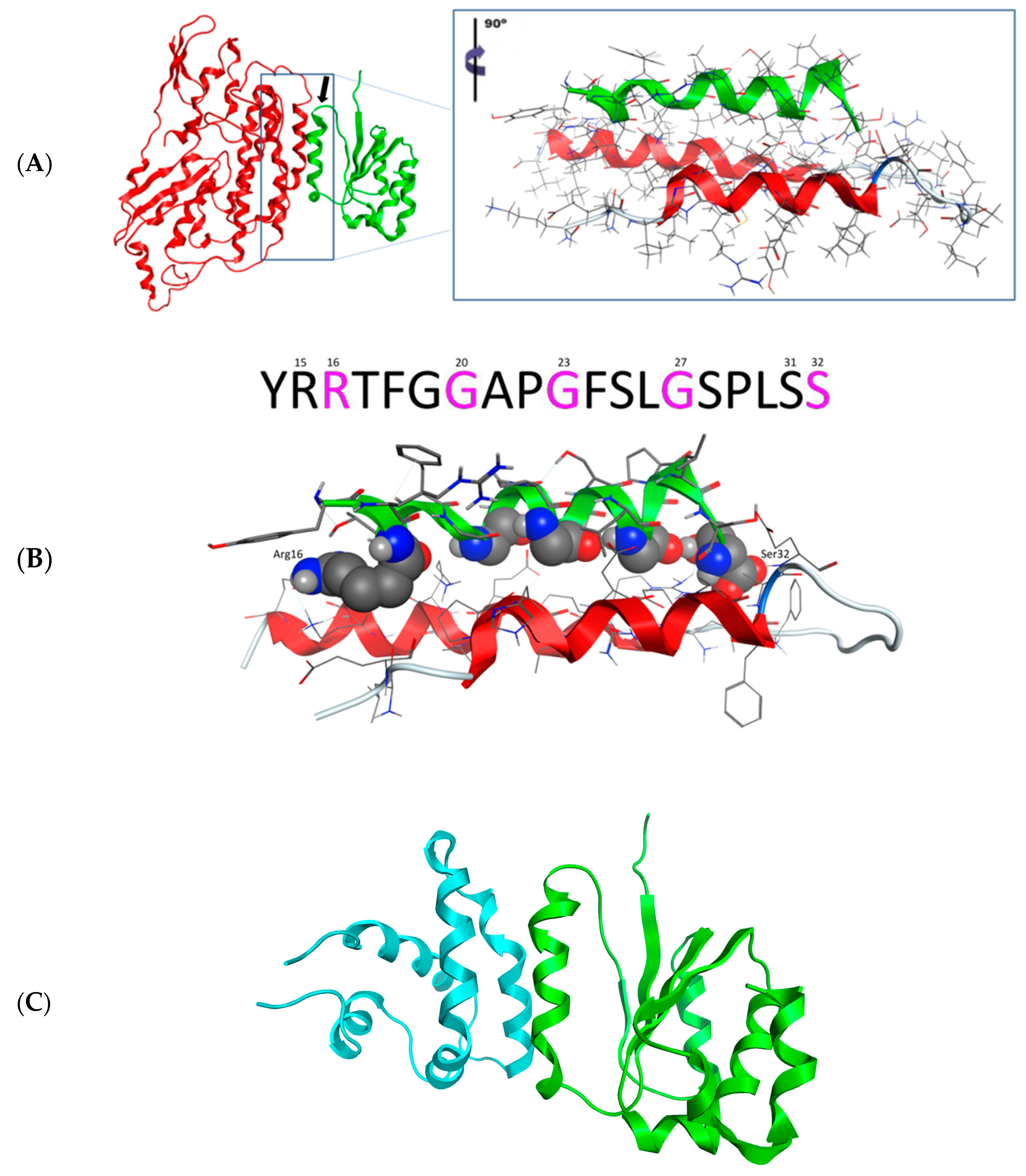
3.2.1. Three-Dimensional Model of Head Domain of Desmin
3.2.2. Three-Dimensional Model of NDUFS2 and the Saposin-D X-ray Crystal Structure
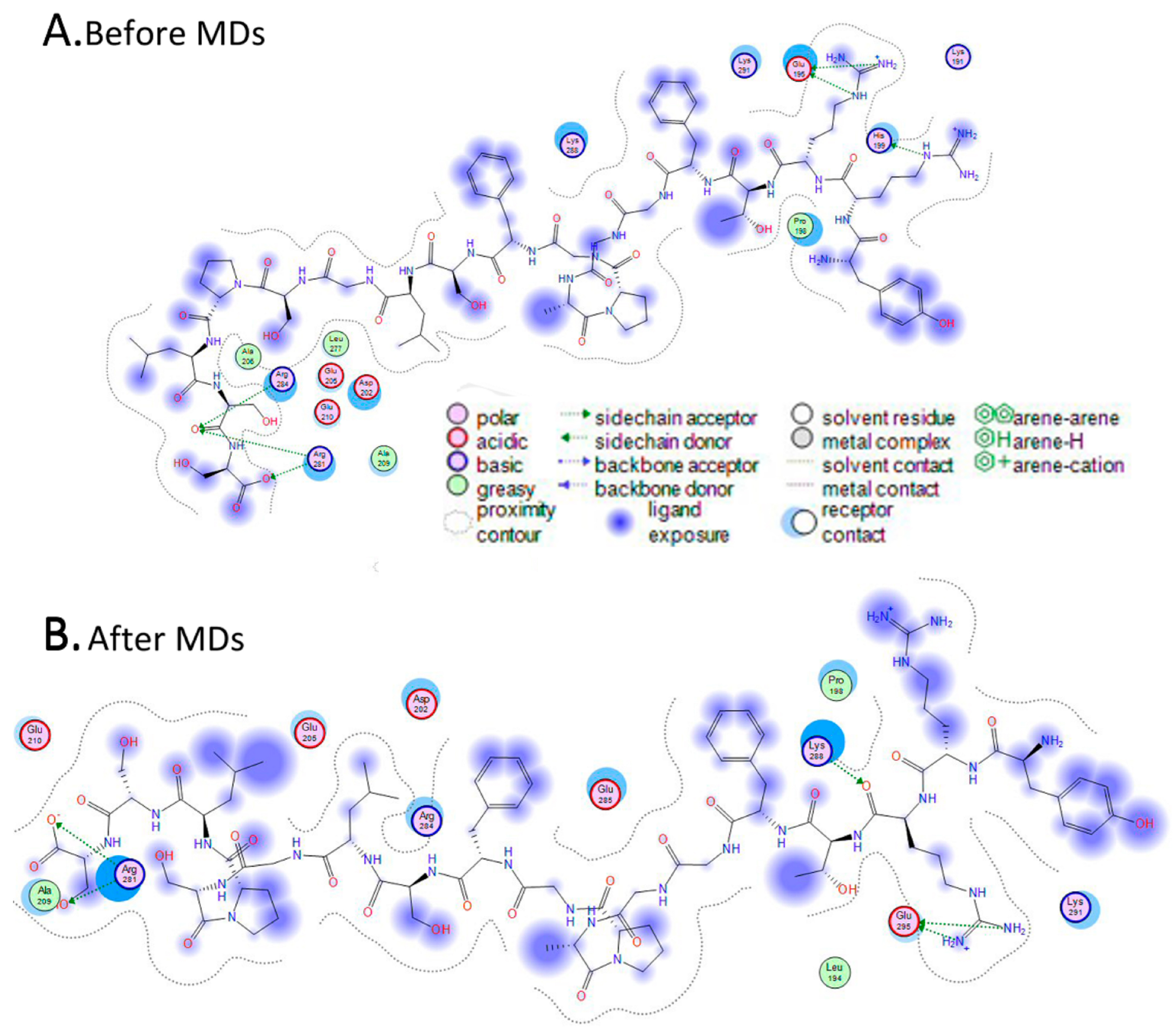
3.2.3. Molecular Docking of the Three-Dimensional Desmin Head Domain and NDUFS2
3.2.4. Molecular Docking of the Three-Dimensional Desmin Head Domain and Saposin D
3.2.5. Both Interactions Occur in a Very Similar Pattern
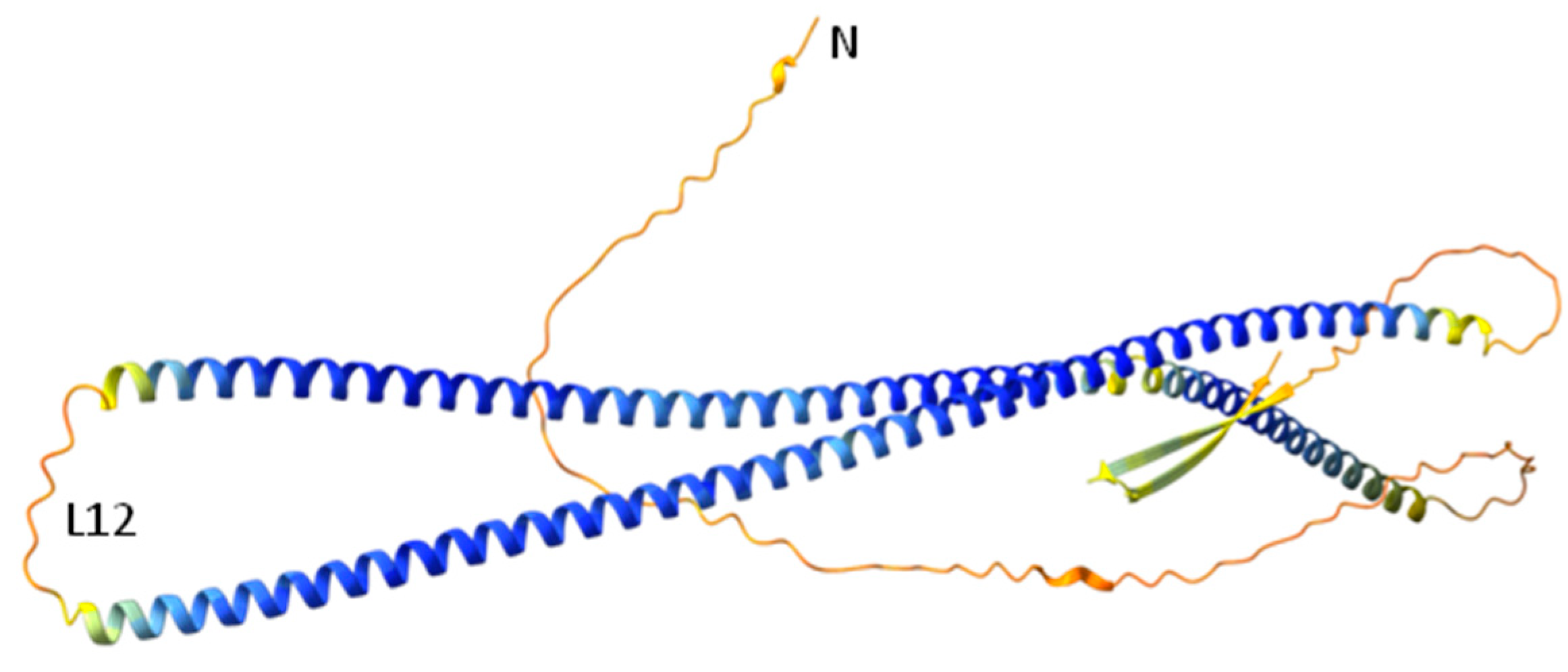
3.2.6. Prediction of Desmin’s Head Domain Three-Dimensional Structure by AlphaFold
3.3. Desmin Head Deletion Abolishes Interaction with NDUFS2 and Saposin-D
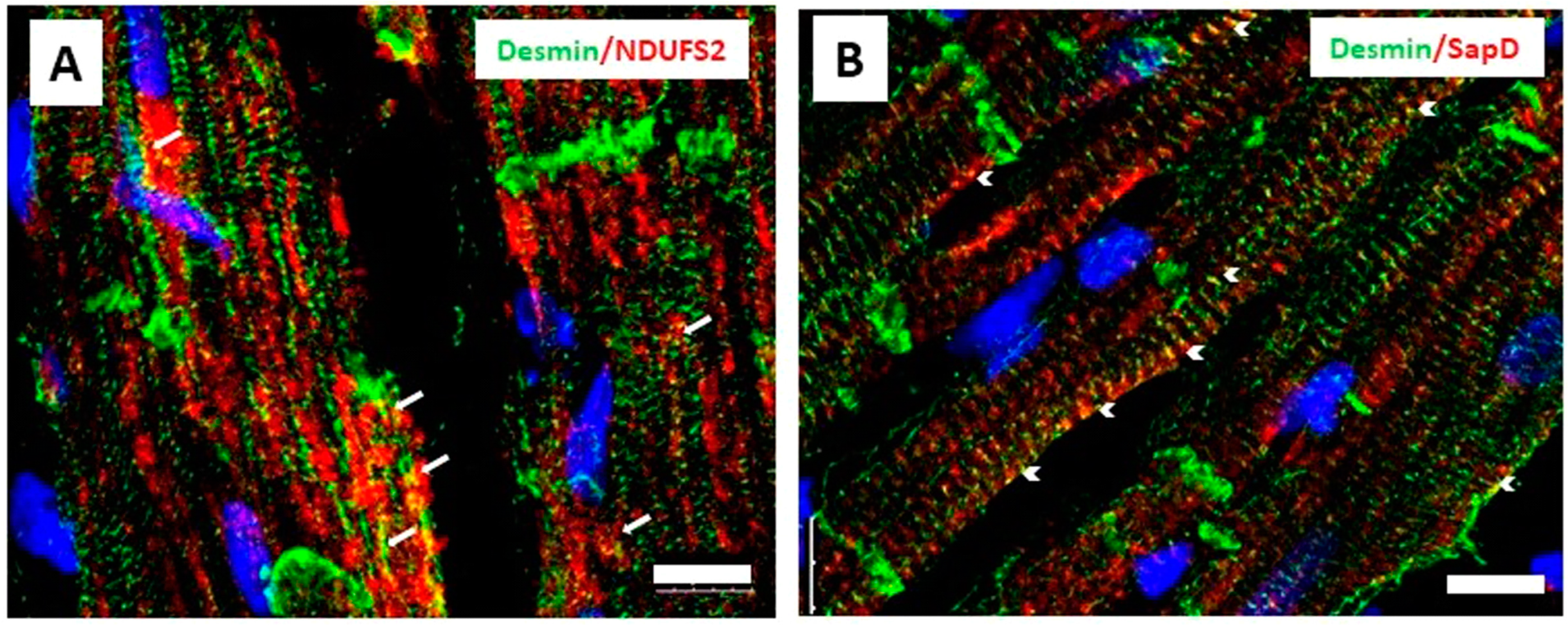
3.4. Desmin Partially Co-Localizes with NDUFS2 and Saposin D in Cardiomyocytes
4. Discussion
Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chernyatina, A.A.; Nicolet, S.; Aebi, U.; Herrmann, H.; Strelkov, S.V. Atomic structure of the vimentin central α-helical domain and its implications for intermediate filament assembly. Proc. Natl. Acad. Sci. USA 2012, 109, 13620–13625. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Aebi, U. Intermediate filaments: Molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu. Rev. Biochem. 2004, 73, 749–789. [Google Scholar] [CrossRef] [PubMed]
- Nicolet, S.; Herrmann, H.; Aebi, U.; Strelkov, S.V. Atomic structure of vimentin coil 2. J. Struct. Biol. 2010, 170, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Strelkov, S.V. History and phylogeny of intermediate filaments: Now in insects. BMC Biol. 2011, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Häner, M.; Brettel, M.; Müller, S.A.; Goldie, K.N.; Fedtke, B.; Lustig, A.; Franke, W.W.; Aebi, U. Structure and Assembly Properties of the Intermediate Filament Protein Vimentin: The Role of its Head, Rod and Tail Domains. J. Mol. Biol. 1996, 264, 933–953. [Google Scholar] [CrossRef]
- Schaffeld, M.; Herrmann, H.; Schultess, J.; Markl, J. Vimentin and desmin of a cartilaginous fish, the shark Scyliorhinus stellaris: Sequence, expression patterns and in vitro assembly. Eur. J. Cell Biol. 2001, 80, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Gruenbaum, Y.; Foisner, R. Lamins: Nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem. 2015, 84, 131–164. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Lee, J.; Jeong, S.; Kang, S.-M.; Park, B.-J.; Ha, N.-C. Beta-strand-mediated dimeric formation of the Ig-like domains of human lamin A/C and B1. Biochem. Biophys. Res. Commun. 2021, 550, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Lin, Y.; Kato, M.; Mori, E.; Liszczak, G.; Sutherland, L.; Sysoev, V.O.; Murray, D.T.; Tycko, R.; McKnight, S.L. Transiently structured head domains control intermediate filament assembly. Proc. Natl. Acad. Sci. USA 2021, 118, e2022121118. [Google Scholar] [CrossRef] [PubMed]
- Eibauer, M.; Weber, M.S.; Kronenberg-Tenga, R.; Beales, C.T.; Boujemaa-Paterski, R.; Turgay, Y.; Sivagurunathan, S.; Kraxner, J.; Köster, S.; Goldman, R.D.; et al. Vimentin filaments integrate low complexity domains in a highly complex helical structure. bioRxiv 2023, preprint. [Google Scholar] [CrossRef]
- Zhou, X.; Kato, M.; McKnight, S.L. How do disordered head domains assist in the assembly of intermediate filaments? Curr. Opin. Cell Biol. 2023, 85, 102262. [Google Scholar] [CrossRef]
- Aziz, A.; Hess, J.F.; Budamagunta, M.S.; Voss, J.C.; FitzGerald, P.G. Site-directed Spin Labeling and Electron Paramagnetic Resonance Determination of Vimentin Head Domain Structure*. J. Biol. Chem. 2010, 285, 15278–15285. [Google Scholar] [CrossRef]
- Aziz, A.; Hess, J.F.; Budamagunta, M.S.; FitzGerald, P.G.; Voss, J.C. Head and Rod 1 Interactions in Vimentin*. J. Biol. Chem. 2009, 284, 7330–7338. [Google Scholar] [CrossRef]
- Guharoy, M.; Szabo, B.; Contreras Martos, S.; Kosol, S.; Tompa, P. Intrinsic structural disorder in cytoskeletal proteins. Cytoskelet 2013, 70, 550–571. [Google Scholar] [CrossRef]
- Kornreich, M.; Avinery, R.; Malka-Gibor, E.; Laser-Azogui, A.; Beck, R. Order and disorder in intermediate filament proteins. FEBS Lett. 2015, 589, 2464–2476. [Google Scholar] [CrossRef]
- Beuttenmüller, M.; Chen, M.; Janetzko, A.; Kühn, S.; Traub, P. Structural elements of the amino-terminal head domain of vimentin essential for intermediate filament formation in vivo and in vitro. Exp. Cell Res. 1994, 213, 128–142. [Google Scholar] [CrossRef]
- Kaufmann, E.; Weber, K.; Geisler, N. Intermediate filament forming ability of desmin derivatives lacking either the amino-terminal 67 or the carboxy-terminal 27 residues. J. Mol. Biol. 1985, 185, 733–742. [Google Scholar] [CrossRef]
- Goldman, R.D.; Cleland, M.M.; Murthy, P.; Mahammad, S.; Kuczmarski, E.R. Inroads into the Structure and Function of Intermediate Filament Networks. J. Struct. Biol. 2012, 177, 14–23. [Google Scholar] [CrossRef]
- Agnetti, G.; Halperin, V.L.; Kirk, J.A.; Chakir, K.; Guo, Y.; Lund, L.; Nicolini, F.; Gherli, T.; Guarnieri, C.; Caldarera, C.M.; et al. Desmin modifications associate with amyloid-like oligomers deposition in heart failure. Cardiovasc. Res. 2014, 102, 24–34. [Google Scholar] [CrossRef]
- Sugden, P.H.; Fuller, S.J.; Weiss, S.C.; Clerk, A. Glycogen synthase kinase 3 (GSK3) in the heart: A point of integration in hypertrophic signalling and a therapeutic target? A critical analysis. Br. J. Pharmacol. 2008, 153 (Suppl. S1), S137–S153. [Google Scholar] [CrossRef]
- Rainer, P.P.; Dong, P.; Sorge, M.; Fert-Bober, J.; Holewinski, R.J.; Wang, Y.; Foss, C.A.; An, S.S.; Baracca, A.; Solaini, G.; et al. Desmin Phosphorylation Triggers Preamyloid Oligomers Formation and Myocyte Dysfunction in Acquired Heart Failure. Circ. Res. 2018, 122, e75–e83. [Google Scholar] [CrossRef]
- Geisler, N.; Weber, K. Phosphorylation of desmin in vitro inhibits formation of intermediate filaments; identification of three kinase A sites in the aminoterminal head domain. EMBO J. 1988, 7, 15–20. [Google Scholar] [CrossRef]
- Nelson, W.J.; Traub, P. Proteolysis of Vimentin and Desmin by the Ca2+-Activated Proteinase Specific for These Intermediate Filament Proteins. Mol. Cell. Biol. 1983, 3, 1146–1156. [Google Scholar] [CrossRef]
- Sharma, S.; Mücke, N.; Katus, H.A.; Herrmann, H.; Bär, H. Disease mutations in the “head” domain of the extra-sarcomeric protein desmin distinctly alter its assembly and network-forming properties. J. Mol. Med. Berl. Ger. 2009, 87, 1207–1219. [Google Scholar] [CrossRef]
- Pica, E.C.; Kathirvel, P.; Pramono, Z.A.D.; Lai, P.-S.; Yee, W.-C. Characterization of a novel S13F desmin mutation associated with desmin myopathy and heart block in a Chinese family. Neuromuscul. Disord. 2008, 18, 178–182. [Google Scholar] [CrossRef]
- van Tintelen, J.P.; Van Gelder, I.C.; Asimaki, A.; Suurmeijer, A.J.H.; Wiesfeld, A.C.P.; Jongbloed, J.D.H.; Wijngaard, A.v.D.; Kuks, J.B.; van Spaendonck-Zwarts, K.Y.; Notermans, N.; et al. Severe cardiac phenotype with right ventricular predominance in a large cohort of patients with a single missense mutation in the DES gene. Heart Rhythm 2009, 6, 1574–1583. [Google Scholar] [CrossRef]
- Arbustini, E.; Pasotti, M.; Pilotto, A.; Pellegrini, C.; Grasso, M.; Previtali, S.; Repetto, A.; Bellini, O.; Azan, G.; Scaffino, M.; et al. Desmin accumulation restrictive cardiomyopathy and atrioventricular block associated with desmin gene defects. Eur. J. Heart Fail. 2006, 8, 477–483. [Google Scholar] [CrossRef]
- Kouloumenta, A.; Mavroidis, M.; Capetanaki, Y. Proper perinuclear localization of the TRIM-like protein myospryn requires its binding partner desmin. J. Biol. Chem. 2007, 282, 35211–35221. [Google Scholar] [CrossRef]
- Diokmetzidou, A.; Tsikitis, M.; Nikouli, S.; Kloukina, I.; Tsoupri, E.; Papathanasiou, S.; Mavroidis, M.; Capetanaki, Y. Strategies to Study Desmin in Cardiac Muscle and Culture Systems. Methods Enzymol. 2016, 568, 427–459. [Google Scholar] [CrossRef]
- Pierce, B.G.; Hourai, Y.; Weng, Z. Accelerating Protein Docking in ZDOCK Using an Advanced 3D Convolution Library. PLoS ONE 2011, 6, e24657. [Google Scholar] [CrossRef]
- Hess, B.; van der Vegt, N.F.A. Hydration thermodynamic properties of amino acid analogues: A systematic comparison of biomolecular force fields and water models. J. Phys. Chem. B 2006, 110, 17616–17626. [Google Scholar] [CrossRef]
- Sellis, D.; Vlachakis, D.; Vlassi, M. Gromita: A Fully Integrated Graphical User Interface to Gromacs 4. Bioinforma. Biol. Insights 2009, 3, 99–102. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Bowie, J.U.; Lüthy, R.; Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Lefrancois, S.; Zeng, J.; Hassan, A.J.; Canuel, M.; Morales, C.R. The lysosomal trafficking of sphingolipid activator proteins (SAPs) is mediated by sortilin. EMBO J. 2003, 22, 6430–6437. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Mimaki, M.; Wang, X.; McKenzie, M.; Thorburn, D.R.; Ryan, M.T. Understanding mitochondrial complex I assembly in health and disease. Biochim. Biophys. Acta BBA-Bioenerg. 2012, 1817, 851–862. [Google Scholar] [CrossRef]
- Diokmetzidou, A.; Soumaka, E.; Kloukina, I.; Tsikitis, M.; Makridakis, M.; Varela, A.; Davos, C.H.; Georgopoulos, S.; Anesti, V.; Vlahou, A.; et al. Desmin and αB-crystallin interplay in maintenance of mitochondrial homeostasis and cardiomyocyte survival. J. Cell Sci. 2016, 129, jcs.192203. [Google Scholar] [CrossRef]
- Dayal, A.A.; Medvedeva, N.V.; Nekrasova, T.M.; Duhalin, S.D.; Surin, A.K.; Minin, A.A. Desmin Interacts Directly with Mitochondria. Int. J. Mol. Sci. 2020, 21, 8122. [Google Scholar] [CrossRef]
- Milner, D.J.; Mavroidis, M.; Weisleder, N.; Capetanaki, Y. Desmin Cytoskeleton Linked to Muscle Mitochondrial Distribution and Respiratory Function. J. Cell Biol. 2000, 150, 1283–1298. [Google Scholar] [CrossRef]
- Capetanaki, Y.; Bloch, R.J.; Kouloumenta, A.; Mavroidis, M.; Psarras, S. Muscle intermediate filaments and their links to membranes and membranous organelles. Exp. Cell Res. 2007, 313, 2063–2076. [Google Scholar] [CrossRef]
- Capetanaki, Y.; Papathanasiou, S.; Diokmetzidou, A.; Vatsellas, G.; Tsikitis, M. Desmin related disease: A matter of cell survival failure. Curr. Opin. Cell Biol. 2015, 32, 113–120. [Google Scholar] [CrossRef]
- Tsikitis, M.; Galata, Z.; Mavroidis, M.; Psarras, S.; Capetanaki, Y. Intermediate filaments in cardiomyopathy. Biophys. Rev. 2018, 10, 1007–1031. [Google Scholar] [CrossRef]
- Tuppen, H.A.L.; Hogan, V.E.; He, L.; Blakely, E.L.; Worgan, L.; Al-Dosary, M.; Saretzki, G.; Alston, C.L.; Morris, A.A.; Clarke, M.; et al. The p.M292T NDUFS2 mutation causes complex I-deficient Leigh syndrome in multiple families. Brain J. Neurol. 2010, 133, 2952–2963. [Google Scholar] [CrossRef]
- Loeffen, J.; Elpeleg, O.; Smeitink, J.; Smeets, R.; Stöckler-Ipsiroglu, S.; Mandel, H.; Sengers, R.; Trijbels, F.; van den Heuvel, L. Mutations in the complex I NDUFS2 gene of patients with cardiomyopathy and encephalomyopathy. Ann. Neurol. 2001, 49, 195–201. [Google Scholar] [CrossRef]
- Dunham-Snary, K.J.; Wu, D.; Potus, F.; Sykes, E.A.; Mewburn, J.D.; Charles, R.L.; Eaton, P.; Sultanian, R.A.; Archer, S.L. Ndufs2, a Core Subunit of Mitochondrial Complex I, Is Essential for Acute Oxygen-Sensing and Hypoxic Pulmonary Vasoconstriction. Circ. Res. 2019, 124, 1727–1746. [Google Scholar] [CrossRef]
- Liu, L.; Qi, L.; Knifley, T.; Piecoro, D.W.; Rychahou, P.; Liu, J.; Mitov, M.I.; Martin, J.; Wang, C.; Wu, J.; et al. S100A4 alters metabolism and promotes invasion of lung cancer cells by up-regulating mitochondrial complex I protein NDUFS2. J. Biol. Chem. 2019, 294, 7516–7527. [Google Scholar] [CrossRef]
- Jain, S.; Hu, C.; Kluza, J.; Ke, W.; Tian, G.; Giurgiu, M.; Bleilevens, A.; Campos, A.R.; Charbono, A.; Stickeler, E.; et al. Metabolic targeting of cancer by a ubiquinone uncompetitive inhibitor of mitochondrial complex I. Cell Chem. Biol. 2022, 29, 436–450.e15. [Google Scholar] [CrossRef]
- Timón-Gómez, A.; Nývltová, E.; Abriata, L.A.; Vila, A.J.; Hosler, J.; Barrientos, A. Mitochondrial Cytochrome c Oxidase Biogenesis: Recent Developments. Semin. Cell Dev. Biol. 2018, 76, 163–178. [Google Scholar] [CrossRef]
- Brischigliaro, M.; Zeviani, M. Cytochrome c oxidase deficiency. Biochim. Biophys. Acta BBA-Bioenerg. 2021, 1862, 148335. [Google Scholar] [CrossRef]
- Yoshida, A.; Rzhetsky, A.; Hsu, L.C.; Chang, C. Human aldehyde dehydrogenase gene family. Eur. J. Biochem. 1998, 251, 549–557. [Google Scholar] [CrossRef]
- Lorenzo, C.; Delgado, P.; Busse, C.E.; Sanz-Bravo, A.; Martos-Folgado, I.; Bonzon-Kulichenko, E.; Ferrarini, A.; Gonzalez-Valdes, I.B.; Mur, S.M.; Roldán-Montero, R.; et al. ALDH4A1 is an atherosclerosis auto-antigen targeted by protective antibodies. Nature 2021, 589, 287–292. [Google Scholar] [CrossRef]
- Schwarz, N.; Leube, R.E. Intermediate Filaments as Organizers of Cellular Space: How They Affect Mitochondrial Structure and Function. Cells 2016, 5, 30. [Google Scholar] [CrossRef]
- Matsuda, J.; Kido, M.; Tadano-Aritomi, K.; Ishizuka, I.; Tominaga, K.; Toida, K.; Takeda, E.; Suzuki, K.; Kuroda, Y. Mutation in saposin D domain of sphingolipid activator protein gene causes urinary system defects and cerebellar Purkinje cell degeneration with accumulation of hydroxy fatty acid-containing ceramide in mouse. Hum. Mol. Genet. 2004, 13, 2709–2723. [Google Scholar] [CrossRef]
- Fujita, N.; Suzuki, K.; Vanier, M.T.; Popko, B.; Maeda, N.; Klein, A.; Henseler, M.; Sandhoff, K.; Nakayasu, H.; Suzuki, K. Targeted disruption of the mouse sphingolipid activator protein gene: A complex phenotype, including severe leukodystrophy and wide-spread storage of multiple sphingolipids. Hum. Mol. Genet. 1996, 5, 711–725. [Google Scholar] [CrossRef]
- Oji, Y.; Hatano, T.; Ueno, S.-I.; Funayama, M.; Ishikawa, K.; Okuzumi, A.; Noda, S.; Sato, S.; Satake, W.; Toda, T.; et al. Variants in saposin D domain of prosaposin gene linked to Parkinson’s disease. Brain 2020, 143, 1190–1205. [Google Scholar] [CrossRef]
- Koochekpour, S.; Lee, T.-J.; Sun, Y.; Hu, S.; Grabowski, G.A.; Liu, Z.; Garay, J. Prosaposin is an AR-target gene and its neurotrophic domain upregulates AR expression and activity in prostate stromal cells. J. Cell. Biochem. 2008, 104, 2272–2285. [Google Scholar] [CrossRef]
- Wu, Y.; Sun, L.; Zou, W.; Xu, J.; Liu, H.; Wang, W.; Yun, X.; Gu, J. Prosaposin, a regulator of estrogen receptor alpha, promotes breast cancer growth. Cancer Sci. 2012, 103, 1820–1825. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhou, J.; Luo, P.; Gao, H.; Ma, Y.; Chen, Y.-S.; Li, L.; Zou, D.; Zhang, Y.; Jing, Z. Prosaposin promotes the proliferation and tumorigenesis of glioma through toll-like receptor 4 (TLR4)-mediated NF-κB signaling pathway. EBioMedicine 2018, 37, 78–90. [Google Scholar] [CrossRef]
- Lefrancois, S.; May, T.; Knight, C.; Bourbeau, D.; Morales, C.R. The lysosomal transport of prosaposin requires the conditional interaction of its highly conserved d domain with sphingomyelin. J. Biol. Chem. 2002, 277, 17188–17199. [Google Scholar] [CrossRef]
- Benson, M.A.; Tinsley, C.L.; Blake, D.J. Myospryn is a novel binding partner for dysbindin in muscle. J. Biol. Chem. 2004, 279, 10450–10458. [Google Scholar] [CrossRef]
- Benson, M.A.; Newey, S.E.; Martin-Rendon, E.; Hawkes, R.; Blake, D.J. Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J. Biol. Chem. 2001, 276, 24232–24241. [Google Scholar] [CrossRef]
- Reynolds, J.G.; McCalmon, S.A.; Donaghey, J.A.; Naya, F.J. Deregulated Protein Kinase A Signaling and Myospryn Expression in Muscular Dystrophy. J. Biol. Chem. 2008, 283, 8070–8074. [Google Scholar] [CrossRef]
- Tsoupri, E.; Kostavasili, I.; Kloukina, I.; Tsikitis, M.; Miliou, D.; Vasilaki, E.; Varela, A.; Nakos-Bimpos, M.; Davos, C.; Mavroidis, M.; et al. Myospryn deficiency leads to impaired cardiac structure and function and schizophrenia-associated symptoms. Cell Tissue Res. 2021, 385, 675–696. [Google Scholar] [CrossRef]
- Bunk, J.; Prieto Huarcaya, S.; Drobny, A.; Dobert, J.P.; Walther, L.; Rose-John, S.; Arnold, P.; Zunke, F. Cathepsin D Variants Associated with Neurodegenerative Diseases Show Dysregulated Functionality and Modified α-Synuclein Degradation Properties. Front. Cell Dev. Biol. 2021, 9, 581805. [Google Scholar] [CrossRef]
- Hiraiwa, M.; Martin, B.M.; Kishimoto, Y.; Conner, G.E.; Tsuji, S.; O’Brien, J.S. Lysosomal proteolysis of prosaposin, the precursor of saposins (sphingolipid activator proteins): Its mechanism and inhibition by ganglioside. Arch. Biochem. Biophys. 1997, 341, 17–24. [Google Scholar] [CrossRef]
- Tayebi, N.; Lopez, G.; Do, J.; Sidransky, E. Pro-cathepsin D, Prosaposin, and Progranulin: Lysosomal Networks in Parkinsonism. Trends Mol. Med. 2020, 26, 913–923. [Google Scholar] [CrossRef]
- Scally, S.W.; Petersen, J.; Law, S.C.; Dudek, N.L.; Nel, H.J.; Loh, K.L.; Wijeyewickrema, L.C.; Eckle, S.B.; van Heemst, J.; Pike, R.N.; et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J. Exp. Med. 2013, 210, 2569–2582. [Google Scholar] [CrossRef]
- Hess, J.F.; Budamagunta, M.S.; Aziz, A.; FitzGerald, P.G.; Voss, J.C. Electron paramagnetic resonance analysis of the vimentin tail domain reveals points of order in a largely disordered region and conformational adaptation upon filament assembly. Protein Sci. 2013, 22, 47–55. [Google Scholar] [CrossRef]
- Guzenko, D.; Chernyatina, A.A.; Strelkov, S.V. Crystallographic Studies of Intermediate Filament Proteins. In Fibrous Proteins: Structures and Mechanisms; Parry, D.A.D., Squire, J.M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 151–170. [Google Scholar] [CrossRef]
- Usman, S.; Aldehlawi, H.; Nguyen, T.K.N.; Teh, M.-T.; Waseem, A. Impact of N-Terminal Tags on De Novo Vimentin Intermediate Filament Assembly. Int. J. Mol. Sci. 2022, 23, 6349. [Google Scholar] [CrossRef]
- Parry, D.A.; Steinert, P.M. Intermediate filaments: Molecular architecture, assembly, dynamics and polymorphism. Q. Rev. Biophys. 1999, 32, 99–187. [Google Scholar] [CrossRef]
- Herrmann, H.; Aebi, U. Intermediate Filaments: Structure and Assembly. Cold Spring Harb. Perspect. Biol. 2016, 8, a018242. [Google Scholar] [CrossRef]
- Mücke, N.; Kämmerer, L.; Winheim, S.; Kirmse, R.; Krieger, J.; Mildenberger, M.; Baßler, J.; Hurt, E.; Goldmann, W.H.; Aebi, U.; et al. Assembly Kinetics of Vimentin Tetramers to Unit-Length Filaments: A Stopped-Flow Study. Biophys. J. 2018, 114, 2408–2418. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vlachakis, D.; Tsilafakis, K.; Kostavasili, I.; Kossida, S.; Mavroidis, M. Unraveling Desmin’s Head Domain Structure and Function. Cells 2024, 13, 603. https://doi.org/10.3390/cells13070603
Vlachakis D, Tsilafakis K, Kostavasili I, Kossida S, Mavroidis M. Unraveling Desmin’s Head Domain Structure and Function. Cells. 2024; 13(7):603. https://doi.org/10.3390/cells13070603
Chicago/Turabian StyleVlachakis, Dimitrios, Konstantinos Tsilafakis, Ioanna Kostavasili, Sophia Kossida, and Manolis Mavroidis. 2024. "Unraveling Desmin’s Head Domain Structure and Function" Cells 13, no. 7: 603. https://doi.org/10.3390/cells13070603





