The Potential Mechanism of Cuproptosis in Hemocytes of the Pacific Oyster Crassostrea gigas upon Elesclomol Treatment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Elesclomol Treatment and Sample Collection
2.3. RNA Extraction, cDNA Synthesis and Sequence Analysis
2.4. Real-Time Quantitative PCR (RT-qPCR) Analysis of CgFDX1 and CgDLAT
2.5. The Cell Mortality After Elesclomol Treatment
2.6. Contents of Copper, MDA, Pyruvic Acid, Citric Acid and Reactive Oxygen Species (ROS) After Elesclomol Treatment
2.7. Observation of Mitochondrial Morphology
2.8. Observation of Cell Morphology
2.9. Immunocytochemical Assay of CgDLAT and CgFDX1
2.10. Flow Cytometry (FCM) Analysis of Hemocytes
2.11. Preparation of Antibody and Western Blot Assay
2.12. Statistical Analysis
3. Results
3.1. Sequence Characteristics and Phylogenetic Relationship of CgFDX1 and CgDLAT
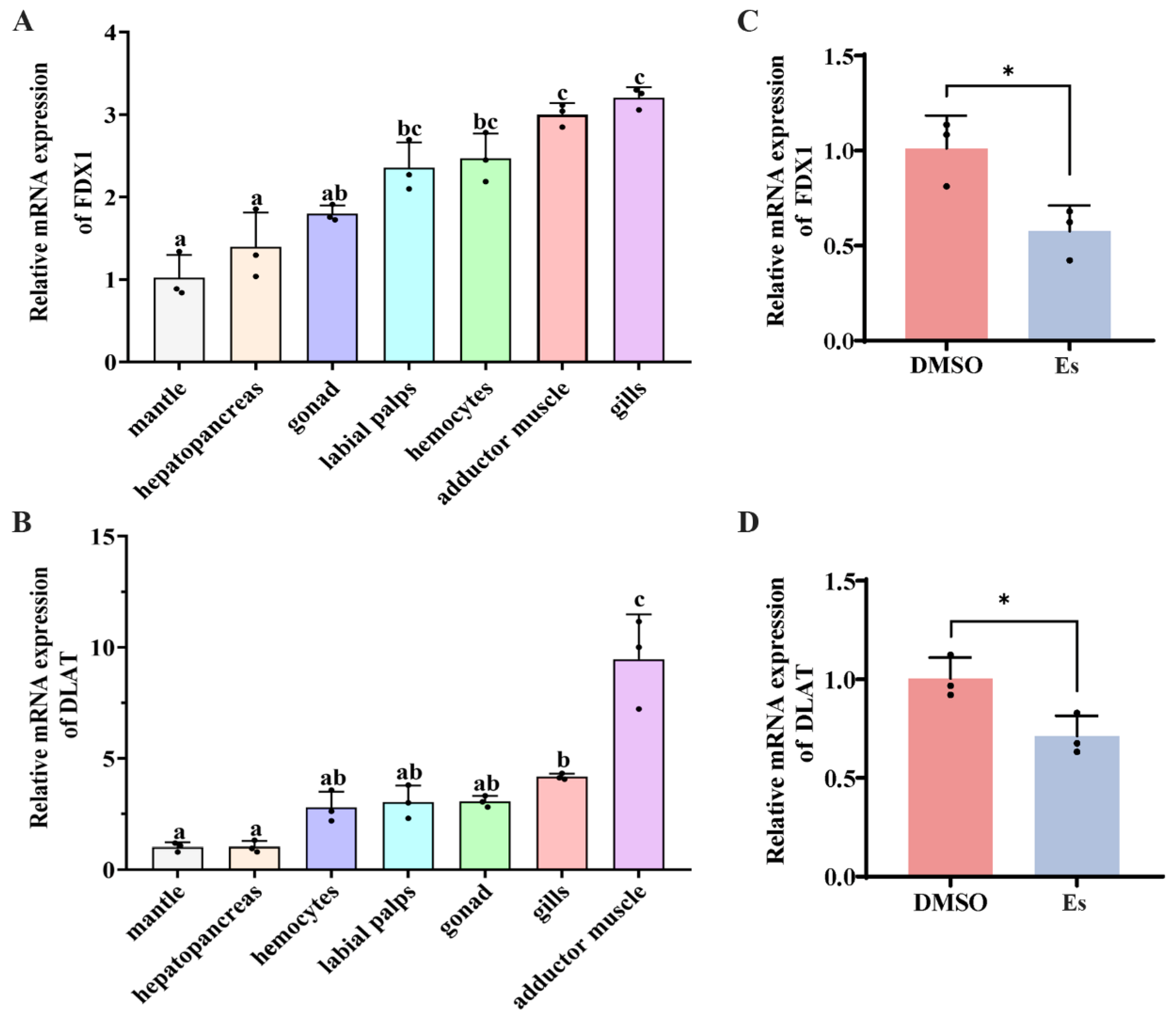
3.2. Expression Pattern of CgFDX1 and CgDLAT mRNA in Different Tissues
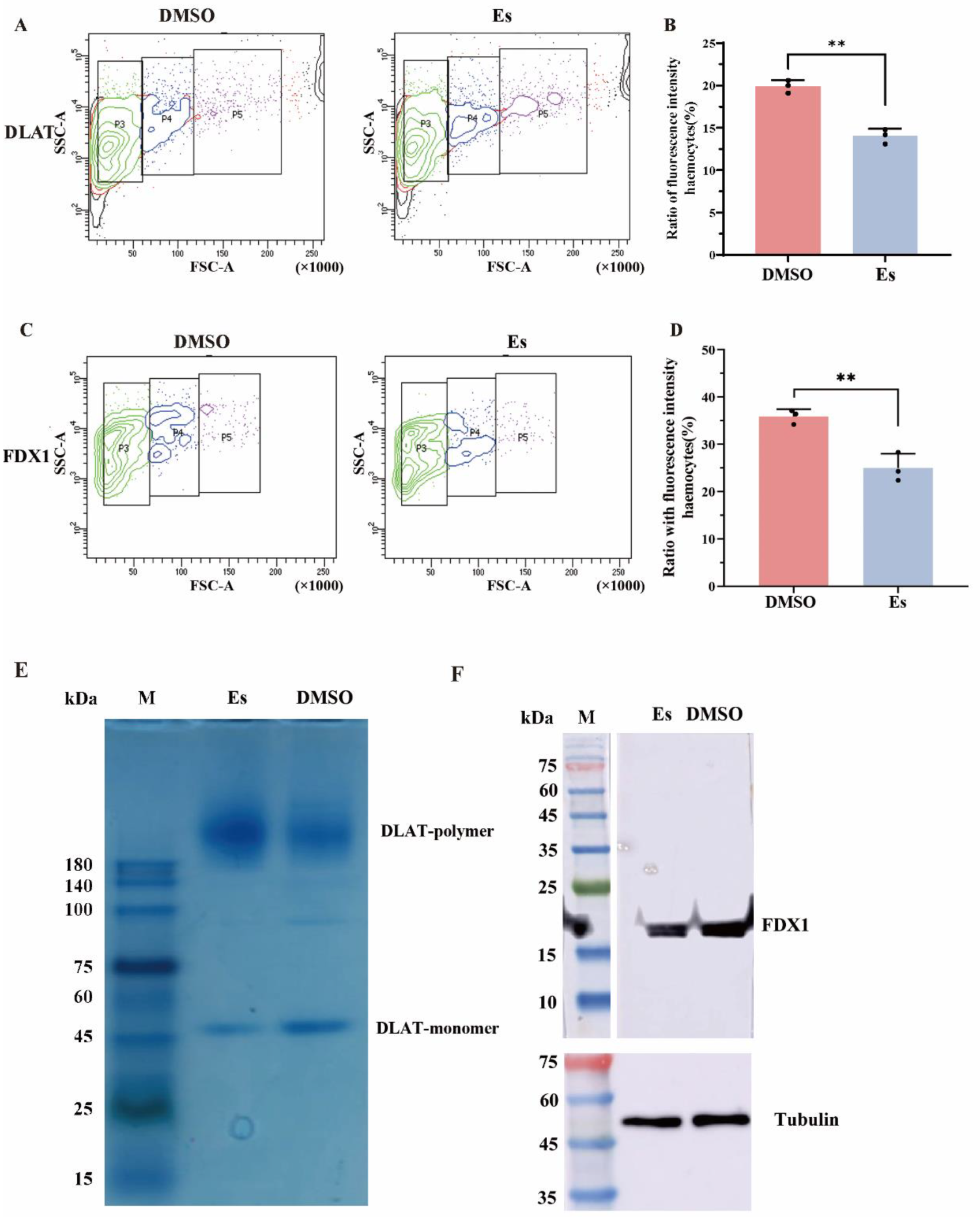
3.3. Expressions of CgFDX1 and CgDLAT After Elesclomol Treatment
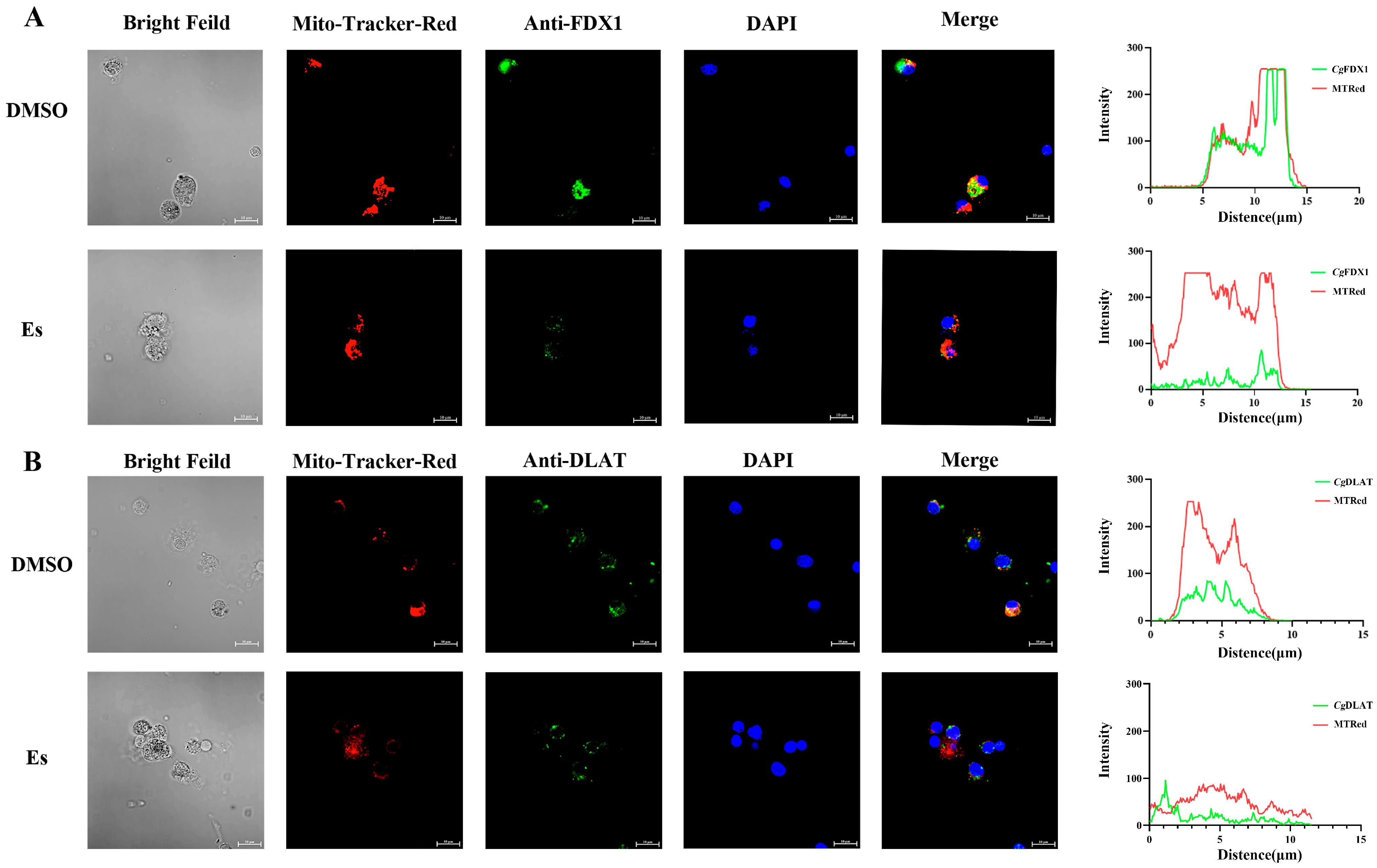
3.4. Colocalization of CgFDX1 and CgDLAT with Mitochondria After Elesclomol Treatment
3.5. The Contents of Copper, MDA, Citric Acid and Pyruvic Acid After Elesclomol Treatment
3.6. FCM Analysis of Hemocytes After Elesclomol Treatment
3.7. Morphology of Hemocytes and the Morphology of Mitochondria in Hemocytes After Elesclomol Treatment
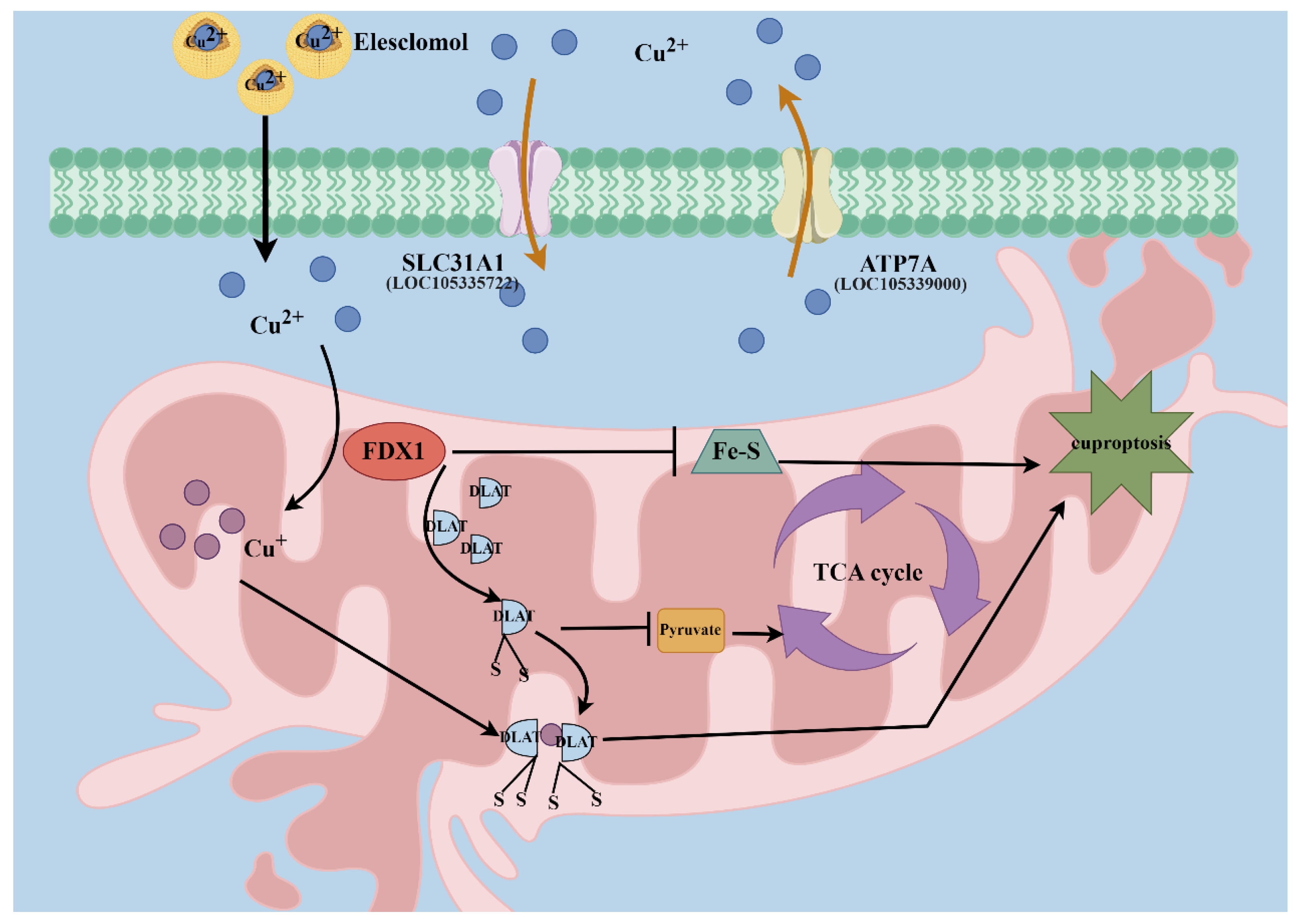
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, L.; Zhou, F. Cuproptosis: A new form of programmed cell death. Cell. Mol. Immunol. 2022, 19, 867–868. [Google Scholar] [CrossRef]
- Guo, B.; Yang, F.; Zhang, L.; Zhao, Q.; Wang, W.; Yin, L.; Chen, D.; Wang, M.; Han, S.; Xiao, H.; et al. Cuproptosis Induced by ROS Responsive Nanoparticles with Elesclomol and Copper Combined with αPD-L1 for Enhanced Cancer Immunotherapy. Adv. Mater. 2023, 35, e2212267. [Google Scholar] [CrossRef]
- Wu, H.; Guo, H.; Liu, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Copper sulfate-induced endoplasmic reticulum stress promotes hepatic apoptosis by activating CHOP, JNK and caspase-12 signaling pathways. Ecotoxicol. Environ. Saf. 2020, 191, 110236. [Google Scholar] [CrossRef]
- Chen, L.; Min, J.; Wang, F. Copper homeostasis and cuproptosis in health and disease. Signal Transduct. Target. Ther. 2022, 7, 378. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, X.; Kroemer, G. Cuproptosis: A copper-triggered modality of mitochondrial cell death. Cell Res. 2022, 32, 417–418. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Jiang, S.; Zhou, J.; Zhang, W. Copper homeostasis and cuproptosis in mitochondria. Life Sci. 2023, 334, 122223. [Google Scholar] [CrossRef]
- Schulz, V.; Basu, S.; Freibert, S.A.; Webert, H.; Boss, L.; Mühlenhoff, U.; Pierrel, F.; Essen, L.O.; Warui, D.M.; Booker, S.J.; et al. Functional spectrum and specificity of mitochondrial ferredoxins FDX1 and FDX2. Nat. Chem. Biol. 2023, 19, 206–217. [Google Scholar] [CrossRef]
- Zhou, P.; Wang, N.; Lu, S.; Xiong, J.; Zhang, Y.; Jiang, Q.; Qian, Q.; Zhou, Q.; Liu, J.; Chen, S. Dihydrolipoamide S-acetyltransferase activation alleviates diabetic kidney disease via AMPK-autophagy axis and mitochondrial protection. Transl. Res. J. Lab. Clin. Med. 2024, 274, 81–100. [Google Scholar] [CrossRef]
- Takano, S.; Tanimizu, M.; Hirata, T.; Sohrin, Y. Isotopic constraints on biogeochemical cycling of copper in the ocean. Nat. Commun. 2014, 5, 5663. [Google Scholar] [CrossRef] [PubMed]
- Festa, R.A.; Thiele, D.J. Copper: An essential metal in biology. Curr. Biol. 2011, 21, R877–R883. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, H.; Liang, J.; Liu, H.; Wang, X.; Gao, L.; Chen, L.; Li, Y.; Guo, Y.; Liang, S.J.R.S.i.M.S. The role of marine bivalves in the oceanic carbon cycle: Physiological processes, carbon budgets and ecosystem perspectives. Reg. Stud. Mar. Sci. 2024, 78, 103815. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, W.X. Lysosomal Cu(I)/Cu(II) Dependence of Antimicrobial Ability of Oyster Hemocytes and Regulation of Phagolysosomal System. Environ. Sci. Technol. 2023, 57, 20219–20227. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Wang, W.X. Highly Sensitive and Specific Responses of Oyster Hemocytes to Copper Exposure: Single-Cell Transcriptomic Analysis of Different Cell Populations. Environ. Sci. Technol. 2022, 56, 2497–2510. [Google Scholar] [CrossRef] [PubMed]
- Öhrvik, H.; Thiele, D.J. The role of Ctr1 and Ctr2 in mammalian copper homeostasis and platinum-based chemotherapy. J. Trace Elem. Med. Biol. 2015, 31, 178–182. [Google Scholar] [CrossRef]
- Chen, X.; Cai, Q.; Liang, R.; Zhang, D.; Liu, X.; Zhang, M.; Xiong, Y.; Xu, M.; Liu, Q.; Li, P.; et al. Copper homeostasis and copper-induced cell death in the pathogenesis of cardiovascular disease and therapeutic strategies. Cell Death Dis. 2023, 14, 105. [Google Scholar] [CrossRef]
- Zheng, P.; Zhou, C.; Lu, L.; Liu, B.; Ding, Y. Elesclomol: A copper ionophore targeting mitochondrial metabolism for cancer therapy. J. Exp. Clin. Cancer Res. 2022, 41, 271. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Korolnek, T.; Kim, B.E. Oral Elesclomol Treatment Alleviates Copper Deficiency in Animal Models. Front. Cell Dev. Biol. 2022, 10, 856300. [Google Scholar] [CrossRef]
- Zhao, G.; Sun, H.; Zhang, T.; Liu, J.X. Copper induce zebrafish retinal developmental defects via triggering stresses and apoptosis. Cell Commun. Signal. 2020, 18, 45. [Google Scholar] [CrossRef]
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef]
- Luo, Y.; Pezacki, A.T.; Matier, C.D.; Wang, W.X. A novel route of intercellular copper transport and detoxification in oyster hemocytes. J. Hazard. Mater. 2024, 476, 135003. [Google Scholar] [CrossRef] [PubMed]
- Weng, N.; Jiang, H.; Wang, W.X. In Situ Subcellular Imaging of Copper and Zinc in Contaminated Oysters Revealed by Nanoscale Secondary Ion Mass Spectrometry. Environ. Sci. Technol. 2017, 51, 14426–14435. [Google Scholar] [CrossRef]
- Li, H.; Zhang, H.; Jiang, S.; Wang, W.; Xin, L.; Wang, H.; Wang, L.; Song, L. A single-CRD C-type lectin from oyster Crassostrea gigas mediates immune recognition and pathogen elimination with a potential role in the activation of complement system. Fish Shellfish Immunol. 2015, 44, 566–575. [Google Scholar] [CrossRef]
- Thomas, R.H. Molecular Evolution and Phylogenetics. Heredity 2001, 86, 385. [Google Scholar] [CrossRef]
- Sun, J.; Wang, L.; Yang, W.; Wang, L.; Fu, Q.; Song, L. IgIT-Mediated Signaling Inhibits the Antimicrobial Immune Response in Oyster Hemocytes. J. Immunol. 2020, 205, 2402–2413. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Crowley, L.C.; Marfell, B.J.; Christensen, M.E.; Waterhouse, N.J. Measuring Cell Death by Trypan Blue Uptake and Light Microscopy. Cold Spring Harb. Protoc. 2016, 2016, prot087155. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liu, Z.; Wang, L.; Li, M.; Zhang, Y.; Zong, Y.; Li, Y.; Song, L. A novel tumor necrosis factor in the Pacific oyster Crassostrea gigas mediates the antibacterial response by triggering the synthesis of lysozyme and nitric oxide. Fish Shellfish Immunol. 2020, 98, 334–341. [Google Scholar] [CrossRef]
- Liu, Z.; Zhou, Z.; Zhang, Y.; Wang, L.; Song, X.; Wang, W.; Zheng, Y.; Zong, Y.; Lv, Z.; Song, L. Ocean acidification inhibits initial shell formation of oyster larvae by suppressing the biosynthesis of serotonin and dopamine. Sci. Total Environ. 2020, 735, 139469. [Google Scholar] [CrossRef]
- Wang, W.; Li, M.; Wang, L.; Chen, H.; Liu, Z.; Jia, Z.; Qiu, L.; Song, L. The granulocytes are the main immunocompetent hemocytes in Crassostrea gigas. Dev. Comp. Immunol. 2017, 67, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Yang, P.C. Western blot: Technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012, 4, 429–434. [Google Scholar] [CrossRef]
- Li, Y.; Lian, X.; Yang, W.; Leng, J.; Sun, J.; Liu, Y.; Fan, S.; Wang, L.; Song, L. A novel DM9-containing protein 7 involved in regulating the expression of CgMyD88 and CgIL-17 in oyster Crassostrea gigas. Dev. Comp. Immunol. 2024, 150, 105076. [Google Scholar] [CrossRef] [PubMed]
- Solmonson, A.; DeBerardinis, R.J. Lipoic acid metabolism and mitochondrial redox regulation. J. Biol. Chem. 2018, 293, 7522–7530. [Google Scholar] [CrossRef] [PubMed]
- Rowland, E.A.; Snowden, C.K.; Cristea, I.M. Protein lipoylation: An evolutionarily conserved metabolic regulator of health and disease. Curr. Opin. Chem. Biol. 2018, 42, 76–85. [Google Scholar] [CrossRef]
- Zulkifli, M.; Spelbring, A.N.; Zhang, Y.; Soma, S.; Chen, S.; Li, L.; Le, T.; Shanbhag, V.; Petris, M.J.; Chen, T.Y.; et al. FDX1-dependent and independent mechanisms of elesclomol-mediated intracellular copper delivery. Proc. Natl. Acad. Sci. USA 2023, 120, e2216722120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ma, Y.; Guo, X.; Du, Y.; Zhu, Q.; Wang, X.; Duan, C. FDX1 can Impact the Prognosis and Mediate the Metabolism of Lung Adenocarcinoma. Front. Pharmacol. 2021, 12, 749134. [Google Scholar] [CrossRef]
- Zhang, J.; Kong, X.; Zhang, Y.; Sun, W.; Wang, J.; Chen, M.; Chen, X. FDXR regulates TP73 tumor suppressor via IRP2 to modulate aging and tumor suppression. J. Pathol. 2020, 251, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Birsoy, K.; Wang, T.; Chen, W.W.; Freinkman, E.; Abu-Remaileh, M.; Sabatini, D.M. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 2015, 162, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Goh, W.Q.; Ow, G.S.; Kuznetsov, V.A.; Chong, S.; Lim, Y.P. DLAT subunit of the pyruvate dehydrogenase complex is upregulated in gastric cancer-implications in cancer therapy. Am. J. Transl. Res. 2015, 7, 1140–1151. [Google Scholar] [PubMed]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Yue, F.; Cheng, Y.; Breschi, A.; Vierstra, J.; Wu, W.; Ryba, T.; Sandstrom, R.; Ma, Z.; Davis, C.; Pope, B.D.; et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 2014, 515, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Zhang, G.; Wang, W.-X. Functional heterogeneity of immune defenses in molluscan oysters Crassostrea hongkongensis revealed by high-throughput single-cell transcriptome. Fish Shellfish Immunol. 2022, 120, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Yang, Y.; Gao, Y.; He, J. Cuproptosis: Mechanisms and links with cancers. Mol. Cancer 2023, 22, 46. [Google Scholar] [CrossRef]
- Vakifahmetoglu-Norberg, H.; Ouchida, A.T.; Norberg, E. The role of mitochondria in metabolism and cell death. Biochem. Biophys. Res. Commun. 2017, 482, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Schieber, M.; Chandel, N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef] [PubMed]
- Perelman, A.; Wachtel, C.; Cohen, M.; Haupt, S.; Shapiro, H.; Tzur, A. JC-1: Alternative excitation wavelengths facilitate mitochondrial membrane potential cytometry. Cell Death Dis. 2012, 3, e430. [Google Scholar] [CrossRef] [PubMed]
- Ivanina, A.V.; Falfushynska, H.I.; Beniash, E.; Piontkivska, H.; Sokolova, I.M. Biomineralization-related specialization of hemocytes and mantle tissues of the Pacific oyster Crassostrea gigas. J. Exp. Biol. 2017, 220, 3209–3221. [Google Scholar] [CrossRef]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef]
- Goguet-Rubio, P.; Seyran, B.; Gayte, L.; Bernex, F.; Sutter, A.; Delpech, H.; Linares, L.K.; Riscal, R.; Repond, C.; Rodier, G.; et al. E4F1-mediated control of pyruvate dehydrogenase activity is essential for skin homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, 11004–11009. [Google Scholar] [CrossRef] [PubMed]
- Kamzolova, S.V.; Morgunov, I.G. Biosynthesis of pyruvic acid from glycerol-containing substrates and its regulation in the yeast Yarrowia lipolytica. Bioresour. Technol. 2018, 266, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.; Detappe, A.; Cai, K.; Keys, H.R.; Brune, Z.; Ying, W.; Thiru, P.; Reidy, M.; Kugener, G.; Rossen, J.; et al. Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat. Chem. Biol. 2019, 15, 681–689. [Google Scholar] [CrossRef]




| Primer | Sequence (5′-3′) |
|---|---|
| RT-PCR primers | |
| CgFDX1-RT-F | ATCAAAGACAAGCCCACAGACGA |
| CgFDX1-RT-R | CTTGGTAACAATCACTTGACAGCCTAAT |
| CgDLAT-RT-F | TTGTAGTCACACCTGGGGCAGAGTA |
| CgDLAT-RT-R | ACCGCTTGGCTATTGTCTTTCTCAT |
| CgEF-RT-F | AGTCACCAAGGCTGCACAGAAAG |
| CgEF-RT-R | TCCGACGTATTTCTTTGCGATGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Sun, J.; Li, S.; Wang, L.; Song, L. The Potential Mechanism of Cuproptosis in Hemocytes of the Pacific Oyster Crassostrea gigas upon Elesclomol Treatment. Cells 2025, 14, 199. https://doi.org/10.3390/cells14030199
Zhang Y, Sun J, Li S, Wang L, Song L. The Potential Mechanism of Cuproptosis in Hemocytes of the Pacific Oyster Crassostrea gigas upon Elesclomol Treatment. Cells. 2025; 14(3):199. https://doi.org/10.3390/cells14030199
Chicago/Turabian StyleZhang, Yuxin, Jiejie Sun, Shurong Li, Lingling Wang, and Linsheng Song. 2025. "The Potential Mechanism of Cuproptosis in Hemocytes of the Pacific Oyster Crassostrea gigas upon Elesclomol Treatment" Cells 14, no. 3: 199. https://doi.org/10.3390/cells14030199
APA StyleZhang, Y., Sun, J., Li, S., Wang, L., & Song, L. (2025). The Potential Mechanism of Cuproptosis in Hemocytes of the Pacific Oyster Crassostrea gigas upon Elesclomol Treatment. Cells, 14(3), 199. https://doi.org/10.3390/cells14030199





