Genetic and Molecular Characterization of H9c2 Rat Myoblast Cell Line
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature Search
2.2. Cell Culture
2.3. Mycoplasma Testing
2.4. Short Tandem Repeat (STR) Profiling
2.5. Preparation of H9c2 Metaphase Chromosomes, Karyotyping, and Molecular Cytogenetics
2.6. Virtual Comparative Genomic Hybridization
2.7. Next-Generation Sequencing and Data Analysis
2.8. Electron Microscopic Cell Analysis
2.9. Western Blot Analysis
2.10. Phalloidin Stain
3. Results
3.1. Usage of H9c2 Cells in Biomedical Research
3.2. Phenotypic Appearance of H9c2 Cells
3.3. Electron Microscopic Analysis of H9c2 Cells
3.4. Expression of Typical Fibroblast Markers in H9c2 Cells
3.4.1. Next-Generation Sequencing
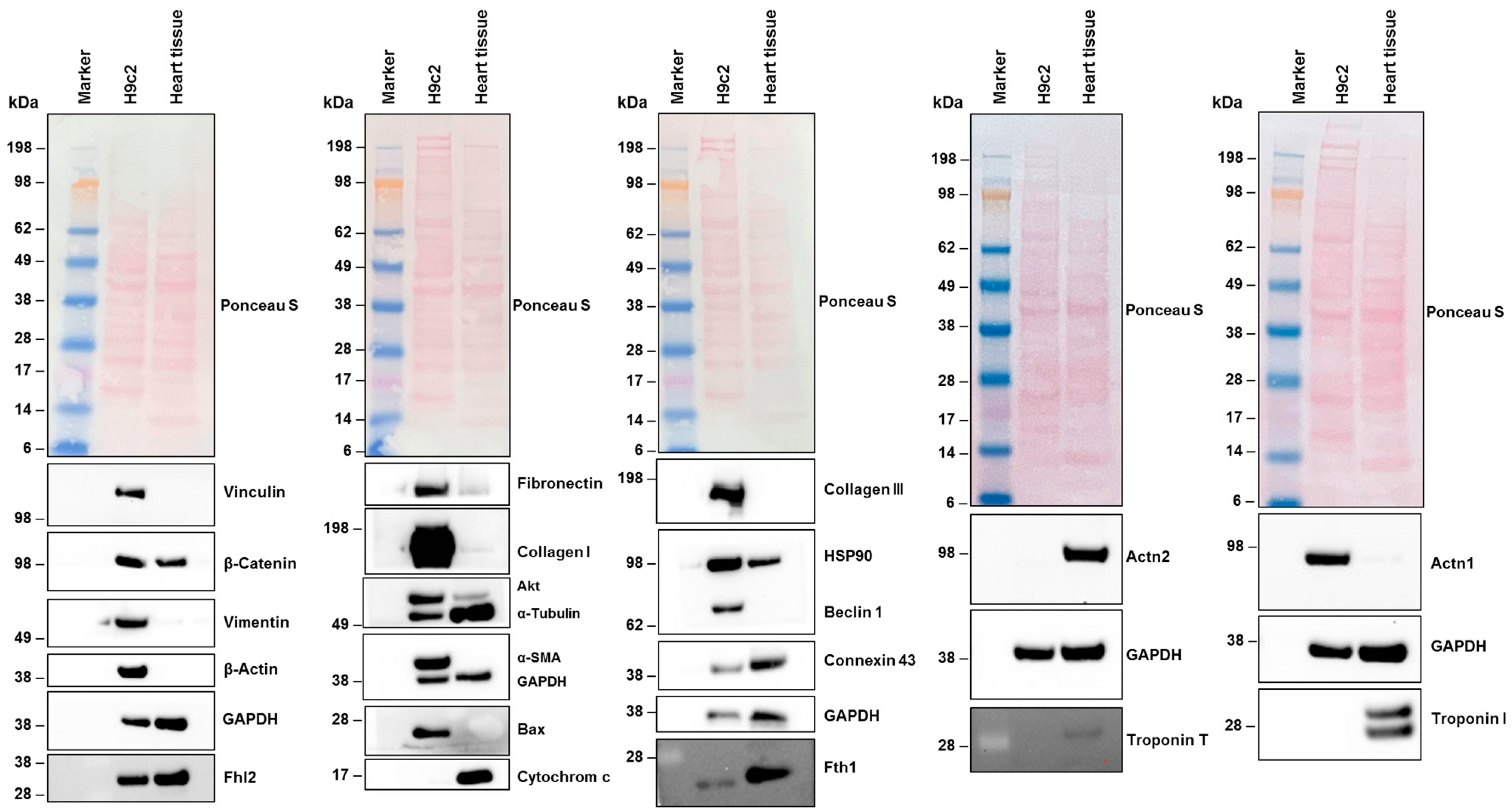
3.4.2. Analysis of Protein Expression and Cytoskeletal Organization in H9c2 Cells
3.5. Karyotype Based on Molecular Cytogenetic Analyses
3.6. Virtual Comparative Genomic Hybridization
3.7. Short Tandem Repeat Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DAPI | 4′,6-diamidino-2-phenylindole |
| mFISH | multicolor fluorescence in situ hybridization |
| mRNA-Seq | mRNA sequencing |
| NGS | next-generation sequencing |
| STR | short tandem repeat |
| TPM | Transcripts Per Million |
| vCGH | virtual Comparative Genomic Hybridization |
References
- Kimes, B.W.; Brandt, B.L. Properties of a clonal muscle cell line from rat heart. Exp. Cell Res. 1976, 98, 367–381. [Google Scholar] [CrossRef]
- Suhaeri, M.; Subbiah, R.; Van, S.Y.; Du, P.; Kim, I.G.; Lee, K.; Park, K. Cardiomyoblast (h9c2) differentiation on tunable extracellular matrix microenvironment. Tissue Eng. Part A 2015, 21, 1940–1951. [Google Scholar] [CrossRef]
- Branco, A.F.; Pereira, S.P.; Gonzalez, S.; Gusev, O.; Rizvanov, A.A.; Oliveira, P.J. Gene expression profiling of H9c2 myoblast differentiation towards a cardiac-like phenotype. PLoS ONE 2015, 10, e0129303. [Google Scholar] [CrossRef]
- Rishiq, A.; Islam, O.; Golomb, E.; Gilon, D.; Smith, Y.; Savchenko, I.; Eliaz, R.; Foo, R.S.; Razin, E.; Tshori, S. The role played by transcription factor E3 in modulating cardiac hypertrophy. Int. Heart J. 2021, 62, 1358–1368. [Google Scholar] [CrossRef] [PubMed]
- Li, G.M.; Chen, J.R.; Zhang, H.Q.; Sun, C.; Chen, G.R.; Xiong, Q.Y.; Cao, X.Y.; Yu, L.; Lin, Z.W.; Qin, J.Y.; et al. Rhein activated Fas-induced apoptosis pathway causing cardiotoxicity in vitro and in vivo. Toxicol. Lett. 2022, 363, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, W.; Yang, X.; Song, Y.; Sun, X.; Tao, G.; Wang, H.; Zhao, N.; Huang, Y.; Chai, E.; et al. Inhibition of miRNA-1-mediated inflammation and autophagy by Astragaloside IV improves lipopolysaccharide-induced cardiac dysfunction in rats. J. Inflamm. Res. 2022, 15, 2617–2629. [Google Scholar] [CrossRef]
- Huang, J.P.; Cheng, M.L.; Wang, C.H.; Huang, S.S.; Hsieh, P.S.; Chang, C.C.; Kuo, C.Y.; Chen, K.H.; Hung, L.M. Therapeutic potential of cPLA2 inhibitor to counteract dilated-cardiomyopathy in cholesterol-treated H9C2 cardiomyocyte and MUNO rat. Pharmacol. Res. 2020, 160, 105201. [Google Scholar] [CrossRef]
- Isa, H.I.; Ferreira, G.C.H.; Crafford, J.E.; Botha, C.J. Epoxyscillirosidine induced cytotoxicity and ultrastructural changes in a rat embryonic cardiomyocyte (H9c2) cell line. Toxins 2019, 11, 284. [Google Scholar] [CrossRef]
- Zheng, M.; Zhu, J.; Lu, T.; Liu, L.; Sun, H.; Liu, Z.; Tian, J. p300-mediated histone acetylation is essential for the regulation of GATA4 and MEF2C by BMP2 in H9c2 cells. Cardiovasc. Toxicol. 2013, 13, 316–322. [Google Scholar] [CrossRef]
- Si, L.; Shi, J.; Gao, W.; Zheng, M.; Liu, L.; Zhu, J.; Tian, J. Smad4 mediated BMP2 signal is essential for the regulation of GATA4 and Nkx2.5 by affecting the histone H3 acetylation in H9c2 cells. Biochem. Biophys. Res. Commun. 2014, 450, 81–86. [Google Scholar] [CrossRef]
- He, Z.; Zhou, Y.; Li, S.; Li, W.; Zhang, Y.; Guo, C.; Guo, Z.; Wei, B.; Bi, Y. Bioactive peptides and evaluation of cardiac cytoprotective effects of red millet yellow wine as functional food. Foods 2024, 13, 4111. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Yao, Y.; Chen, Y.; Li, Y.; Sun, X.; Zhu, X. TRPC5 promotes intermittent hypoxia-induced cardiomyocyte injury through oxidative stress. Nat. Sci. Sleep 2024, 16, 2125–2141. [Google Scholar] [CrossRef]
- Cai, C.; Sang, C.; Du, J.; Jia, H.; Tu, J.; Wan, Q.; Bao, B.; Xie, S.; Huang, Y.; Li, A.; et al. Knockout of tnni1b in zebrafish causes defects in atrioventricular valve development via the inhibition of the myocardial wnt signaling pathway. FASEB J. 2019, 33, 696–710. [Google Scholar] [CrossRef]
- Wen, X.; Iwata, K.; Ikuta, K.; Zhang, X.; Zhu, K.; Ibi, M.; Matsumoto, M.; Asaoka, N.; Liu, J.; Katsuyama, M.; et al. NOX1/NADPH oxidase regulates the expression of multidrug resistance-associated protein 1 and maintains intracellular glutathione levels. FEBS J. 2019, 286, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Lenčo, J.; Lenčová-Popelová, O.; Link, M.; Jirkovská, A.; Tambor, V.; Potůčková, E.; Stulík, J.; Šimůnek, T.; Štěrba, M. Proteomic investigation of embryonic rat heart-derived H9c2 cell line sheds new light on the molecular phenotype of the popular cell model. Exp. Cell Res. 2015, 339, 174–186, Erratum in Exp. Cell Res. 2016, 343, 267. https://doi.org/10.1016/j.yexcr.2016.02.007. [Google Scholar] [CrossRef] [PubMed]
- Hamta, A.; Adamovic, T.; Samuelson, E.; Helou, K.; Behboudi, A.; Levan, G. Chromosome ideograms of the laboratory rat (Rattus norvegicus) based on high-resolution banding, and anchoring of the cytogenetic map to the DNA sequence by FISH in sample chromosomes. Cytogenet. Genome. Res. 2006, 115, 158–168. [Google Scholar] [CrossRef]
- Liehr, T. Multicolor-FISH—Methods and applications. In Cytogenetics and Molecular Cytogenetics; Liehr, T., Ed.; Taylor and Francis: Milton Park, UK, 2022; pp. 151–156, eBook; ISBN 9781003223658. [Google Scholar]
- MetaSystems Probes. Available online: https://metasystems-probes.com/en/probes/mfish/d-1525-060-di/ (accessed on 11 January 2025).
- Liehr, T.; Kankel, S.; Buhl, E.M.; Schröder-Lange, S.K.; Weiskirchen, R. Genetic characteristics of the rat fibroblast cell line Rat-1. Cells 2024, 14, 21. [Google Scholar] [CrossRef]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- Krueger, F.; James, F.; Ewels, P.; Afyounian Schuster-Boeckler, B. FelixKrueger/TrimGalore: v0.6.7—DOI via Zenodo. Available online: https://zenodo.org/records/5127899 (accessed on 20 February 2025).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Nanda, I.; Schröder, S.K.; Steinlein, C.; Haaf, T.; Buhl, E.M.; Grimm, D.G.; Weiskirchen, R. Rat hepatic stellate cell line CFSC-2G: Genetic markers and short tandem repeat profile useful for cell line authentication. Cells 2022, 11, 2900. [Google Scholar] [CrossRef]
- Schröder, S.K.; Tag, C.G.; Kessel, J.C.; Antonson, P.; Weiskirchen, R. Immunohistochemical detection of estrogen receptor-beta (ERβ) with PPZ0506 antibody in murine tissue: From pitfalls to optimization. Biomedicines 2022, 10, 3100. [Google Scholar] [CrossRef] [PubMed]
- Schröder, S.K.; Tag, C.G.; Weiskirchen, S.; Weiskirchen, R. Phalloidin staining for F-actin in hepatic stellate cells. Methods Mol. Biol. 2023, 2669, 55–66. [Google Scholar] [CrossRef]
- Kodama, M.; Furutani, K.; Kimura, R.; Ando, T.; Sakamoto, K.; Nagamori, S.; Ashihara, T.; Kurachi, Y.; Sekino, Y.; Furukawa, T.; et al. Systematic expression analysis of genes related to generation of action potentials in human iPS cell-derived cardiomyocytes. J. Pharmacol. Sci. 2019, 140, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Man, J.; Barnett, P.; Christoffels, V.M. Structure and function of the Nppa-Nppb cluster locus during heart development and disease. Cell Mol. Life Sci. 2018, 75, 1435–1444. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, Y.A.; Moyes, A.J.; Hobbs, A.J. C-type natriuretic peptide (CNP): The cardiovascular system and beyond. Pharmacol. Ther. 2024, 262, 108708. [Google Scholar] [CrossRef]
- Chen, Y.; Lüttmann, F.F.; Schoger, E.; Schöler, H.R.; Zelarayán, L.C.; Kim, K.P.; Haigh, J.J.; Kim, J.; Braun, T. Reversible reprogramming of cardiomyocytes to a fetal state drives heart regeneration in mice. Science 2021, 373, 1537–1540. [Google Scholar] [CrossRef]
- Okubo, C.; Narita, M.; Inagaki, A.; Nishikawa, M.; Hotta, A.; Yamanaka, S.; Yoshida, Y. Expression dynamics of HAND1/2 in in vitro human cardiomyocyte differentiation. Stem Cell Rep. 2021, 16, 1906–1922. [Google Scholar] [CrossRef]
- Guo, H.; Hang, C.; Lin, B.; Lin, Z.; Xiong, H.; Zhang, M.; Lu, R.; Liu, J.; Shi, D.; Xie, D.; et al. HAND factors regulate cardiac lineage commitment and differentiation from human pluripotent stem cells. Stem Cell Res. Ther. 2024, 15, 31. [Google Scholar] [CrossRef]
- Eitner, F.; Richter, B.; Schwänen, S.; Szaroszyk, M.; Vogt, I.; Grund, A.; Thum, T.; Heineke, J.; Haffner, D.; Leifheit-Nestler, M. Comprehensive expression analysis of cardiac fibroblast growth factor 23 in health and pressure-induced cardiac hypertrophy. Front. Cell Dev. Biol. 2022, 9, 791479. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, U.; Ghalayini, W.; Dong, F.; Weber, K.; Zou, Y.R.; Rabbany, S.Y.; Rafii, S.; Penn, M.S. Role of cardiac myocyte CXCR4 expression in development and left ventricular remodeling after acute myocardial infarction. Circ. Res. 2010, 107, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Merkel, C.D.; Li, Y.; Raza, Q.; Stolz, D.B.; Kwiatkowski, A.V. Vinculin anchors contractile actin to the cardiomyocyte adherens junction. Mol. Biol. Cell. 2019, 30, 2639–2650. [Google Scholar] [CrossRef]
- Quaife-Ryan, G.A.; Mills, R.J.; Lavers, G.; Voges, H.K.; Vivien, C.J.; Elliott, D.A.; Ramialison, M.; Hudson, J.E.; Porrello, E.R. β-Catenin drives distinct transcriptional networks in proliferative and nonproliferative cardiomyocytes. Development 2020, 147, dev193417. [Google Scholar] [CrossRef]
- Kondo, T.; Takahashi, M.; Yamasaki, G.; Sugimoto, M.; Kuse, A.; Morichika, M.; Nakagawa, K.; Sakurada, M.; Asano, M.; Ueno, Y. Immunohistochemical analysis of vimentin expression in myocardial tissue from autopsy cases of ischemic heart disease. Leg. Med. 2022, 54, 102003. [Google Scholar] [CrossRef]
- Grimes, K.M.; Prasad, V.; McNamara, J.W. Supporting the heart: Functions of the cardiomyocyte’s non-sarcomeric cytoskeleton. J. Mol. Cell Cardiol. 2019, 131, 187–196. [Google Scholar] [CrossRef]
- Stathopoulou, K.; Schnittger, J.; Raabe, J.; Fleischer, F.; Mangels, N.; Piasecki, A.; Findlay, J.; Hartmann, K.; Krasemann, S.; Schlossarek, S.; et al. CMYA5 is a novel interaction partner of FHL2 in cardiac myocytes. FEBS J. 2022, 289, 4622–4645. [Google Scholar] [CrossRef]
- Ong, L.P.; Bargehr, J.; Knight-Schrijver, V.R.; Lee, J.; Colzani, M.; Bayraktar, S.; Bernard, W.G.; Marchiano, S.; Bertero, A.; Murry, C.E.; et al. Epicardially secreted fibronectin drives cardiomyocyte maturation in 3D-engineered heart tissues. Stem Cell Rep. 2023, 18, 936–951. [Google Scholar] [CrossRef] [PubMed]
- Heras-Bautista, C.O.; Mikhael, N.; Lam, J.; Shinde, V.; Katsen-Globa, A.; Dieluweit, S.; Molcanyi, M.; Uvarov, V.; Jütten, P.; Sahito, R.G.A.; et al. Cardiomyocytes facing fibrotic conditions re-express extracellular matrix transcripts. Acta Biomater. 2019, 89, 180–192. [Google Scholar] [CrossRef]
- Komati, H.; Nemer, M. Repairing hearts with AKT. Proc. Natl. Acad. Sci. USA 2015, 112, 13131–13132. [Google Scholar] [CrossRef]
- Vite, A.; Caporizzo, M.A.; Corbin, E.A.; Brandimarto, J.; McAfee, Q.; Livingston, C.E.; Prosser, B.L.; Margulies, K.B. Extracellular stiffness induces contractile dysfunction in adult cardiomyocytes via cell-autonomous and microtubule-dependent mechanisms. Basic Res. Cardiol. 2022, 117, 41. [Google Scholar] [CrossRef] [PubMed]
- Potta, S.P.; Liang, H.; Winkler, J.; Doss, M.X.; Chen, S.; Wagh, V.; Pfannkuche, K.; Hescheler, J.; Sachinidis, A. Isolation and functional characterization of alpha-smooth muscle actin expressing cardiomyocytes from embryonic stem cells. Cell Physiol. Biochem. 2010, 25, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Hsu, Y.T. Bax translocates from cytosol to mitochondria in cardiac cells during apoptosis: Development of a GFP-Bax-stable H9c2 cell line for apoptosis analysis. Am. J. Physiol. Heart Circ. Physiol. 2005, 289, H477–H487. [Google Scholar] [CrossRef]
- van Beek-Harmsen, B.J.; van der Laarse, W.J. Immunohistochemical determination of cytosolic cytochrome C concentration in cardiomyocytes. J. Histochem. Cytochem. 2005, 53, 803–807. [Google Scholar] [CrossRef] [PubMed]
- Dabravolski, S.A.; Sukhorukov, V.N.; Kalmykov, V.A.; Orekhov, N.A.; Grechko, A.V.; Orekhov, A.N. Heat Shock Protein 90 as Therapeutic Target for CVDs and Heart Ageing. Int. J. Mol. Sci. 2022, 23, 649. [Google Scholar] [CrossRef]
- Zheng, D.; Zhang, M.; Liu, T.; Zhou, T.; Shen, A. Osteoprotegerin prompts cardiomyocyte hypertrophy via autophagy inhibition mediated by FAK/BECLIN1 pathway. Life Sci. 2021, 264, 118550. [Google Scholar] [CrossRef]
- Basheer, W.A.; Fu, Y.; Shimura, D.; Xiao, S.; Agvanian, S.; Hernandez, D.M.; Hitzeman, T.C.; Hong, T.; Shaw, R.M. Stress response protein GJA1-20k promotes mitochondrial biogenesis, metabolic quiescence, and cardioprotection against ischemia/reperfusion injury. JCI Insight 2018, 3, e121900. [Google Scholar] [CrossRef]
- Mohr, M.E.; Li, S.; Trouten, A.M.; Stairley, R.A.; Roddy, P.L.; Liu, C.; Zhang, M.; Sucov, H.M.; Tao, G. Cardiomyocyte-fibroblast interaction regulates ferroptosis and fibrosis after myocardial injury. iScience 2024, 27, 109219. [Google Scholar] [CrossRef]
- ISCN. ISCN 2020: An International System for Human Cytogenomic Nomenclature (2020) Reprint of Cytogenetic and Genome Research 2020, Vol. 160, No. 7–8; McGowan-Jordan, J., Hastings, R.J., Moore, S., Eds.; Karger: Basel, Switzerland, 2020; ISBN 978-3-318-06706-4. [Google Scholar] [CrossRef]
- ISCN. ISCN 2024: An International System for Human Cytogenomic Nomenclature (2024); McGowan-Jordan, J., Hastings, R.J., Moore, S., Eds.; Karger: Basel, Switzerland, 2024; ISBN 978-3-318-07330-0. [Google Scholar] [CrossRef]
- Koelsche, C.; Benhamida, J.K.; Kommoss, F.K.F.; Stichel, D.; Jones, D.T.W.; Pfister, S.M.; Heilig, C.E.; Fröhling, S.; Stenzinger, A.; Buslei, R.; et al. Intimal sarcomas and undifferentiated cardiac sarcomas carry mutually exclusive MDM2, MDM4, and CDK6 amplifications and share a common DNA methylation signature. Mod. Pathol. 2021, 34, 2122–2129. [Google Scholar] [CrossRef]
- Lamszus, K.; Kluwe, L.; Matschke, J.; Meissner, H.; Laas, R.; Westphal, M. Allelic losses at 1p, 9q, 10q, 14q, and 22q in the progression of aggressive meningiomas and undifferentiated meningeal sarcomas. Cancer Genet. Cytogenet. 1999, 110, 103–110. [Google Scholar] [CrossRef]
- Carneiro, A.; Francis, P.; Bendahl, P.O.; Fernebro, J.; Akerman, M.; Fletcher, C.; Rydholm, A.; Borg, A.; Nilbert, M. Indistinguishable genomic profiles and shared prognostic markers in undifferentiated pleomorphic sarcoma and leiomyosarcoma: Different sides of a single coin? Lab. Investig. 2009, 89, 668–675, Erratum in Lab. Investig. 2009, 89, 840. [Google Scholar] [CrossRef] [PubMed]
- Nanda, I.; Steinlein, C.; Haaf, T.; Buhl, E.M.; Grimm, D.G.; Friedman, S.L.; Meurer, S.K.; Schröder, S.K.; Weiskirchen, R. Genetic characterization of rat hepatic stellate cell line HSC-T6 for in vitro cell line authentication. Cells 2022, 11, 1783. [Google Scholar] [CrossRef]
- Gäberlein, K.; Schröder, S.K.; Nanda, I.; Steinlein, C.; Haaf, T.; Buhl, E.M.; Sauvant, P.; Sapin, V.; Abergel, A.; Weiskirchen, R. Genetic characterization of rat hepatic stellate cell line PAV-1. Cells 2023, 12, 1603. [Google Scholar] [CrossRef]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Zaaijer, S.; Groen, S.C.; Sanjana, N.E. Tracking cell lineages to improve research reproducibility. Nat. Biotechnol. 2021, 39, 666–670. [Google Scholar] [CrossRef]
- Koenig, A.L.; Shchukina, I.; Amrute, J.; Andhey, P.S.; Zaitsev, K.; Lai, L.; Bajpai, G.; Bredemeyer, A.; Smith, G.; Jones, C.; et al. Single-cell transcriptomics reveals cell-type-specific diversification in human heart failure. Nat. Cardiovasc. Res. 2022, 1, 263–280. [Google Scholar] [CrossRef] [PubMed]
- Salzillo, C.; La Verde, M.; Imparato, A.; Molitierno, R.; Lucà, S.; Pagliuca, F.; Marzullo, A. Cardiovascular diseases in public health: Chromosomal abnormalities in congenital heart disease causing sudden cardiac death in children. Medicina 2024, 60, 1976. [Google Scholar] [CrossRef]
- Goetze, J.P.; Bruneau, B.G.; Ramos, H.R.; Ogawa, T.; de Bold, M.K.; de Bold, A.J. Cardiac natriuretic peptides. Nat. Rev. Cardiol. 2020, 17, 698–717. [Google Scholar] [CrossRef]
- Zhu, S.; Li, W.; Zhou, H.; Wei, W.; Ambasudhan, R.; Lin, T.; Kim, J.; Zhang, K.; Ding, S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010, 7, 651–655. [Google Scholar] [CrossRef]
- Patel, I.; Parchem, R.J. Regulation of Oct4 in stem cells and neural crest cells. Birth Defects Res. 2022, 114, 983–1002. [Google Scholar] [CrossRef]
- Wang, H.; Cao, N.; Spencer, C.I.; Nie, B.; Ma, T.; Xu, T.; Zhang, Y.; Wang, X.; Srivastava, D.; Ding, S. Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep. 2014, 6, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Hescheler, J.; Meyer, R.; Plant, S.; Krautwurst, D.; Rosenthal, W.; Schultz, G. Morphological, biochemical, and electrophysiological characterization of a clonal cell (H9c2) line from rat heart. Circ. Res. 1991, 69, 1476–1486. [Google Scholar] [CrossRef] [PubMed]
- Watkins, S.J.; Borthwick, G.M.; Arthur, H.M. The H9C2 cell line and primary neonatal cardiomyocyte cells show similar hypertrophic responses in vitro. Vitr. Cell. Dev. Biol. Anim. 2011, 47, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Zordoky, B.N.; El-Kadi, A.O. H9c2 cell line is a valuable in vitro model to study the drug metabolizing enzymes in the heart. J. Pharmacol. Toxicol. Methods 2007, 56, 317–322. [Google Scholar] [CrossRef]





| Antibody | Cat. no. | Company | Dilution | Clonality |
|---|---|---|---|---|
| α-Actinin 1 | 6487 | Cell Signaling Technology, Leiden, The Netherlands | 1:1000 | r mAb |
| α-Actinin 2 | A7811 | Sigma-Aldrich, Merck, Taufkirchen, Germany | 1:10,000 | m mAb |
| α-SMA | CBL171-I | Sigma-Aldrich, Merck, Taufkirchen, Germany | 1:1000 | m mAb |
| α-Tubulin (B-7) | sc-5286 | Santa Cruz, Santa Cruz, CA, USA | 1:1000 | m mAb |
| β-Actin | A5441 | Sigma-Aldrich, Merck, Taufkirchen, Germany | 1:10,000 | m mAb |
| β-Catenin (E-5) | sc-7963 | Santa Cruz, Santa Cruz, CA, USA | 1:1000 | m mAb |
| Acsl1 | 4047S | Cell Signaling Technology, Leiden, The Netherlands | 1:1000 | r pAb |
| Akt (pan) (11E7) | 4685 | Cell Signaling Technology, Leiden, The Netherlands | 1:1000 | r mAb |
| Bax | 2772S | Cell Signaling Technology, Leiden, The Netherlands | 1:1000 | r pAb |
| Beclin-1 (D40C5) | 3495 | Cell Signaling Technology, Leiden, The Netherlands | 1:1000 | r mAb |
| Collagen I | 14695-1-AP | Proteintech, Chromo Tek GmbH, Planegg-Martinsried, Germany | 1:1000 | r pAb |
| Collagen III | 22734-1-AP | Proteintech, Chromo Tek GmbH, Planegg-Martinsried, Germany | 1:1000 | r pAb |
| Connexin 43 (C-20) | sc-6560-R | Santa Cruz, Santa Cruz, CA, USA | 1:1000 | r pAb |
| Cytochrome c (D18C7) | 11940 | Cell Signaling Technology, Leiden, The Netherlands | 1:1000 | r mAb |
| Ferritin heavy chain (B-12) | sc-376594 | Santa Cruz, Santa Cruz, CA, USA | 1:500 | m mAb |
| Fhl2 | AF4758 | R&D Systems, Bio-Techne, Abingdon, UK | 1:500 | g pAb |
| Fibronectin | AB1954 | Sigma-Aldrich, Merck, Taufkirchen, Germany | 1:3000 | r pAb |
| GAPDH (6C5) | sc-32233 | Santa Cruz, Santa Cruz, CA, USA | 1:1000 | m mAb |
| HSP90 (C45G5) | 4877 | Cell Signaling Technology, Leiden, The Netherlands | 1:1000 | r mAb |
| Troponin I (cardiac) | ab209809 | Abcam, Cambridge, UK | 1:1000 | r mAb |
| Troponin T (cardiac) | 5593 | Cell Signaling Technology, Leiden, The Netherlands | 1:1000 | r pAb |
| Vimentin | ab92547 | Abcam, Cambridge, UK | 1:3000 | r mAb |
| Vinculin | 66305-1-Ig | Proteintech, Chromo Tek GmbH, Planegg-Martinsried, Germany | 1:5000 | m mAb |
| Goat anti-rabbit IgG (H+L), HRP | 31460 | Invitrogen, Thermo Fisher Scientific, Schwerte, Germany | 1:5000 | g |
| Goat anti-mouse IgG (H+L), HRP | 31430 | Invitrogen, Thermo Fisher Scientific, Schwerte, Germany | 1:5000 | r |
| Mouse anti-goat IgG (H+L), HRP | 31400 | Invitrogen, Thermo Fisher Scientific, Schwerte, Germany | 1:5000 | m |
| Transcript Id 1 | Gene Id | Gene | Gene Description | TPM |
|---|---|---|---|---|
| ENSRNOT00000018097 ENSRNOT00000119402 ENSRNOT00000112334 ENSRNOT00000017838 ENSRNOT00000119402 | ENSRNOG00000013262 | Myl1 | myosin light chain 1 | 5.163359 0.648313 1.223487 1.103736 0.648313 |
| ENSRNOT00000072480 ENSRNOT00000101539 | ENSRNOG00000050675 | Myl4 | myosin light chain 4 | 42.672105 0.14434 |
| ENSRNOT00000089074 ENSRNOT00000099810 ENSRNOT00000082518 ENSRNOT00000094248 ENSRNOT00000107790 ENSRNOT00000085644 | ENSRNOG00000054140 | Myl6 | myosin light chain 6 | 1846.647426 874.627113 25.302537 24.381243 19.012288 11.635463 |
| ENSRNOT00000048453 | ENSRNOG00000028837 | LOC120093525 | myosin light chain 6B | 43.718117 |
| ENSRNOT00000027445 | ENSRNOG00000020246 | Myl9 | myosin light chain 9 | 1155.386796 |
| ENSRNOT00000023944 ENSRNOT00000112797 | ENSRNOG00000017645 | Myl11 | myosin light chain 11 | 285.690465 0.824397 |
| ENSRNOT00000021048 ENSRNOT00000111518 ENSRNOT00000105340 ENSRNOT00000117541 ENSRNOT00000108694 ENSRNOT00000048125 | ENSRNOG00000015278 | Myl12b | myosin light chain 12B | 624.478983 396.879829 166.614506 95.549881 0.898159 0.864107 |
| ENSRNOT00000004236 | ENSRNOG00000065740 | Myh2 | myosin heavy chain 2 | 0.672346 |
| ENSRNOT00000004147 ENSRNOT00000115161 | ENSRNOG00000046276 | Myh3 | myosin heavy chain 3 | 103.894988 0.387361 |
| ENSRNOT00000004295 ENSRNOT00000082871 ENSRNOT00000045718 | ENSRNOG00000049695 | Myh4 | myosin heavy chain 4 | 33.85847 0.142894 0.086715 |
| ENSRNOT00000115198 | ENSRNOG00000025757 | Myh6 | myosin heavy chain 6 | 0.050162 |
| ENSRNOT00000024186 | ENSRNOG00000016983 | Myh7 | myosin heavy chain 7 | 0.048688 |
| ENSRNOT00000025859 | ENSRNOG00000018997 | Myh7b | myosin heavy chain 7B | 0.608836 |
| ENSRNOT00000105953 | ENSRNOG00000068010 | Myh8 | myosin heavy chain 8 | 0.080643 |
| ENSRNOT00000037611 ENSRNOT00000007398 ENSRNOT00000116925 ENSRNOT00000119854 | ENSRNOG00000049236 | Myh9 | myosin heavy chain 9 | 859.802126 397.860041 10.572224 0.960109 |
| ENSRNOT00000105761 ENSRNOT00000113616 ENSRNOT00000065895 | ENSRNOG00000002886 | Myh10 | myosin heavy chain 10 | 83.625491 31.943467 19.761232 |
| ENSRNOT00000084608 ENSRNOT00000112644 | ENSRNOG00000057880 | Myh11 | myosin heavy chain 11 | 4.104926 1.264626 |
| ENSRNOT00000118051 | ENSRNOG00000067378 | Myh13 | myosin heavy chain 13 | 0.010706 |
| ENSRNOT00000091760 | ENSRNOG00000020014 | Myh14 | myosin heavy chain 14 | 0.04009 |
| ENSRNOT00000090307 | ENSRNOG00000061038 | Myh15 | myosin heavy chain 15 | 1.364068 |
| ENSRNOT00000106031 | ENSRNOG00000004177 | Myo1a | myosin IA | 0.158265 |
| ENSRNOT00000068433 ENSRNOT00000108379 ENSRNOT00000118845 | ENSRNOG00000048152 | Myo1b | myosin Ib | 1.012463 0.817926 0.124786 |
| ENSRNOT00000117022 ENSRNOT00000036666 ENSRNOT00000108756 ENSRNOT00000101210 | ENSRNOG00000004072 | Myo1c | myosin 1C | 256.059864 57.901436 15.070173 5.120976 |
| ENSRNOT00000004609 ENSRNOT00000108255 | ENSRNOG00000003276 | Myo1d | myosin ID | 92.963065 0.537064 |
| ENSRNOT00000104863 | ENSRNOG00000061928 | Myo1e | myosin IE | 56.688608 |
| ENSRNOT00000011513 ENSRNOT00000110236 | ENSRNOG00000008409 | Myo1f | myosin IF | 1.427103 0.040403 |
| ENSRNOT00000119331 | ENSRNOG00000059140 | Myo1g | myosin IG | 0.101252 |
| ENSRNOT00000078807 | ENSRNOG00000047191 | Myo1h | myosin IH | 0.037109 |
| ENSRNOT00000109142 ENSRNOT00000082288 ENSRNOT00000102636 ENSRNOT00000091789 | ENSRNOG00000058866 | Myo5a | myosin VA | 25.016567 9.28073 8.944404 0.299799 |
| ENSRNOT00000094389 ENSRNOT00000019512 | ENSRNOG00000014104 | Myo5b | myosin Vb | 5.440433 1.151928 |
| ENSRNOT00000120263 ENSRNOT00000108142 ENSRNOT00000112254 | ENSRNOG00000011852 | Myo6 | myosin VI | 25.109277 1.384369 0.20236 |
| ENSRNOT00000019053 ENSRNOT00000103282 | ENSRNOG00000013641 | Myo7a | myosin VIIA | 26.382815 11.975166 |
| ENSRNOT00000046864 | ENSRNOG00000015035 | Myo7b | myosin VIIb | 0.009858 |
| ENSRNOT00000104304 ENSRNOT00000103923 ENSRNOT00000015963 ENSRNOT00000118561 | ENSRNOG00000011619 | Myo9a | myosin IXA | 6.65948 4.352163 3.013147 0.953695 |
| ENSRNOT00000045099 ENSRNOT00000083651 ENSRNOT00000081321 | ENSRNOG00000016256 | Myo9b | myosin IXb | 16.306978 15.395789 1.48221 |
| ENSRNOT00000065897 ENSRNOT00000102421 | ENSRNOG00000010161 | Myo10 | myosin X | 61.874959 1.946731 |
| ENSRNOT00000079133 | ENSRNOG00000059219 | Myo15a | myosin XVA | 0.017051 |
| ENSRNOT00000035001 | ENSRNOG00000042445 | Myo15b | myosin XVB | 0.023935 |
| ENSRNOT00000088919 ENSRNOT00000109794 ENSRNOT00000102240 ENSRNOT00000110549 ENSRNOT00000100376 | ENSRNOG00000033101 | Myo18a | myosin XVIIIa | 17.10711 14.92682 13.895175 0.924955 0.563736 |
| ENSRNOT00000098133 | ENSRNOG00000048430 | Myo18b | myosin XVIIIb | 0.048917 |
| ENSRNOT00000003886 ENSRNOT00000119574 | ENSRNOG00000002852 | Myo19 | myosin XIX | 8.602543 1.680903 |
| ENSRNOT00000050443 ENSRNOT00000110793 ENSRNOT00000041328 ENSRNOT00000114924 | ENSRNOG00000018630 | LOC108351137 | glyceraldehyde-3-phosphate dehydrogenase | 2955.510839 902.439425 147.63458 41.959217 |
| Transcript Id 1 | Gene Id | Gene | Gene Description | TPM |
|---|---|---|---|---|
| ENSRNOT00000091560 ENSRNOT00000079824 ENSRNOT00000088795 ENSRNOT00000112260 | ENSRNOG00000056756 | Actn1 | actinin, alpha 1 | 360.869674 121.742056 95.274999 1.456574 |
| ENSRNOT00000101663 ENSRNOT00000101075 | ENSRNOG00000017833 | Actn2 | actinin, alpha 2 | 0.988042 0.866778 |
| ENSRNOT00000011773 ENSRNOT00000116592 | ENSRNOG00000008536 | Actc1 | actin, alpha, cardiac muscle 1 | 29.680643 0.241854 |
| ENSRNOT00000089988 | ENSRNOG00000001821 | Adipoq | adiponectin, C1Q and collagen domain containing | 0.088185 |
| ENSRNOT00000005551 ENSRNOT00000119052 ENSRNOT00000102094 | ENSRNOG00000004143 | Adipor1 | adiponectin receptor 1 | 219.61476 0.5584 0.233142 |
| ENSRNOT00000010556 | ENSRNOG00000007990 | Adipor2 | adiponectin receptor 2 | 91.021116 |
| ENSRNOT00000104562 ENSRNOT00000002738 | ENSRNOG00000001989 | Alcam | activated leukocyte cell adhesion molecule | 34.73353 10.607857 |
| ENSRNOT00000108356 ENSRNOT00000025258 ENSRNOT00000120144 ENSRNOT00000097402 ENSRNOT00000117221 ENSRNOT00000108831 | ENSRNOG00000018598 | Ankrd1 | ankyrin repeat domain 1 | 1099.57145 696.594054 536.462968 127.375767 125.63322 0.202132 |
| ENSRNOT00000024347 ENSRNOT00000001738 | ENSRNOG00000001285 | Atp2a2 | ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 | 163.17185 128.264822 |
| ENSRNOT00000019554 ENSRNOT00000111490 ENSRNOT00000109811 ENSRNOT00000108710 ENSRNOT00000113833 ENSRNOT00000116052 | ENSRNOG00000014453 | Anxa5 | annexin A5 | 326.883008 326.883008 7.961338 1.032791 0.881536 0.076392 |
| ENSRNOT00000106165 ENSRNOT00000111975 ENSRNOT00000096029 ENSRNOT00000014464 ENSRNOT00000119845 | ENSRNOG00000010668 | Anxa6 | annexin A6 | 292.28195 104.941243 68.292397 2.736565 1.174227 |
| ENSRNOT00000073636 ENSRNOT00000080190 | ENSRNOG00000047466 | Bdnf | brain-derived neurotrophic factor | 22.063067 2.770632 |
| ENSRNOT00000083268 ENSRNOT00000012957 | ENSRNOG00000009694 | Bmp4 | bone morphogenetic protein 4 | 41.067185 28.953671 |
| ENSRNOT00000080271 | ENSRNOG00000057713 | Cav2 | caveolin 2 | 38.392081 |
| ENSRNOT00000007601 | ENSRNOG00000005798 | Cav3 | caveolin 3 | 35.815516 |
| ENSRNOT00000115561 | ENSRNOG00000015602 | Cdh2 | cadherin 2 | 0.034055 |
| ENSRNOT00000019652 | ENSRNOG00000014254 | Cpt1a | carnitine palmitoyltransferase 1A | 21.367628 |
| ENSRNOT00000080598 ENSRNOT00000067011 ENSRNOT00000097951 | ENSRNOG00000003772 | Csrp2 | cysteine and glycine-rich protein 2 | 46.077184 15.957459 5.717219 |
| ENSRNOT00000101345 ENSRNOT00000079085 | ENSRNOG00000054172 | Ctnnb1 | catenin beta 1 | 148.736322 133.524514 |
| ENSRNOT00000026860 | ENSRNOG00000019810 | Des | desmin | 4.863506 |
| ENSRNOT00000081061 ENSRNOT00000034372 ENSRNOT00000094721 ENSRNOT00000091467 ENSRNOT00000109716 | ENSRNOG00000046366 | Dmd | dystrophin | 5.83409 4.877158 4.242989 3.755658 2.561635 |
| ENSRNOT00000005612 ENSRNOT00000118138 ENSRNOT00000096738 | ENSRNOG00000004078 | Eno3 | enolase 3 | 33.28236 24.245848 0.551336 |
| ENSRNOT00000017325 | ENSRNOG00000012879 | Fabp3 (H-Fabp) | fatty acid binding protein 3 | 13.928079 |
| ENSRNOT00000023388 | ENSRNOG00000017392 | Fgf2 | fibroblast growth factor 2 | 1.080419 |
| ENSRNOT00000114387 ENSRNOT00000101412 ENSRNOT00000023014 | ENSRNOG00000016866 | Fhl2 | four and a half LIM domains 2 | 77.911289 59.661397 45.106886 |
| ENSRNOT00000014320 | ENSRNOG00000010708 | Gata4 | GATA binding protein 4 | 4.261875 |
| ENSRNOT00000081399 | ENSRNOG00000023433 | Gata6 | GATA binding protein 6 | 11.70926 |
| ENSRNOT00000001054 ENSRNOT00000100494 | ENSRNOG00000000805 | Gja1 | gap junction protein, alpha 1 | 99.918567 17.905309 |
| ENSRNOT00000079552 | ENSRNOG00000060448 | Hand2 (dHand) | heart and neural crest derivatives expressed 2 | 70.427748 |
| ENSRNOT00000118307 ENSRNOT00000085680 ENSRNOT00000005995 | ENSRNOG00000004517 | Igf1 | insulin-like growth factor 1 | 22.187026 1.599373 1.449521 |
| ENSRNOT00000020885 ENSRNOT00000117668 | ENSRNOG00000015068 | Il11ra | interleukin 11 receptor subunit alpha 1 | 28.250421 0.130904 |
| ENSRNOT00000019844 | ENSRNOG00000014426 | Lox | lysyl oxidase | 279.086949 |
| ENSRNOT00000076230 ENSRNOT00000076992 ENSRNOT00000076481 ENSRNOT00000075931 ENSRNOT00000076136 | ENSRNOG00000033134 | Mef2c | myocyte enhancer factor 2C | 21.461792 10.307472 2.878599 2.158897 0.445899 |
| ENSRNOT00000055680 | ENSRNOG00000046424 | Mfn2 | mitofusin 2 | 65.615575 |
| ENSRNOT00000051121 | ENSRNOG00000008658 | Mitf | Melanocyte inducing transcription factor | 3.362071 |
| ENSRNOT00000042686 | ENSRNOG00000031093 | Mov10l1 | Mov10 like RISC complex RNA helicase 1 | 0.030013 |
| ENSRNOT00000028155 | ENSRNOG00000020747 | Nkx2-5 | NK2 homeobox 5 | 0.319419 |
| ENSRNOT00000026212 ENSRNOT00000104296 | ENSRNOG00000019322 | Notch1 | notch receptor 1 | 8.838129 0.095737 |
| ENSRNOT00000090547 ENSRNOT00000102686 | ENSRNOG00000054212 | Pde1a | phosphodiesterase 1A | 1.752889 0.660082 |
| ENSRNOT00000111781 ENSRNOT00000101684 ENSRNOT00000113369 ENSRNOT00000066384 ENSRNOT00000110594 ENSRNOT00000112056 | ENSRNOG00000042536 | Pde4d | phosphodiesterase 4D | 9.706262 7.996515 2.912791 1.736483 0.588956 0.028455 |
| ENSRNOT00000073486 | ENSRNOG00000046057 | Pnmt | phenylethanolamine-N-methyltransferase | 0.161158 |
| ENSRNOT00000028636 | ENSRNOG00000021090 | Pygm | glycogen phosphorylase, muscle associated | 20.312461 |
| ENSRNOT00000008319 ENSRNOT00000075962 ENSRNOT00000067543 ENSRNOT00000091249 | ENSRNOG00000005906 | Rgd1565355 (CD36) | similar to fatty acid translocase/CD36 | 0.680231 0.346115 0.162213 0.109615 |
| ENSRNOT00000111439 | ENSRNOG00000017060 | Ryr2 | ryanodine receptor 2 | 0.00405 |
| ENSRNOT00000084391 | ENSRNOG00000000565 | Sgpl1 | sphingosine-1-phosphate lyase 1 | 122.518127 |
| ENSRNOT00000025606 ENSRNOT00000094646 | ENSRNOG00000018943 | Tnnc1 | troponin C1, slow skeletal and cardiac type | 140.282686 69.212851 |
| ENSRNOT00000110513 | ENSRNOG00000018250 | Tnni3 | troponin I3, cardiac type | 0.528838 |
| ENSRNOT00000034957 ENSRNOT00000058843 | ENSRNOG00000028041 | Tnnt1 | troponin T1, slow skeletal type | 86.297291 4.163693 |
| ENSRNOT00000084986 ENSRNOT00000108522 ENSRNOT00000047682 ENSRNOT00000050284 | ENSRNOG00000033734 | Tnnt2 | troponin T2, cardiac type | 385.200438 161.390324 130.206578 6.04521 |
| ENSRNOT00000057641 ENSRNOT00000024575 ENSRNOT00000048044 ENSRNOT00000090288 ENSRNOT00000085894 ENSRNOT00000024617 ENSRNOT00000099012 ENSRNOT00000040808 ENSRNOT00000112475 ENSRNOT00000024493 | ENSRNOG00000018184 | Tpm1 | tropomyosin 1 | 609.160335 394.939461 348.84129 309.385315 250.542706 70.487194 62.960197 23.306842 9.964889 9.432292 |
| ENSRNOT00000026493 | ENSRNOG00000019486 | Trpv1 | transient receptor potential cation channel, subfamily V, member 1 | 0.210286 |
| ENSRNOT00000108121 ENSRNOT00000101577 ENSRNOT00000107188 ENSRNOT00000114553 | ENSRNOG00000069271 | Ttn | titin | 2.315625 1.775349 0.597153 0.459324 |
| ENSRNOT00000027487 ENSRNOT00000076187 | ENSRNOG00000020276 | Tnnt2 | troponin I2, fast skeletal type | 10.186619 0.503748 |
| ENSRNOT00000014127 ENSRNOT00000079275 | ENSRNOG00000010390 | Hmbs | hydroxymethylbilane synthase | 27.1177 0.615926 |
| ENSRNOT00000096774 ENSRNOT00000035628 | ENSRNOG00000008195 | Ywhaz | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta | 529.169941 197.545795 |
| Rat RGSC 5.0/rn5 | Human GRCh37/hg19 | |
|---|---|---|
| Gain | ||
| 1pter->1q?12 (+2) => 1pter->1q12 | 1-75515806 | 6q22.31q27 5pterp15.31 5q15q15 |
| 1q12->1qter (+1) | 75515807-290094216 | 10q23.2q26.3 11q12.1q14.3 15q25.1qter 11p15.4p15.2 19q12q13.33 9q21.11q21.31 9pterp24.1 16p12.3p11.2 |
| 2pter->q?13 => (+1) 2pter->2q13 | 1-40882813 | 5q14.1q15 |
| 6pter->6qter (+1) | 1-156897508 | 14q12q32.33 2pterp16.3 7p21.2p15.3 7q22.3q31.1 |
| 9pter->9qter (+1) | 1-121549591 | 2q32.1qter 6p21.2q13 2q11.2q12.2 18p11.32p11.22 5q21.1q22.1 |
| +12pter->q1?2 (+4) =12pter->q12 | 1-31247709 | 13q12.13q13.2 7pterp22.1 7q11.21q11.22 7q21.2q22.1 7q11.23q11.23 |
| 14pter->14qter (+1) | 1-115151701 | 4pter->q22.1 2p16.2p14 7p13p12.1 22q12.1q12.3 1p22.2p22.1 |
| Xq3?->Xqter (+1) => Xq34->Xqter | 111295344-154597545 | Xq22.3q37.3 |
| Loss | ||
| 3pter->3q11 (−3) | 1-22900371 | 9q33.2q34.3 2q13q13 |
| Xq2?->Xq3? (−1) => Xq22->Xq34 | 57322686-111295344 | Xp22.11q22.3 |
| Allele Sizes (bp) in | |||||||
|---|---|---|---|---|---|---|---|
| SN | Marker Name 1 | Location on Chromosome | H9c2 | CFSC-2G | HSC-T6 | PAV-1 | Rat-1 |
| 1 | 73 | 1 | 211 | 194, 203 | 194 | 194 | 211, 213 |
| 2 | 8 | 2 | 195 | 236 | 234 | 234, 238 | 232, 236 |
| 3 | 2 | 2 | 127 | 126 | 127 | 128 | 129 |
| 4 | 4 | 3 | 250, 252 | 268, 270 | 238 | 236, 238 | 250, 252 |
| 5 | 3 | 3 | 160 | 160, 182 | 160, 162 | 162 | 178, 182 |
| 6 | 26 | 4 | 148 | 150 | 166 | 154 | 162 |
| 7 | 19 | 4 | 174 | 180 | 175 | 179 | 176, 178 |
| 8 | 81 | 5 | 127 | 130, 134 | 130, 132 | 130 | 128 |
| 9 | 34 | 6 | 193 | 184, 189 | 188 | 182, 187 | 184, 189 |
| 10 | 30 | 7 | 192, 194 | 188, 192 | 186, 192 | 192 | 186 |
| 11 | 24 | 8 | 258, 260 | 260 | 249, 253 | 254, 259 | 247, 249 |
| 12 | 59 | 9 | 143, 145 | 145 | 143, 146, 180 | 145, 148 | 176, 178 |
| 13 | 62 | 9 | 154 | 166 | 177 | 166 | 154 |
| 14 | 1 | 10 | 104 | 105 | 96 | 96, 105 | 96 |
| 15 | 55 | 10 | 205, 207 | 210, 214 | 210, 218 | 210, 218 | 203, 205 |
| 16 | 36 | 11 | 263 | 222 | 234 | 222 | 228 |
| 17 | 67 | 11 | 161 | 154, 156 | 165 | 165 | 165, 167 |
| 18 | 13 | 12 | 121 | 121 | 121, 135 | 121 | 121 |
| 19 | 35 | 13 | 203 | 197 | 197, 203 | 203 | 203 |
| 20 | 42 | 13 | 149 | 125 | 127 | 144, 156 | 154, 156 |
| 21 | 70 | 14 | 175 | 158, 175 | 175, 179 | 158, 175 | 158 |
| 22 | 61 | 15 | 128 | 128 | 128 | 128 | 110 |
| 23 | 79 | 15 | 172 | 172, 180 | 172 | 172 | 172 |
| 24 | 90 | 16 | 172, 174 | 159, 161 | 174 | 175 | 159, 161 |
| 25 | 69 | 16 | 143 | 138 | 139 | 136, 139 | 148 |
| 26 | 78 | 17 | 151, 153 | 136, 151 | 147, 151 | 147, 149 | 136, 140 |
| 27 | 15 | 18 | 232 | 232 | 232 | 232 | 238 |
| 28 | 16 | 18 | 243 | 251, 260 | 247, 251 | 251 | 247, 251 |
| 29 | 75 | 19 | 140 | 144 | 144, 184 | 144, 184 | 144 |
| 30 | 96 | 20 | 240 | 210 | 210, 212 | 210 | 208, 210 |
| 31 | 91 | 20 | 221 | 221 | 205, 211 | 211, 225 | 219, 221 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liehr, T.; Kankel, S.; Hardt, K.S.; Buhl, E.M.; Noels, H.; Keller, D.T.; Schröder-Lange, S.K.; Weiskirchen, R. Genetic and Molecular Characterization of H9c2 Rat Myoblast Cell Line. Cells 2025, 14, 502. https://doi.org/10.3390/cells14070502
Liehr T, Kankel S, Hardt KS, Buhl EM, Noels H, Keller DT, Schröder-Lange SK, Weiskirchen R. Genetic and Molecular Characterization of H9c2 Rat Myoblast Cell Line. Cells. 2025; 14(7):502. https://doi.org/10.3390/cells14070502
Chicago/Turabian StyleLiehr, Thomas, Stefanie Kankel, Katharina S. Hardt, Eva M. Buhl, Heidi Noels, Diandra T. Keller, Sarah K. Schröder-Lange, and Ralf Weiskirchen. 2025. "Genetic and Molecular Characterization of H9c2 Rat Myoblast Cell Line" Cells 14, no. 7: 502. https://doi.org/10.3390/cells14070502
APA StyleLiehr, T., Kankel, S., Hardt, K. S., Buhl, E. M., Noels, H., Keller, D. T., Schröder-Lange, S. K., & Weiskirchen, R. (2025). Genetic and Molecular Characterization of H9c2 Rat Myoblast Cell Line. Cells, 14(7), 502. https://doi.org/10.3390/cells14070502









