Enrichment, Characterization, and Proteomic Profiling of Small Extracellular Vesicles Derived from Human Limbal Mesenchymal Stromal Cells and Melanocytes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Enrichment and Characterization of sEVs
2.1.1. Cell Isolation
2.1.2. sEV Enrichment
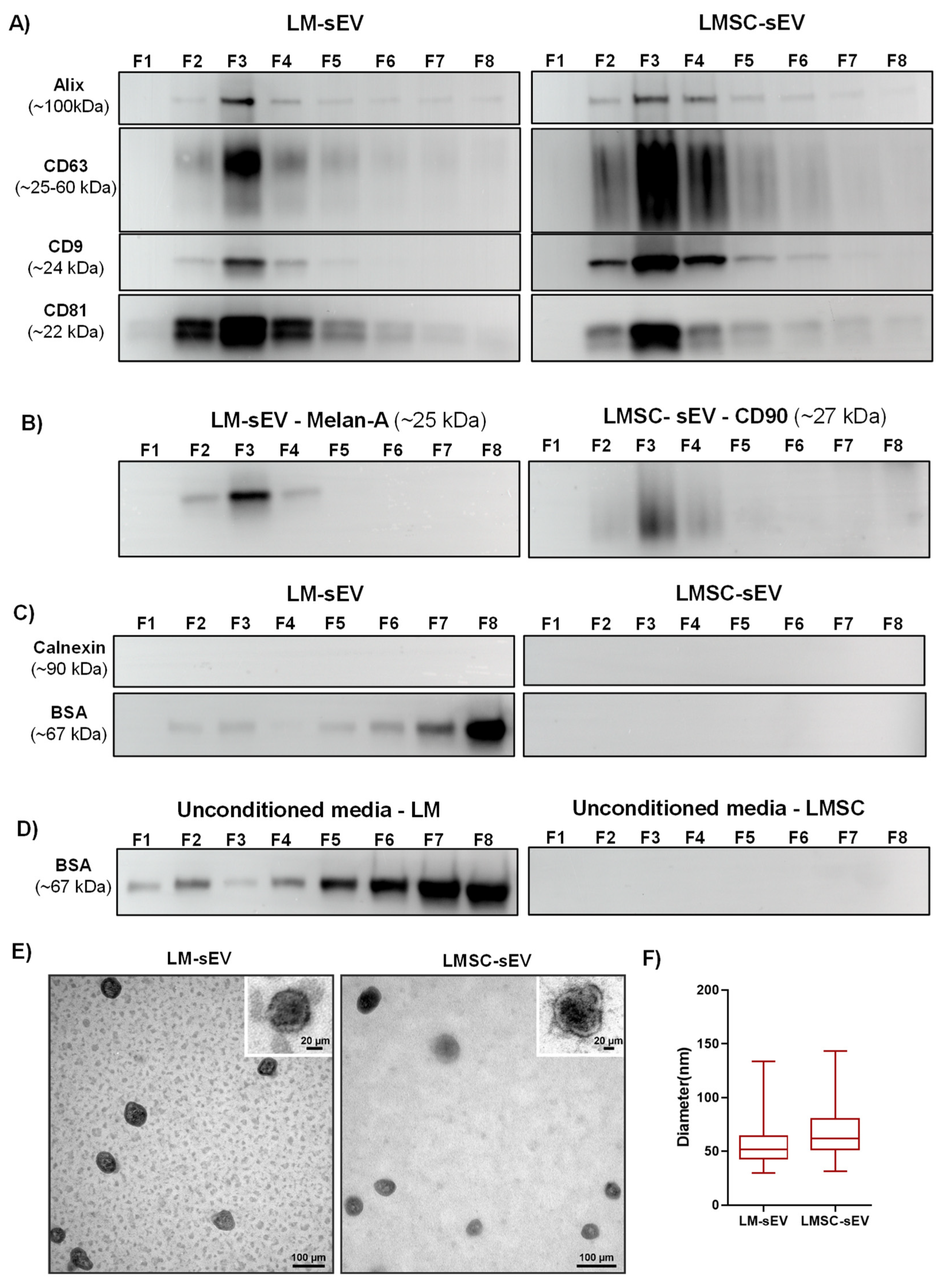
2.1.3. Biophysical Properties
2.1.4. Western Blot Analysis
2.1.5. Electron Microscopy
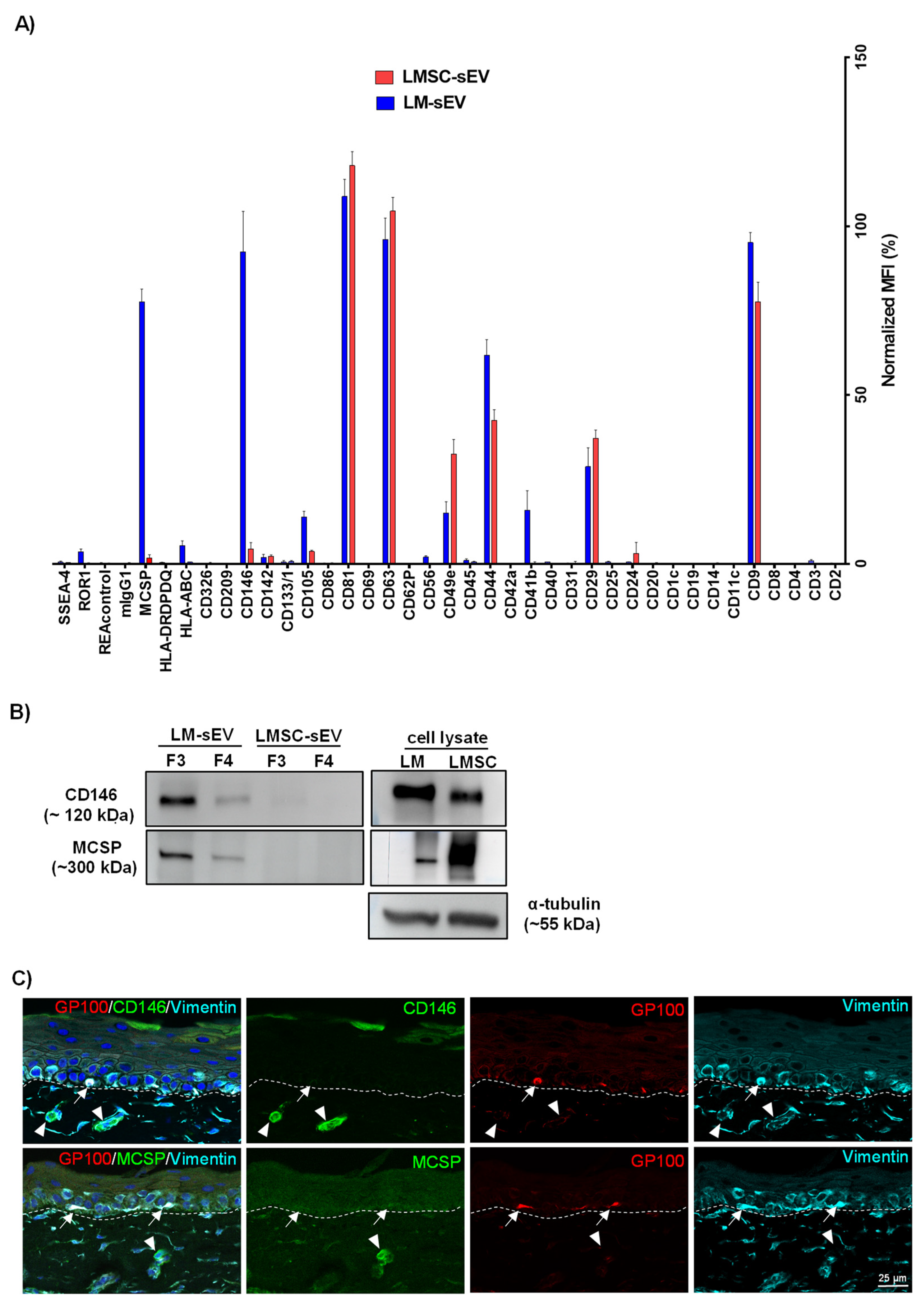
2.2. Flow Cytometry Bead Assays
2.3. Proteomics Analysis
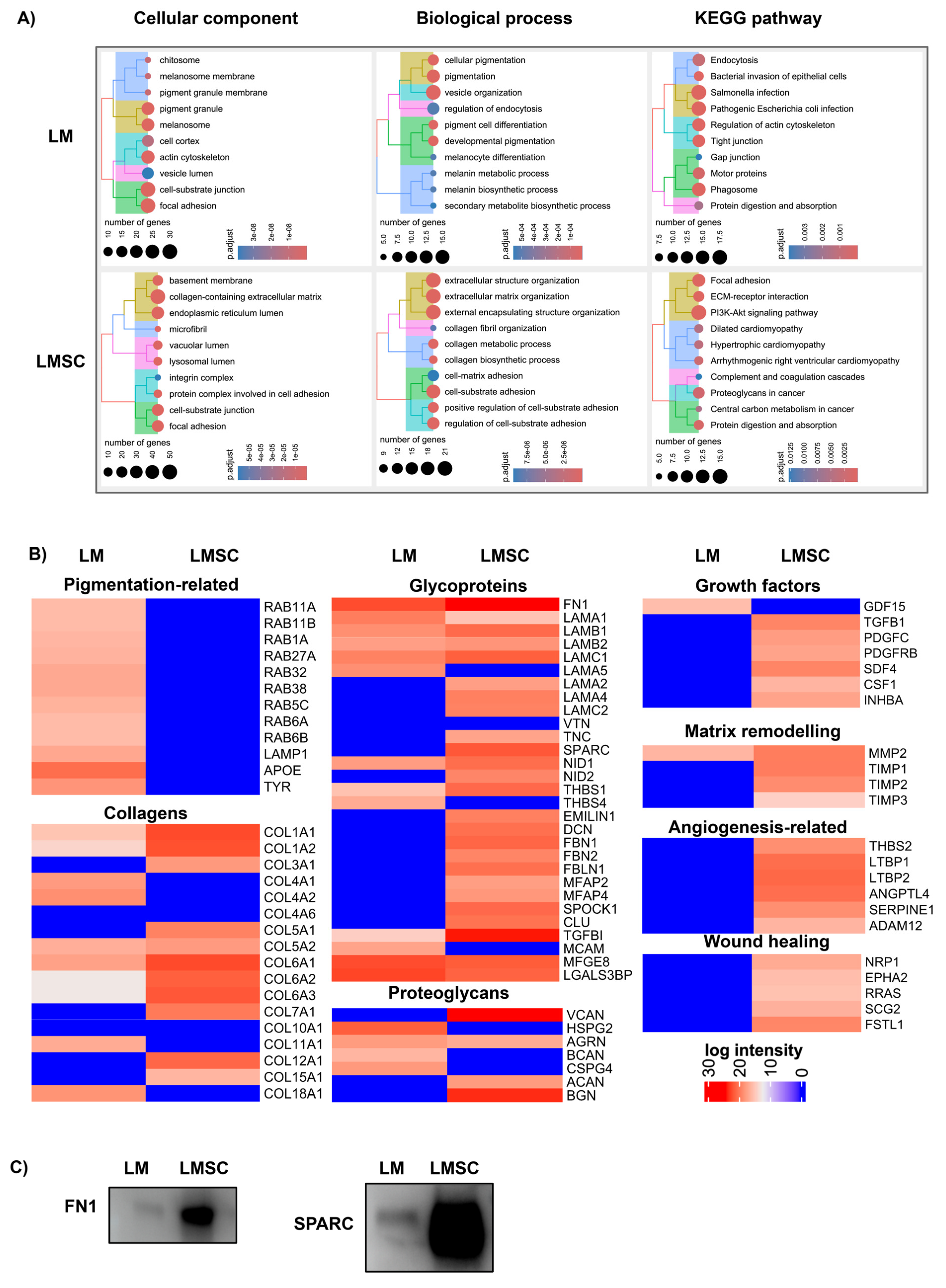
2.3.1. General sEV Proteomes
Assessment of sEV Purity
Functional Enrichment Analysis
2.3.2. Comparative sEV Proteome Profiling
Melanosome-Related Proteins and Pigmentation
Extracellular Matrix Proteins and Niche Regulation
Growth Factors, Cytokines, and Matrix Remodeling Proteins
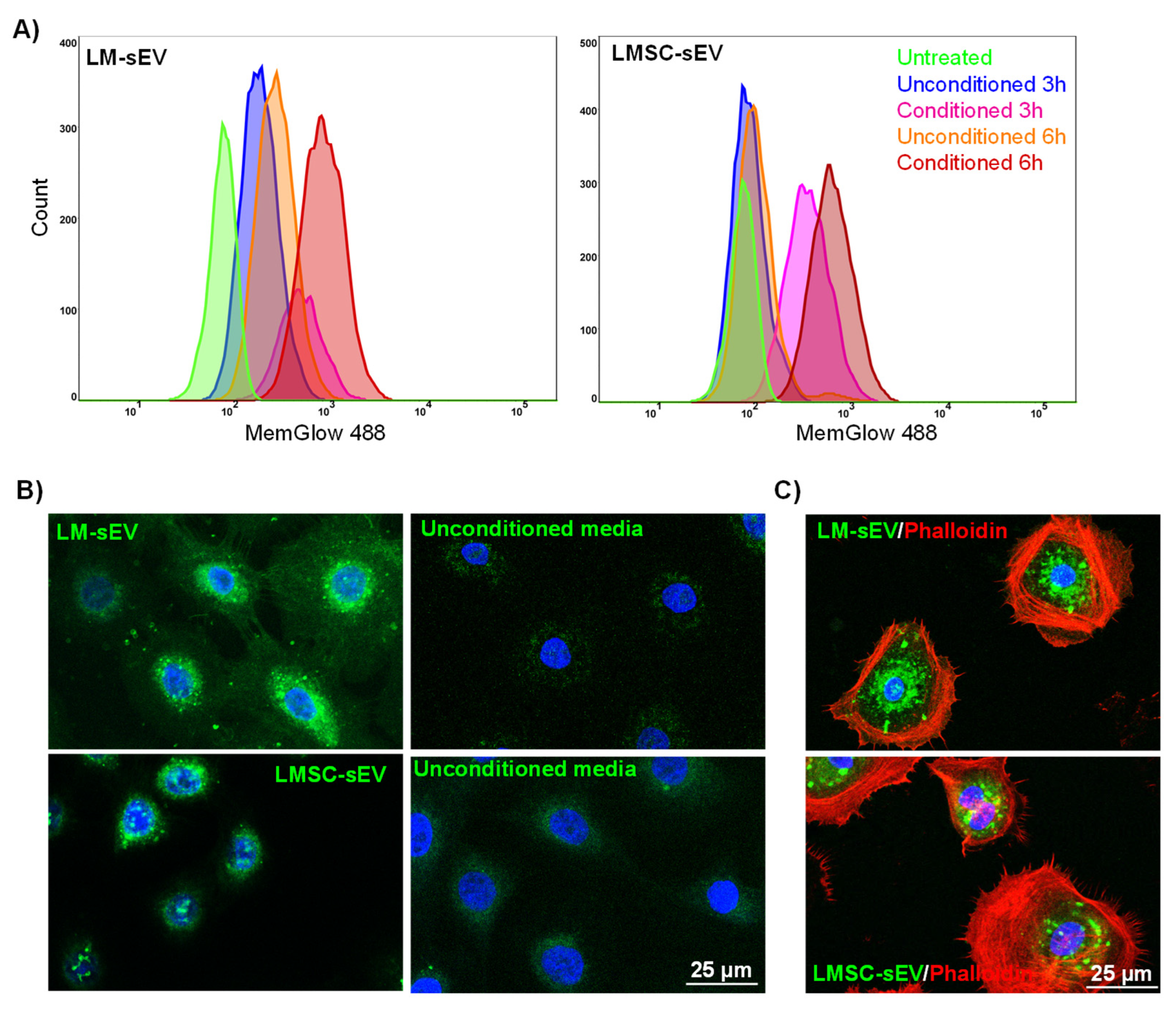
2.4. Uptake of Isolated sEVs by Recipient Cells
3. Conclusions
4. Limitations of the Study
5. Materials and Methods
5.1. Cell Isolation and Culturing
5.2. Preparation of Conditioned Media and EV Isolation
5.3. Characterization of LMSC and LM sEVs
5.3.1. Measurement of Particle Count and Total Protein Concentration
5.3.2. Transmission Electron Microscopy (TEM)
5.3.3. Western Blot Analysis
5.4. EV Analysis with the Multiplex Bead-Based Platform
5.5. Proteomics
5.6. Bioinformatics
5.7. EV Internalization Assay
5.8. Immunohistochemistry
5.9. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cotsarelis, G.; Cheng, S.Z.; Dong, G.; Sun, T.T.; Lavker, R.M. Existence of Slow-Cycling Limbal Epithelial Basal Cells That Can Be Preferentially Stimulated to Proliferate: Implications on Epithelial Stem Cells. Cell 1989, 57, 201–209. [Google Scholar] [CrossRef]
- Li, W.; Hayashida, Y.; Chen, Y.-T.; Tseng, S.C.G. Niche Regulation of Corneal Epithelial Stem Cells at the Limbus. Cell Res. 2007, 17, 26–36. [Google Scholar] [CrossRef]
- Ordonez, P.; Di Girolamo, N. Limbal Epithelial Stem Cells: Role of the Niche Microenvironment. Stem Cells 2012, 30, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Mei, H.; Gonzalez, S.; Deng, S.X. Extracellular Matrix Is an Important Component of Limbal Stem Cell Niche. J. Funct. Biomater. 2012, 3, 879–894. [Google Scholar] [CrossRef] [PubMed]
- Dziasko, M.A.; Tuft, S.J.; Daniels, J.T. Limbal Melanocytes Support Limbal Epithelial Stem Cells in 2D and 3D Microenvironments. Exp. Eye Res. 2015, 138, 70–79. [Google Scholar] [CrossRef]
- Polisetti, N.; Zenkel, M.; Menzel-Severing, J.; Kruse, F.E.; Schlötzer-Schrehardt, U. Cell Adhesion Molecules and Stem Cell-Niche-Interactions in the Limbal Stem Cell Niche. Stem Cells 2016, 34, 203–219. [Google Scholar] [CrossRef] [PubMed]
- Polisetti, N.; Gießl, A.; Li, S.; Sorokin, L.; Kruse, F.E.; Schlötzer-Schrehardt, U. Laminin-511-E8 Promotes Efficient in Vitro Expansion of Human Limbal Melanocytes. Sci. Rep. 2020, 10, 11074. [Google Scholar] [CrossRef]
- Polisetti, N.; Gießl, A.; Zenkel, M.; Heger, L.; Dudziak, D.; Naschberger, E.; Stich, L.; Steinkasserer, A.; Kruse, F.E.; Schlötzer-Schrehardt, U. Melanocytes as Emerging Key Players in Niche Regulation of Limbal Epithelial Stem Cells. Ocul. Surf. 2021, 22, 172–189. [Google Scholar] [CrossRef]
- Polisetti, N.; Sharaf, L.; Schlötzer-Schrehardt, U.; Schlunck, G.; Reinhard, T. Efficient Isolation and Functional Characterization of Niche Cells from Human Corneal Limbus. Int. J. Mol. Sci. 2022, 23, 2750. [Google Scholar] [CrossRef]
- Duan, C.-Y.; Xie, H.-T.; Zhao, X.-Y.; Xu, W.-H.; Zhang, M.-C. Limbal Niche Cells Can Reduce the Angiogenic Potential of Cultivated Oral Mucosal Epithelial Cells. Cell. Mol. Biol. Lett. 2019, 24, 3. [Google Scholar] [CrossRef]
- Amirjamshidi, H.; Milani, B.Y.; Sagha, H.M.; Movahedan, A.; Shafiq, M.A.; Lavker, R.M.; Yue, B.Y.T.; Djalilian, A.R. Limbal Fibroblast Conditioned Media: A Non-Invasive Treatment for Limbal Stem Cell Deficiency. Mol. Vis. 2011, 17, 658–666. [Google Scholar]
- An, S.; Anwar, K.; Ashraf, M.; Lee, H.; Jung, R.; Koganti, R.; Ghassemi, M.; Djalilian, A.R. Wound-Healing Effects of Mesenchymal Stromal Cell Secretome in the Cornea and the Role of Exosomes. Pharmaceutics 2023, 15, 1486. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Raposo, G. Exosomes—Vesicular Carriers for Intercellular Communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Amina, S.J.; Azam, T.; Dagher, F.; Guo, B. A Review on the Use of Extracellular Vesicles for the Delivery of Drugs and Biological Therapeutics. Expert Opin. Drug Deliv. 2024, 21, 45–70. [Google Scholar] [CrossRef] [PubMed]
- Belting, M.; Wittrup, A. Nanotubes, Exosomes, and Nucleic Acid-Binding Peptides Provide Novel Mechanisms of Intercellular Communication in Eukaryotic Cells: Implications in Health and Disease. J. Cell Biol. 2008, 183, 1187–1191. [Google Scholar] [CrossRef]
- Ratajczak, J.; Wysoczynski, M.; Hayek, F.; Janowska-Wieczorek, A.; Ratajczak, M.Z. Membrane-Derived Microvesicles: Important and Underappreciated Mediators of Cell-to-Cell Communication. Leukemia 2006, 20, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Nieuwland, R.; Sturk, A. Why Do Cells Release Vesicles? Thromb. Res. 2010, 125 (Suppl. S1), S49–S51. [Google Scholar] [CrossRef]
- Lee, I.; Choi, Y.; Shin, D.-U.; Kwon, M.; Kim, S.; Jung, H.; Nam, G.-H.; Kwon, M. Small Extracellular Vesicles as a New Class of Medicines. Pharmaceutics 2023, 15, 325. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, R.; Jiao, X.; Wu, L.; Wei, Y.; Shi, F.; Zhong, J.; Xiong, L. Small Extracellular Vesicles: Functions and Potential Clinical Applications as Cancer Biomarkers. Life 2021, 11, 1044. [Google Scholar] [CrossRef]
- Chen, H.; Wang, L.; Zeng, X.; Schwarz, H.; Nanda, H.S.; Peng, X.; Zhou, Y. Exosomes, a New Star for Targeted Delivery. Front. Cell Dev. Biol. 2021, 9, 751079. [Google Scholar] [CrossRef] [PubMed]
- Leszczynska, A.; Kulkarni, M.; Ljubimov, A.V.; Saghizadeh, M. Exosomes from Normal and Diabetic Human Corneolimbal Keratocytes Differentially Regulate Migration, Proliferation and Marker Expression of Limbal Epithelial Cells. Sci. Rep. 2018, 8, 15173. [Google Scholar] [CrossRef] [PubMed]
- Samaeekia, R.; Rabiee, B.; Putra, I.; Shen, X.; Park, Y.J.; Hematti, P.; Eslani, M.; Djalilian, A.R. Effect of Human Corneal Mesenchymal Stromal Cell-Derived Exosomes on Corneal Epithelial Wound Healing. Investig. Ophthalmol. Vis. Sci. 2018, 59, 5194–5200. [Google Scholar] [CrossRef] [PubMed]
- Shojaati, G.; Khandaker, I.; Funderburgh, M.L.; Mann, M.M.; Basu, R.; Stolz, D.B.; Geary, M.L.; Dos Santos, A.; Deng, S.X.; Funderburgh, J.L. Mesenchymal Stem Cells Reduce Corneal Fibrosis and Inflammation via Extracellular Vesicle-Mediated Delivery of MiRNA. Stem Cells Transl. Med. 2019, 8, 1192–1201. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Chen, X.; Cao, H.; Zheng, L.; Li, Q.; Zhang, K.; Han, Z.; Han, Z.-C.; Guo, Z.; Li, Z.; et al. Mesenchymal Stem Cell-Derived Extracellular Vesicles for Corneal Wound Repair. Stem Cells Int. 2019, 2019, 5738510. [Google Scholar] [CrossRef]
- Harting, M.T.; Srivastava, A.K.; Zhaorigetu, S.; Bair, H.; Prabhakara, K.S.; Toledano Furman, N.E.; Vykoukal, J.V.; Ruppert, K.A.; Cox, C.S.; Olson, S.D. Inflammation-Stimulated Mesenchymal Stromal Cell-Derived Extracellular Vesicles Attenuate Inflammation. Stem Cells 2018, 36, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Polisetti, N.; Sharaf, L.; Martin, G.; Schlunck, G.; Reinhard, T. P-Cadherin Is Expressed by Epithelial Progenitor Cells and Melanocytes in the Human Corneal Limbus. Cells 2022, 11, 1975. [Google Scholar] [CrossRef]
- Abramowicz, A.; Widlak, P.; Pietrowska, M. Proteomic Analysis of Exosomal Cargo: The Challenge of High Purity Vesicle Isolation. Mol. Biosyst. 2016, 12, 1407–1419. [Google Scholar] [CrossRef]
- Chetty, V.K.; Ghanam, J.; Anchan, S.; Reinhardt, K.; Brenzel, A.; Gelléri, M.; Cremer, C.; Grueso-Navarro, E.; Schneider, M.; von Neuhoff, N.; et al. Efficient Small Extracellular Vesicles (EV) Isolation Method and Evaluation of EV-Associated DNA Role in Cell-Cell Communication in Cancer. Cancers 2022, 14, 2068. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Dong, L.; Zieren, R.C.; Horie, K.; Kim, C.-J.; Mallick, E.; Jing, Y.; Feng, M.; Kuczler, M.D.; Green, J.; Amend, S.R.; et al. Comprehensive Evaluation of Methods for Small Extracellular Vesicles Separation from Human Plasma, Urine and Cell Culture Medium. J. Extracell. Vesicles 2020, 10, e12044. [Google Scholar] [CrossRef] [PubMed]
- Lou, P.; Liu, S.; Wang, Y.; Lv, K.; Zhou, X.; Li, L.; Zhang, Y.; Chen, Y.; Cheng, J.; Lu, Y.; et al. Neonatal-Tissue-Derived Extracellular Vesicle Therapy (NEXT): A Potent Strategy for Precision Regenerative Medicine. Adv. Mater. 2023, 35, e2300602. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Sun, J.; Shao, J.; Xu, J. Ultraviolet B Irradiation Enhances the Secretion of Exosomes by Human Primary Melanocytes and Changes Their Exosomal MiRNA Profile. PLoS ONE 2020, 15, e0237023. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Polisetti, N.; Schlötzer-Schrehardt, U.; Reinhard, T.; Schlunck, G. Isolation and Enrichment of Melanocytes from Human Corneal Limbus Using CD117 (c-Kit) as Selection Marker. Sci. Rep. 2020, 10, 17588. [Google Scholar] [CrossRef] [PubMed]
- Bachurski, D.; Schuldner, M.; Nguyen, P.-H.; Malz, A.; Reiners, K.S.; Grenzi, P.C.; Babatz, F.; Schauss, A.C.; Hansen, H.P.; Hallek, M.; et al. Extracellular Vesicle Measurements with Nanoparticle Tracking Analysis—An Accuracy and Repeatability Comparison between NanoSight NS300 and ZetaView. J. Extracell. Vesicles 2019, 8, 1596016. [Google Scholar] [CrossRef]
- Kotrbová, A.; Štěpka, K.; Maška, M.; Pálenik, J.J.; Ilkovics, L.; Klemová, D.; Kravec, M.; Hubatka, F.; Dave, Z.; Hampl, A.; et al. TEM ExosomeAnalyzer: A Computer-Assisted Software Tool for Quantitative Evaluation of Extracellular Vesicles in Transmission Electron Microscopy Images. J. Extracell. Vesicles 2019, 8, 1560808. [Google Scholar] [CrossRef]
- Mangahas, C.R.; dela Cruz, G.V.; Schneider, R.J.; Jamal, S. Endothelin-1 Upregulates MCAM in Melanocytes. J. Investig. Dermatol. 2004, 123, 1135–1139. [Google Scholar] [CrossRef]
- Ferrone, S.; Whiteside, T.L. Targeting CSPG4 for Isolation of Melanoma Cell-Derived Exosomes from Body Fluids. HNO 2020, 68, 100–105. [Google Scholar] [CrossRef]
- Fukushi, J.; Makagiansar, I.T.; Stallcup, W.B. NG2 Proteoglycan Promotes Endothelial Cell Motility and Angiogenesis via Engagement of Galectin-3 and Alpha3beta1 Integrin. Mol. Biol. Cell 2004, 15, 3580–3590. [Google Scholar] [CrossRef]
- D’Arcy, C.; Kiel, C. Cell Adhesion Molecules in Normal Skin and Melanoma. Biomolecules 2021, 11, 1213. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, J.B. What Are We Looking At? Extracellular Vesicles, Lipoproteins, or Both? Circ. Res. 2017, 121, 920–922. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, S.; Hirano, M.; Isoyama, J.; Nagayama, S.; Tomonaga, T.; Adachi, J. Comprehensive Proteomic Profiling of Plasma and Serum Phosphatidylserine-Positive Extracellular Vesicles Reveals Tissue-Specific Proteins. iScience 2022, 25, 104012. [Google Scholar] [CrossRef] [PubMed]
- Veerman, R.E.; Teeuwen, L.; Czarnewski, P.; Güclüler Akpinar, G.; Sandberg, A.; Cao, X.; Pernemalm, M.; Orre, L.M.; Gabrielsson, S.; Eldh, M. Molecular Evaluation of Five Different Isolation Methods for Extracellular Vesicles Reveals Different Clinical Applicability and Subcellular Origin. J. Extracell. Vesicles 2021, 10, e12128. [Google Scholar] [CrossRef]
- Villatoro, A.J.; Alcoholado, C.; Martín-Astorga, M.D.C.; Rico, G.; Fernández, V.; Becerra, J. Characterization of the Secretory Profile and Exosomes of Limbal Stem Cells in the Canine Species. PLoS ONE 2020, 15, e0244327. [Google Scholar] [CrossRef]
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M. A Comparison of Methods for the Isolation and Separation of Extracellular Vesicles from Protein and Lipid Particles in Human Serum. Sci. Rep. 2020, 10, 1039. [Google Scholar] [CrossRef] [PubMed]
- Schallreuter, K.U.; Hasse, S.; Rokos, H.; Chavan, B.; Shalbaf, M.; Spencer, J.D.; Wood, J.M. Cholesterol Regulates Melanogenesis in Human Epidermal Melanocytes and Melanoma Cells. Exp. Dermatol. 2009, 18, 680–688. [Google Scholar] [CrossRef]
- Théry, C.; Boussac, M.; Véron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic Analysis of Dendritic Cell-Derived Exosomes: A Secreted Subcellular Compartment Distinct from Apoptotic Vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of Distinct Nanoparticles and Subsets of Extracellular Vesicles by Asymmetric Flow Field-Flow Fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Varela, M.; de Menezes-Neto, A.; Perez-Zsolt, D.; Gámez-Valero, A.; Seguí-Barber, J.; Izquierdo-Useros, N.; Martinez-Picado, J.; Fernández-Becerra, C.; Del Portillo, H.A. Proteomics Study of Human Cord Blood Reticulocyte-Derived Exosomes. Sci. Rep. 2018, 8, 14046. [Google Scholar] [CrossRef] [PubMed]
- Lázaro-Ibáñez, E.; Lässer, C.; Shelke, G.V.; Crescitelli, R.; Jang, S.C.; Cvjetkovic, A.; García-Rodríguez, A.; Lötvall, J. DNA Analysis of Low- and High-Density Fractions Defines Heterogeneous Subpopulations of Small Extracellular Vesicles Based on Their DNA Cargo and Topology. J. Extracell. Vesicles 2019, 8, 1656993. [Google Scholar] [CrossRef] [PubMed]
- Gautier, E.-F.; Ducamp, S.; Leduc, M.; Salnot, V.; Guillonneau, F.; Dussiot, M.; Hale, J.; Giarratana, M.-C.; Raimbault, A.; Douay, L.; et al. Comprehensive Proteomic Analysis of Human Erythropoiesis. Cell Rep. 2016, 16, 1470–1484. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.J.; Ashraf, A.; Chung, E.J. Extracellular Vesicles as Regulators of the Extracellular Matrix. Bioengineering 2023, 10, 136. [Google Scholar] [CrossRef] [PubMed]
- Al Halawani, A.; Mithieux, S.M.; Yeo, G.C.; Hosseini-Beheshti, E.; Weiss, A.S. Extracellular Vesicles: Interplay with the Extracellular Matrix and Modulated Cell Responses. Int. J. Mol. Sci. 2022, 23, 3389. [Google Scholar] [CrossRef]
- van Niel, G.; Carter, D.R.F.; Clayton, A.; Lambert, D.W.; Raposo, G.; Vader, P. Challenges and Directions in Studying Cell–Cell Communication by Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2022, 23, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Wäster, P.; Eriksson, I.; Vainikka, L.; Rosdahl, I.; Öllinger, K. Extracellular Vesicles Are Transferred from Melanocytes to Keratinocytes after UVA Irradiation. Sci. Rep. 2016, 6, 27890. [Google Scholar] [CrossRef]
- Liu, L.; Nielsen, F.M.; Emmersen, J.; Bath, C.; Østergaard Hjortdal, J.; Riis, S.; Fink, T.; Pennisi, C.P.; Zachar, V. Pigmentation Is Associated with Stemness Hierarchy of Progenitor Cells Within Cultured Limbal Epithelial Cells. Stem Cells 2018, 36, 1411–1420. [Google Scholar] [CrossRef]
- Blanc, L.; Vidal, M. New Insights into the Function of Rab GTPases in the Context of Exosomal Secretion. Small GTPases 2018, 9, 95–106. [Google Scholar] [CrossRef]
- Fukuda, M. Rab GTPases: Key Players in Melanosome Biogenesis, Transport, and Transfer. Pigment. Cell Melanoma Res. 2021, 34, 222–235. [Google Scholar] [CrossRef]
- Eskelinen, E.-L.; Tanaka, Y.; Saftig, P. At the Acidic Edge: Emerging Functions for Lysosomal Membrane Proteins. Trends Cell Biol. 2003, 13, 137–145. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, Biogenesis and Function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Palazzolo, G.; Albanese, N.N.; DI Cara, G.; Gygax, D.; Vittorelli, M.L.; Pucci-Minafra, I. Proteomic Analysis of Exosome-like Vesicles Derived from Breast Cancer Cells. Anticancer. Res. 2012, 32, 847–860. [Google Scholar] [PubMed]
- Jimbow, K.; Luo, D.; Chen, H.; Hara, H.; Lee, M.H. Coordinated MRNA and Protein Expression of Human LAMP-1 in Induction of Melanogenesis after UV-B Exposure and Co-Transfection of Human Tyrosinase and TRP-1 CDNAs. Pigment. Cell Res. 1994, 7, 311–319. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; Bergam, P.; Di Cicco, A.; Hurbain, I.; Lo Cicero, A.; Dingli, F.; Palmulli, R.; Fort, C.; Potier, M.C.; Schurgers, L.J.; et al. Apolipoprotein E Regulates Amyloid Formation within Endosomes of Pigment Cells. Cell Rep. 2015, 13, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Volatier, T.; Schumacher, B.; Meshko, B.; Hadrian, K.; Cursiefen, C.; Notara, M. Short-Term UVB Irradiation Leads to Persistent DNA Damage in Limbal Epithelial Stem Cells, Partially Reversed by DNA Repairing Enzymes. Biology 2023, 12, 265. [Google Scholar] [CrossRef] [PubMed]
- Maurizi, E.; Schiroli, D.; Atkinson, S.D.; Mairs, L.; Courtney, D.G.; O’Hagan, B.; McGilligan, V.E.; Pagnamenta, A.T.; Taylor, J.C.; Vasquez, J.J.D.; et al. A Novel Role for CRIM1 in the Corneal Response to UV and Pterygium Development. Exp. Eye Res. 2019, 179, 75–92. [Google Scholar] [CrossRef] [PubMed]
- Ljubimov, A.V.; Burgeson, R.E.; Butkowski, R.J.; Michael, A.F.; Sun, T.T.; Kenney, M.C. Human Corneal Basement Membrane Heterogeneity: Topographical Differences in the Expression of Type IV Collagen and Laminin Isoforms. Lab. Investig. 1995, 72, 461–473. [Google Scholar]
- Schlötzer-Schrehardt, U.; Dietrich, T.; Saito, K.; Sorokin, L.; Sasaki, T.; Paulsson, M.; Kruse, F.E. Characterization of Extracellular Matrix Components in the Limbal Epithelial Stem Cell Compartment. Exp. Eye Res. 2007, 85, 845–860. [Google Scholar] [CrossRef]
- Rilla, K.; Mustonen, A.-M.; Arasu, U.T.; Härkönen, K.; Matilainen, J.; Nieminen, P. Extracellular Vesicles Are Integral and Functional Components of the Extracellular Matrix. Matrix Biol. 2019, 75–76, 201–219. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Stewart, R.; Yung, S.; Kolli, S.; Armstrong, L.; Stojkovic, M.; Figueiredo, F.; Lako, M. Differentiation of Human Embryonic Stem Cells into Corneal Epithelial-Like Cells by In Vitro Replication of the Corneal Epithelial Stem Cell Niche. Stem Cells 2007, 25, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
- Polisetti, N.; Sorokin, L.; Okumura, N.; Koizumi, N.; Kinoshita, S.; Kruse, F.E.; Schlötzer-Schrehardt, U. Laminin-511 and -521-Based Matrices for Efficient Ex Vivo-Expansion of Human Limbal Epithelial Progenitor Cells. Sci. Rep. 2017, 7, 5152. [Google Scholar] [CrossRef] [PubMed]
- Klintworth, G.K. Corneal Dystrophies. Orphanet J. Rare Dis. 2009, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Schlötzer-Schrehardt, U.; Latta, L.; Gießl, A.; Zenkel, M.; Fries, F.N.; Käsmann-Kellner, B.; Kruse, F.E.; Seitz, B. Dysfunction of the Limbal Epithelial Stem Cell Niche in Aniridia-Associated Keratopathy. Ocul. Surf. 2021, 21, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Karnas, E.; Dudek, P.; Zuba-Surma, E.K. Stem Cell-Derived Extracellular Vesicles as New Tools in Regenerative Medicine-Immunomodulatory Role and Future Perspectives. Front. Immunol. 2023, 14, 1120175. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhang, G.; Pittelkow, M.R.; Ramoni, M.; Tsao, H. Expression Profiling of UVB Response in Melanocytes Identifies a Set of P53-Target Genes. J. Investig. Dermatol. 2006, 126, 2490–2506. [Google Scholar] [CrossRef] [PubMed]
- Tavakkoli, F.; Damala, M.; Koduri, M.A.; Gangadharan, A.; Rai, A.K.; Dash, D.; Basu, S.; Singh, V. Transcriptomic Profiling of Human Limbus-Derived Stromal/Mesenchymal Stem Cells-Novel Mechanistic Insights into the Pathways Involved in Corneal Wound Healing. Int. J. Mol. Sci. 2022, 23, 8226. [Google Scholar] [CrossRef]
- Streit, M.; Riccardi, L.; Velasco, P.; Brown, L.F.; Hawighorst, T.; Bornstein, P.; Detmar, M. Thrombospondin-2: A Potent Endogenous Inhibitor of Tumor Growth and Angiogenesis. Proc. Natl. Acad. Sci. USA 1999, 96, 14888–14893. [Google Scholar] [CrossRef]
- Yang, X.; Cheng, Y.; Su, G. A Review of the Multifunctionality of Angiopoietin-like 4 in Eye Disease. Biosci. Rep. 2018, 38, BSR20180557. [Google Scholar] [CrossRef]
- Roy, R.; Dagher, A.; Butterfield, C.; Moses, M.A. ADAM12 Is a Novel Regulator of Tumor Angiogenesis via STAT3 Signaling. Mol. Cancer Res. 2017, 15, 1608–1622. [Google Scholar] [CrossRef]
- Lee, P.S.-Y.; Gao, N.; Dike, M.; Shkilnyy, O.; Me, R.; Zhang, Y.; Yu, F.-S.X. Opposing Effects of Neuropilin-1 and -2 on Sensory Nerve Regeneration in Wounded Corneas: Role of Sema3C in Ameliorating Diabetic Neurotrophic Keratopathy. Diabetes 2019, 68, 807–818. [Google Scholar] [CrossRef]
- Kaplan, N.; Wang, J.; Wray, B.; Patel, P.; Yang, W.; Peng, H.; Lavker, R.M. Single-Cell RNA Transcriptome Helps Define the Limbal/Corneal Epithelial Stem/Early Transit Amplifying Cells and How Autophagy Affects This Population. Investig. Ophthalmol. Vis. Sci. 2019, 60, 3570–3583. [Google Scholar] [CrossRef] [PubMed]
- Polisetti, N.; Schmid, A.; Schlötzer-Schrehardt, U.; Maier, P.; Lang, S.J.; Steinberg, T.; Schlunck, G.; Reinhard, T. A Decellularized Human Corneal Scaffold for Anterior Corneal Surface Reconstruction. Sci. Rep. 2021, 11, 2992. [Google Scholar] [CrossRef]
- Polisetti, N.; Schlunck, G.; Reinhard, T. PAX6 Expression Patterns in the Adult Human Limbal Stem Cell Niche. Cells 2023, 12, 400. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Dessau, R.B.; Pipper, C.B. R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 17 February 2024).
- Yu, G. Enrichplot; Bioconductor: Boston, MA, USA, 2018. [Google Scholar]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor Package for Pathway-Based Data Integration and Visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Gao, C.-H.; Chen, C.; Yu, G.; Dusa, A.; Cao, B.; Cai, P.; Akyol, T.Y. GgVennDiagram: A “ggplot2” Implement of Venn Diagram; iMeta: Southampton, UK, 2024. [Google Scholar]
- Polisetti, N.; Martin, G.; Ulrich, E.; Glegola, M.; Schlötzer-Schrehardt, U.; Schlunck, G.; Reinhard, T. Influence of Organ Culture on the Characteristics of the Human Limbal Stem Cell Niche. Int. J. Mol. Sci. 2023, 24, 16856. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kistenmacher, S.; Schwämmle, M.; Martin, G.; Ulrich, E.; Tholen, S.; Schilling, O.; Gießl, A.; Schlötzer-Schrehardt, U.; Bucher, F.; Schlunck, G.; et al. Enrichment, Characterization, and Proteomic Profiling of Small Extracellular Vesicles Derived from Human Limbal Mesenchymal Stromal Cells and Melanocytes. Cells 2024, 13, 623. https://doi.org/10.3390/cells13070623
Kistenmacher S, Schwämmle M, Martin G, Ulrich E, Tholen S, Schilling O, Gießl A, Schlötzer-Schrehardt U, Bucher F, Schlunck G, et al. Enrichment, Characterization, and Proteomic Profiling of Small Extracellular Vesicles Derived from Human Limbal Mesenchymal Stromal Cells and Melanocytes. Cells. 2024; 13(7):623. https://doi.org/10.3390/cells13070623
Chicago/Turabian StyleKistenmacher, Sebastian, Melanie Schwämmle, Gottfried Martin, Eva Ulrich, Stefan Tholen, Oliver Schilling, Andreas Gießl, Ursula Schlötzer-Schrehardt, Felicitas Bucher, Günther Schlunck, and et al. 2024. "Enrichment, Characterization, and Proteomic Profiling of Small Extracellular Vesicles Derived from Human Limbal Mesenchymal Stromal Cells and Melanocytes" Cells 13, no. 7: 623. https://doi.org/10.3390/cells13070623





