DNA Methylation Mediates the Transcription of STAT4 to Regulate KISS1 During Follicular Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Culture of GCs and Follicles
2.3. Real-Time Quantitative PCR (qRT-PCR)
2.4. EdU Assay
2.5. Cell Counting Kit-8 Viability Assay (CCK-8)
2.6. Flow Cytometry
2.7. ELISA
2.8. Hematoxylin and Eosin Staining (HE)
2.9. Bisulfite Sequencing PCR (BSP)
2.10. Chromatin Immunoprecipitation Assay (ChIP)
2.11. Dual-Luciferase Reporter Gene Assay
2.12. Western Blot Analysis
2.13. Statistical Analysis
3. Results
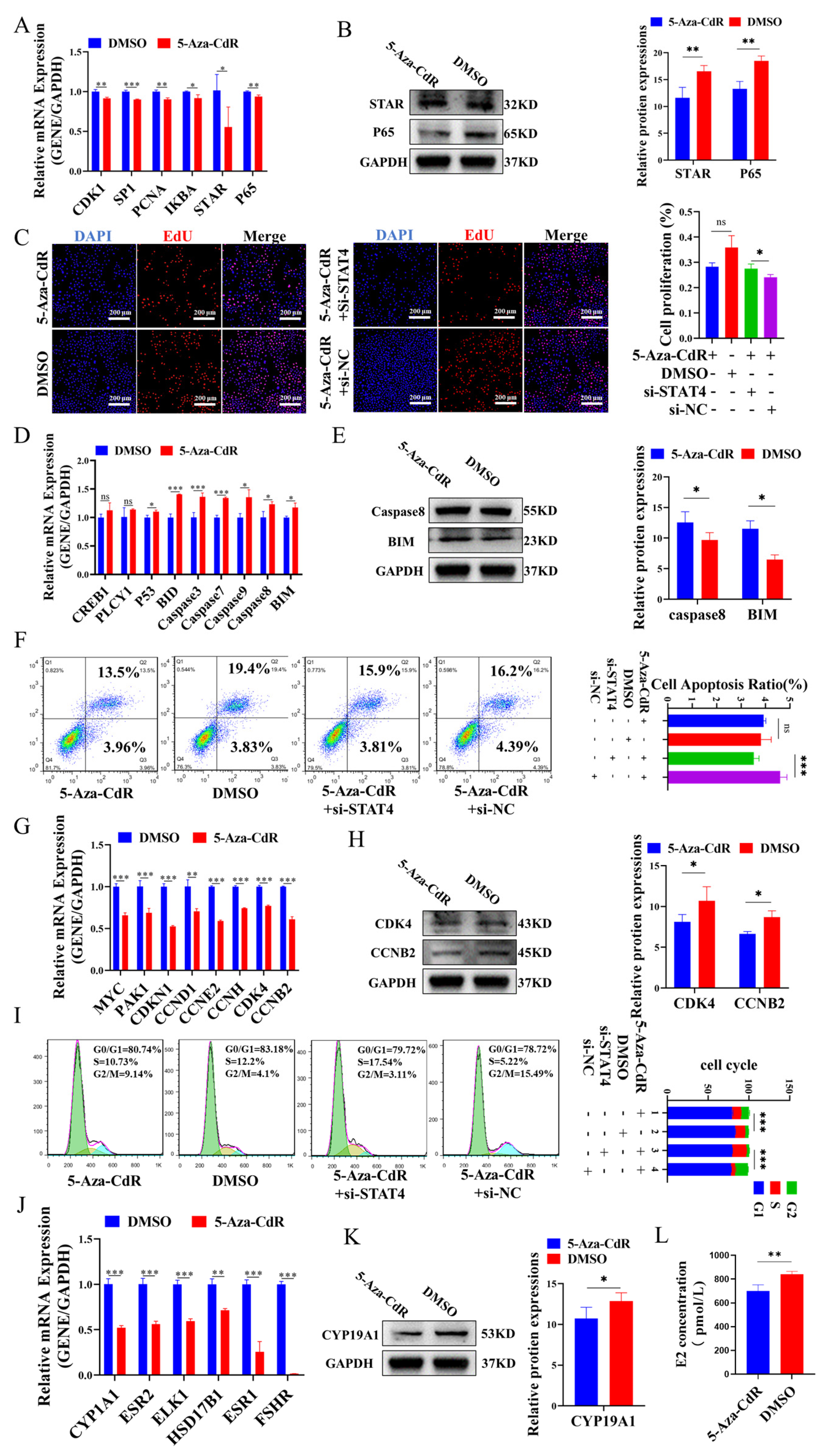
3.1. DNA Methylation May Regulate STAT4 to Increase Apoptosis in GCs
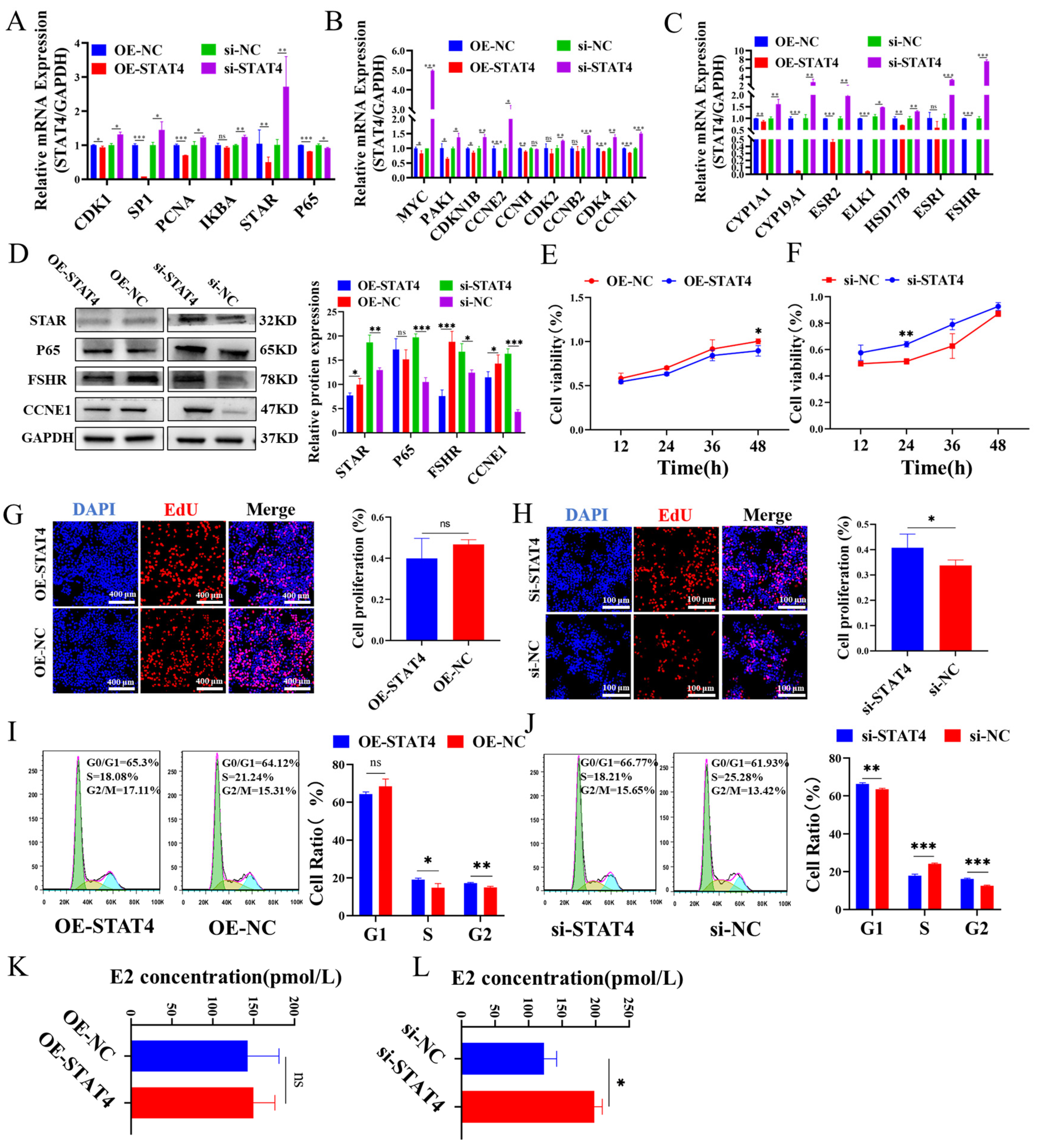
3.2. STAT4 Inhibits the Cellular Proliferation and E2 Secretion in GCs
3.3. DNA Hypomethylation of STAT4 Induces Apoptosis in GCs
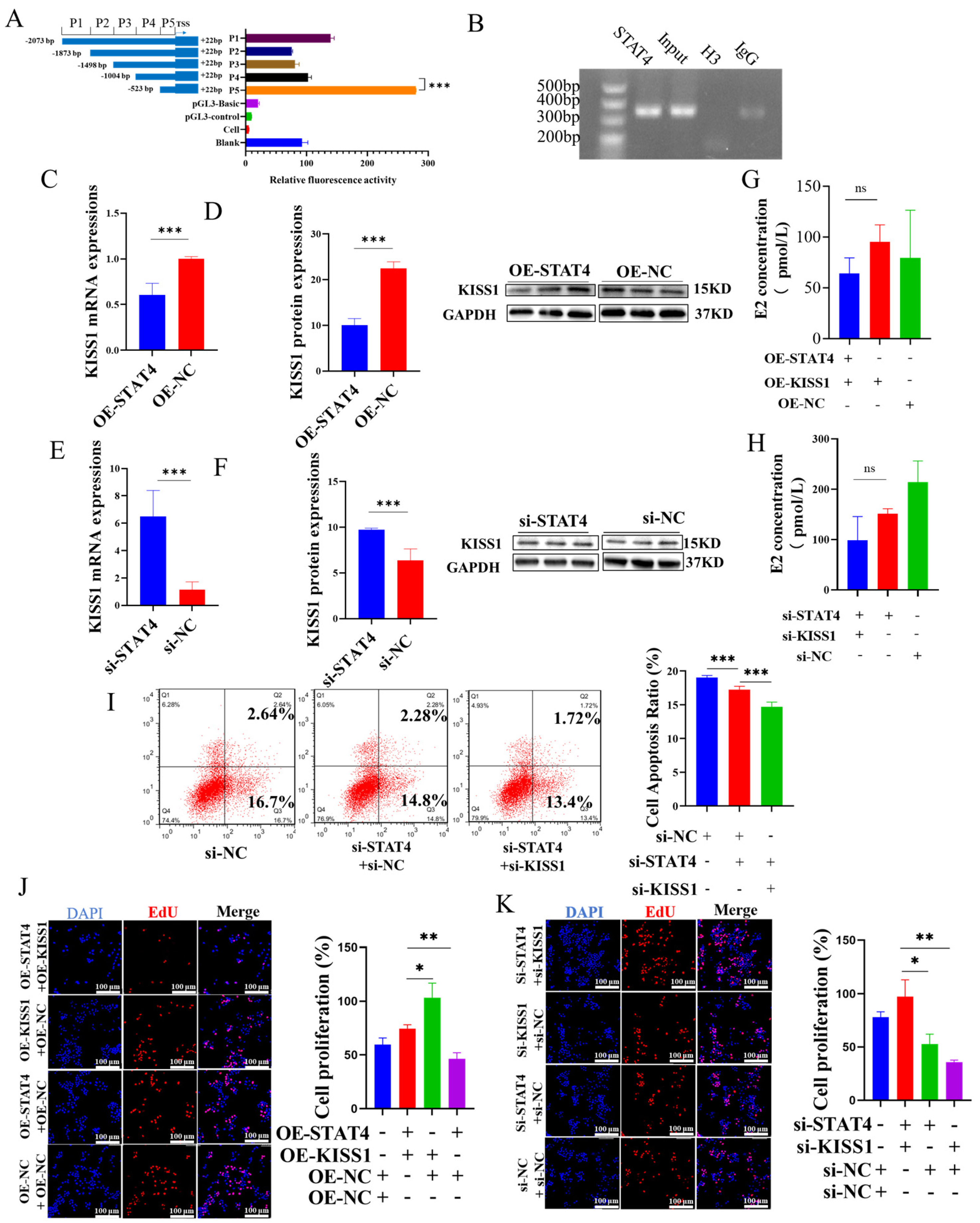
3.4. STAT4 Induces Apoptosis in GCs While Inhibiting the Expression of KISS1
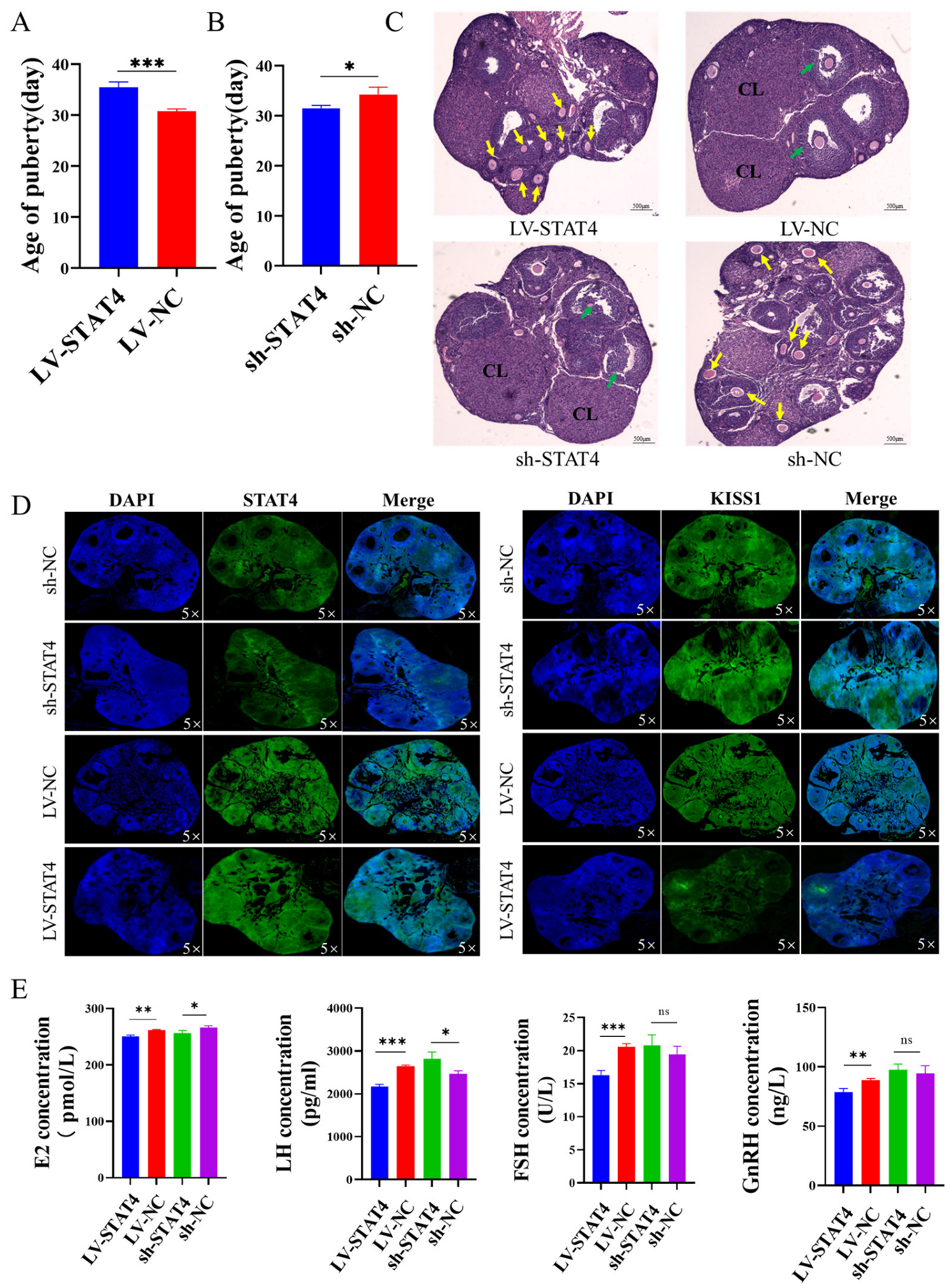
3.5. STAT4 Inhibits Follicular Development in Pigs
3.6. STAT4 Blocks Follicular Development in Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| STAT4 | Signal transducer and activator of transcription 4 |
| KISS1 | Kisspeptin-1 |
| GCs | Granulosa cells |
| 5-Aza-CdR | 5-azacytidine |
| E2 | Estradiol |
| LH | Luteinizing hormone |
| FSH | Follicle-stimulating hormone |
| GnRH | Gonadotropin-releasing hormone |
References
- Christenson, R.K.; Ford, J.J. Puberty and estrus in confinement-reared gilts. J. Anim. Sci. 1979, 49, 743–751. [Google Scholar] [PubMed]
- Stringer, J.M.; Alesi, L.R.; Winship, A.L.; Hutt, K.J. Beyond apoptosis: Evidence of other regulated cell death pathways in the ovary throughout development and life. Hum. Reprod. Update 2023, 29, 434–456. [Google Scholar] [PubMed]
- Field, S.L.; Dasgupta, T.; Cummings, M.; Orsi, N.M. Cytokines in ovarian folliculogenesis, oocyte maturation and luteinisation. Mol. Reprod. Dev. 2014, 81, 284–314. [Google Scholar] [PubMed]
- Jiang, J.Y.; Cheung, C.K.; Wang, Y.; Tsang, B.K. Regulation of cell death and cell survival gene expression during ovarian follicular development and atresia. Front. Biosci. 2003, 8, d222–d237. [Google Scholar]
- Manabe, N.; Goto, Y.; Matsuda-Minehata, F.; Inoue, N.; Maeda, A.; Sakamaki, K.; Miyano, T. Regulation mechanism of selective atresia in porcine follicles: Regulation of granulosa cell apoptosis during atresia. J. Reprod. Dev. 2004, 50, 493–514. [Google Scholar]
- Wang, X.; Jiang, Y.L.; Wang, Z.L.; Kang, B. Mechanism of programmed cell death in follicular atresia. Sheng Li Xue Bao 2023, 75, 82–90. [Google Scholar]
- Engblom, L.; Lundeheim, N.; Dalin, A.; Andersson, K. Sow removal in Swedish commercial herds. Livest. Sci. 2007, 106, 76–86. [Google Scholar]
- Silva, J.R.; Figueiredo, J.R.; van den Hurk, R. Involvement of growth hormone (GH) and insulin-like growth factor (IGF) system in ovarian folliculogenesis. Theriogenology 2009, 71, 1193–1208. [Google Scholar]
- Pan, Z.; Zhang, J.; Li, Q.; Li, Y.; Shi, F.; Xie, Z.; Liu, H. Current advances in epigenetic modification and alteration during mammalian ovarian folliculogenesis. J. Genet. Genom. 2012, 39, 111–123. [Google Scholar]
- Kim, J.M.; Yoon, Y.D.; Tsang, B.K. Involvement of the Fas/Fas ligand system in p53-mediated granulosa cell apoptosis during follicular development and atresia. Endocrinology 1999, 140, 2307–2317. [Google Scholar]
- Cao, L.; Li, S.; Huang, S.; Shi, D.; Li, X. AQP8 participates in oestrogen-mediated buffalo follicular development by regulating apoptosis of granulosa cells. Reprod. Domest. Anim. 2021, 56, 812–820. [Google Scholar] [PubMed]
- Matsuda, F.; Inoue, N.; Maeda, A.; Cheng, Y.; Sai, T.; Gonda, H.; Goto, Y.; Sakamaki, K.; Manabe, N. Expression and function of apoptosis initiator FOXO3 in granulosa cells during follicular atresia in pig ovaries. J. Reprod. Dev. 2011, 57, 151–158. [Google Scholar]
- Mattei, A.L.; Bailly, N.; Meissner, A. DNA methylation: A historical perspective. Trends Genet. 2022, 38, 676–707. [Google Scholar]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [PubMed]
- Qureshi, M.Z.; Sabitaliyevich, U.Y.; Rabandiyarov, M.; Arystanbekuly, A.T. Role of DNA Methyltransferases (DNMTs) in metastasis. Cell. Mol. Biol. 2022, 68, 226–236. [Google Scholar] [PubMed]
- Lomniczi, A.; Wright, H.; Ojeda, S.R. Epigenetic regulation of female puberty. Front. Neuroendocrinol. 2015, 36, 90–107. [Google Scholar]
- Rehnitz, J.; Youness, B.; Nguyen, X.P.; Dietrich, J.E.; Roesner, S.; Messmer, B.; Strowitzki, T.; Vogt, P.H. FMR1 expression in human granulosa cells and variable ovarian response: Control by epigenetic mechanisms. Mol. Hum. Reprod. 2021, 27, gaab001. [Google Scholar]
- Yang, C.; Ye, J.; Liu, Y.; Ding, J.; Liu, H.; Gao, X.; Li, X.; Zhang, Y.; Zhou, J.; Zhang, X.; et al. Methylation pattern variation between goats and rats during the onset of puberty. Reprod. Domest. Anim. 2018, 53, 793–800. [Google Scholar]
- Yang, C.; Gao, X.; Ye, J.; Ding, J.; Liu, Y.; Liu, H.; Li, X.; Zhang, Y.; Zhou, J.; Huang, W.; et al. The interaction between DNA methylation and long non-coding RNA during the onset of puberty in goats. Reprod. Domest. Anim. 2018, 53, 1287–1297. [Google Scholar]
- Li, S.; Zeng, L.; Miao, F.; Li, N.; Liao, W.; Zhou, X.; Chen, Y.; Quan, H.; He, Y.; Zhang, H.; et al. Knockdown of DNMT1 Induces SLCO3A1 to Promote Follicular Growth by Enhancing the Proliferation of Granulosa Cells in Mammals. Int. J. Mol. Sci. 2024, 25, 2468. [Google Scholar] [CrossRef]
- Yuan, X.; Ye, S.; Chen, Z.; Pan, X.; Huang, S.; Li, Z.; Zhong, Y.; Gao, N.; Zhang, H.; Li, J.; et al. Dynamic DNA methylation of ovaries during pubertal transition in gilts. BMC Genom. 2019, 20, 510. [Google Scholar]
- Zhou, X.; He, Y.; Li, N.; Bai, G.; Pan, X.; Zhang, Z.; Zhang, H.; Li, J.; Yuan, X. DNA methylation mediated RSPO2 to promote follicular development in mammals. Cell Death Dis. 2021, 12, 653. [Google Scholar] [PubMed]
- Zhou, X.; Zhou, X.; He, Y.; Pan, X.; Quan, H.; He, B.; Li, Y.; Bai, G.; Li, N.; Zhang, Z.; et al. DNMT1-mediated lncRNA IFFD controls the follicular development via targeting GLI1 by sponging miR-370. Cell Death Differ. 2023, 30, 576–588. [Google Scholar]
- Chen, W.; Daines, M.O.; Khurana, H.G. Turning off signal transducer and activator of transcription (STAT): The negative regulation of STAT signaling. J. Allergy Clin. Immunol. 2004, 114, 476–489; quiz 490. [Google Scholar]
- Hall, S.E.; Upton, R.M.O.; McLaughlin, E.A.; Sutherland, J.M. Phosphoinositide 3-kinase/protein kinase B (PI3K/AKT) and Janus kinase/signal transducer and activator of transcription (JAK/STAT) follicular signalling is conserved in the mare ovary. Reprod. Fertil. Dev. 2018, 30, 624–633. [Google Scholar]
- Ilha, G.F.; Rovani, M.T.; Gasperin, B.G.; Antoniazzi, A.Q.; Gonçalves, P.B.; Bordignon, V.; Duggavathi, R. Lack of FSH support enhances LIF-STAT3 signaling in granulosa cells of atretic follicles in cattle. Reproduction 2015, 150, 395–403. [Google Scholar]
- Gasperin, B.G.; Rovani, M.T.; Ferreira, R.; Ilha, G.F.; Bordignon, V.; Gonçalves, P.B.; Duggavathi, R. Functional status of STAT3 and MAPK3/1 signaling pathways in granulosa cells during bovine follicular deviation. Theriogenology 2015, 83, 353–359. [Google Scholar]
- Yuan, X.; Zhou, X.; He, Y.; Zhong, Y.; Zhang, A.; Zhang, Z.; Zhang, H.; Li, J. C/EBPbeta Promotes STAT3 Expression and Affects Cell Apoptosis and Proliferation in Porcine Ovarian Granulosa Cells. Genes 2018, 9, 295. [Google Scholar]
- Huang, J.; Ma, G.; Zhu, M.; Pan, J.; Zhang, W.; Zhao, S.H. Molecular characterization of the porcine STAT4 and STAT6 genes. Mol. Biol. Rep. 2012, 39, 6959–6965. [Google Scholar]
- Liu, Z.; Li, C.; Chen, Q.; Bai, C.; Wu, G.; Fu, C.; He, T.; Shen, M.; Feng, C.; Liu, H. Follicular fluid meiosis-activating sterol prevents porcine ovarian granulosa cells from hypoxia-induced apoptosis via inhibiting STAT4 expression. J. Anim. Sci. 2024, 102, skae125. [Google Scholar]
- Jiang, Y.; Xin, X.; Pan, X.; Zhang, A.; Zhang, Z.; Li, J.; Yuan, X. STAT4 targets KISS1 to promote the apoptosis of ovarian granulosa cells. J. Ovarian Res. 2020, 13, 135. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zheng, X.; Zhou, S.; Sun, P.; Liu, M.; Yang, L.; Liang, A. IL-11 regulates the biosynthesis of PGE2 through JAK1/STAT3 signaling pathway in bovine granulosa cells. Theriogenology 2025, 237, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jing, Y.; Qu, X.; Yang, J.; Pan, P.; Liu, X.; Gao, H.; Pei, X.; Zhang, C.; Yang, Y. The Activation of Reticulophagy by ER Stress through the ATF4-MAP1LC3A-CCPG1 Pathway in Ovarian Granulosa Cells Is Linked to Apoptosis and Necroptosis. Int. J. Mol. Sci. 2023, 24, 2749. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Dong, L.P.; Cui, Y.G.; Lu, H.Y. MiR-520h inhibits viability and facilitates apoptosis of KGN cells through modulating IL6R and the JAK/STAT pathway. Reprod. Biol. 2022, 22, 100607. [Google Scholar] [CrossRef]
- Li, N.; Yun, B.; Zeng, L.; Lv, Y.; Zhou, Y.; Fang, M.; Li, S.; Chen, Y.; Huang, E.; Zhang, L.; et al. The antisense lncRNA of TAB2 that prevents oxidative stress to enhance the follicular growth in mammals. Commun. Biol. 2024, 7, 1246. [Google Scholar] [CrossRef]
- Son, W.Y.; Das, M.; Shalom-Paz, E.; Holzer, H. Mechanisms of follicle selection and development. Minerva Ginecol. 2011, 63, 89–102. [Google Scholar]
- Zhang, T.; BaSang, W.D.; Chang, W.; Huo, S.; Ma, X.; Ju, X.; Yu, S.; Cui, S. Dynamics of apoptosis-related gene expression during follicular development in yak. J. Anim. Physiol. Anim. Nutr. 2021, 105, 1002–1013. [Google Scholar]
- Yang, M.Y.; Rajamahendran, R. Involvement of apoptosis in the atresia of nonovulatory dominant follicle during the bovine estrous cycle. Biol. Reprod. 2000, 63, 1313–1321. [Google Scholar]
- Macklon, N.S.; Fauser, B.C. Follicle-stimulating hormone and advanced follicle development in the human. Arch. Med. Res. 2001, 32, 595–600. [Google Scholar] [CrossRef]
- McNeilly, A.S. The ovarian follicle and fertility. J. Steroid Biochem. Mol. Biol. 1991, 40, 29–33. [Google Scholar] [CrossRef]
- Adriaens, I.; Cortvrindt, R.; Smitz, J. Differential FSH exposure in preantral follicle culture has marked effects on folliculogenesis and oocyte developmental competence. Hum. Reprod. 2004, 19, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.P.; Yang, J.L.; Zhang, J.; Li, L.; Huang, L.; Ji, S.Y.; Hu, Z.Y.; Gao, F.; Liu, Y.X. Notch signaling is involved in ovarian follicle development by regulating granulosa cell proliferation. Endocrinology 2011, 152, 2437–2447. [Google Scholar] [CrossRef] [PubMed]
- Jolly, P.D.; Tisdall, D.J.; Heath, D.A.; Lun, S.; McNatty, K.P. Apoptosis in bovine granulosa cells in relation to steroid synthesis, cyclic adenosine 3′,5′-monophosphate response to follicle-stimulating hormone and luteinizing hormone, and follicular atresia. Biol. Reprod. 1994, 51, 934–944. [Google Scholar] [CrossRef] [PubMed]
- Guthrie, H.D.; Grimes, R.W.; Cooper, B.S.; Hammond, J.M. Follicular atresia in pigs: Measurement and physiology. J. Anim. Sci. 1995, 73, 2834–2844. [Google Scholar] [CrossRef]
- Edwards, J.R.; Yarychkivska, O.; Boulard, M.; Bestor, T.H. DNA methylation and DNA methyltransferases. Epigenetics Chromatin 2017, 10, 23. [Google Scholar] [CrossRef]
- Li, E.; Zhang, Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef]
- Bestor, T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000, 9, 2395–2402. [Google Scholar] [CrossRef]
- Schermelleh, L.; Spada, F.; Easwaran, H.P.; Zolghadr, K.; Margot, J.B.; Cardoso, M.C.; Leonhardt, H. Trapped in action: Direct visualization of DNA methyltransferase activity in living cells. Nat. Methods 2005, 2, 751–756. [Google Scholar] [CrossRef]
- Kawai, T.; Richards, J.S.; Shimada, M. The Cell Type-Specific Expression of Lhcgr in Mouse Ovarian Cells: Evidence for a DNA-Demethylation-Dependent Mechanism. Endocrinology 2018, 159, 2062–2074. [Google Scholar] [CrossRef]
- Geng, X.; Zhao, J.; Huang, J.; Li, S.; Chu, W.; Wang, W.-S.; Chen, Z.-J.; Du, Y. lnc-MAP3K13-7:1 Inhibits Ovarian GC Proliferation in PCOS via DNMT1 Downregulation-Mediated CDKN1A Promoter Hypomethylation. Mol. Ther. 2021, 29, 1279–1293. [Google Scholar] [CrossRef]
- Shin, H.J.; Park, H.Y.; Jeong, S.J.; Park, H.W.; Kim, Y.K.; Cho, S.H.; Kim, Y.Y.; Cho, M.L.; Kim, H.Y.; Min, K.U.; et al. STAT4 expression in human T cells is regulated by DNA methylation but not by promoter polymorphism. J. Immunol. 2005, 175, 7143–7150. [Google Scholar] [PubMed]
- Kim, S.W.; Kim, E.S.; Moon, C.M.; Kim, T.I.; Kim, W.H.; Cheon, J.H. Abnormal Genetic and Epigenetic Changes in Signal Transducer and Activator of Transcription 4 in the Pathogenesis of Inflammatory Bowel Diseases. Dig. Dis. Sci. 2012, 57, 2600–2607. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Richards, J.S.; Shimada, M. Large-scale DNA demethylation occurs in proliferating ovarian granulosa cells during mouse follicular development. Commun. Biol. 2021, 4, 1334. [Google Scholar]
- Wieczorek, M.; Ginter, T.; Brand, P.; Heinzel, T.; Krämer, O.H. Acetylation modulates the STAT signaling code. Cytokine Growth Factor Rev. 2012, 23, 293–305. [Google Scholar]
- Yu, Q.; Thieu, V.T.; Kaplan, M.H. Stat4 limits DNA methyltransferase recruitment and DNA methylation of the IL-18Ralpha gene during Th1 differentiation. EMBO J. 2007, 26, 2052–2060. [Google Scholar] [CrossRef]
- Zhang, B.; Nie, X.; Miao, X.; Wang, S.; Li, J.; Wang, S. Development and verification of an immune-related gene pairs prognostic signature in ovarian cancer. J. Cell. Mol. Med. 2021, 25, 2918–2930. [Google Scholar]
- Yan, W.; Wei, J.; Deng, X.; Shi, Z.; Zhu, Z.; Shao, D.; Li, B.; Wang, S.; Tong, G.; Ma, Z. Transcriptional analysis of immune-related gene expression in p53-deficient mice with increased susceptibility to influenza A virus infection. BMC Med. Genom. 2015, 8, 52. [Google Scholar]
- Usman, T.; Ali, N.; Wang, Y.; Yu, Y. Association of Aberrant DNA Methylation Level in the CD4 and JAK-STAT-Pathway-Related Genes with Mastitis Indicator Traits in Chinese Holstein Dairy Cattle. Animals 2021, 12, 65. [Google Scholar] [CrossRef]
- Smith, Z.D.; Hetzel, S.; Meissner, A. DNA methylation in mammalian development and disease. Nat. Rev. Genet. 2025, 26, 7–30. [Google Scholar]
- Zhang, X.; Wang, Y.; Lv, J. STAT4 targets KISS1 to inhibit the oxidative damage, inflammation and neuronal apoptosis in experimental PD models by inactivating the MAPK pathway. Neurochem. Int. 2024, 175, 105683. [Google Scholar]
- Bohlen, T.M.; de Paula, D.G.; Teixeira, P.D.S.; da Silva Mansano, N.; Andrade Alves, G.; Donato, J., Jr.; Frazao, R. Socs3 ablation in kisspeptin cells partially prevents lipopolysaccharide-induced body weight loss. Cytokine 2022, 158, 155999. [Google Scholar] [PubMed]
- Cui, P.; Yang, C.; Zhang, K.; Gao, X.; Luo, L.; Tian, Y.; Song, M.; Liu, Y.; Zhang, Y.; Li, Y.; et al. Effect of estrogen on the expression of GnRH and kisspeptin in the hypothalamus of rats during puberty. Theriogenology 2015, 84, 1556–1564. [Google Scholar] [PubMed]
- Marques, P.; De Sousa Lages, A.; Skorupskaite, K.; Rozario, K.S.; Anderson, R.A.; George, J.T. Physiology of GnRH and Gonadotrophin Secretion; National Library of Medicine: Bethesda, MD, USA, 2000. [Google Scholar]
- Xie, Q.; Kang, Y.; Zhang, C.; Xie, Y.; Wang, C.; Liu, J.; Yu, C.; Zhao, H.; Huang, D. The Role of Kisspeptin in the Control of the Hypothalamic-Pituitary-Gonadal Axis and Reproduction. Front. Endocrinol. 2022, 13, 925206. [Google Scholar]
- Parkening, T.A.; Collins, T.J.; Smith, E.R. Plasma and pituitary concentrations of LH, FSH, and prolactin in aging C57BL/6 mice at various times of the estrous cycle. Neurobiol. Aging 1982, 3, 31–35. [Google Scholar]
- Bergman, M.D.; Karelus, K.; Felicio, L.S.; Nelson, J.F. Age-related alterations in estrogen receptor dynamics are independent of cycling status in middle-aged C57BL/6J mice. J. Steroid Biochem. Mol. Biol. 1991, 38, 127–133. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, D.; Fang, M.; Huang, E.; Quan, H.; Zhang, L.; He, Y.; Zhou, X.; Ma, B.; Yuan, X.; Li, J. DNA Methylation Mediates the Transcription of STAT4 to Regulate KISS1 During Follicular Development. Cells 2025, 14, 523. https://doi.org/10.3390/cells14070523
Chen D, Fang M, Huang E, Quan H, Zhang L, He Y, Zhou X, Ma B, Yuan X, Li J. DNA Methylation Mediates the Transcription of STAT4 to Regulate KISS1 During Follicular Development. Cells. 2025; 14(7):523. https://doi.org/10.3390/cells14070523
Chicago/Turabian StyleChen, Danxia, Ming Fang, Enyuan Huang, Hongyan Quan, Liuhong Zhang, Yingting He, Xiaofeng Zhou, Bin Ma, Xiaolong Yuan, and Jiaqi Li. 2025. "DNA Methylation Mediates the Transcription of STAT4 to Regulate KISS1 During Follicular Development" Cells 14, no. 7: 523. https://doi.org/10.3390/cells14070523
APA StyleChen, D., Fang, M., Huang, E., Quan, H., Zhang, L., He, Y., Zhou, X., Ma, B., Yuan, X., & Li, J. (2025). DNA Methylation Mediates the Transcription of STAT4 to Regulate KISS1 During Follicular Development. Cells, 14(7), 523. https://doi.org/10.3390/cells14070523






