Metabolic Signature of Dietary Iron Overload in a Mouse Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Animal Models
2.3. Blood VAMS
2.4. Blood, Plasma and Liver Metabolite Extraction
2.5. Ultra-High Performance Liquid Chromatography (UHPLC) Combined with MASS Spectrometry (MS)
2.6. Data Analysis
3. Results
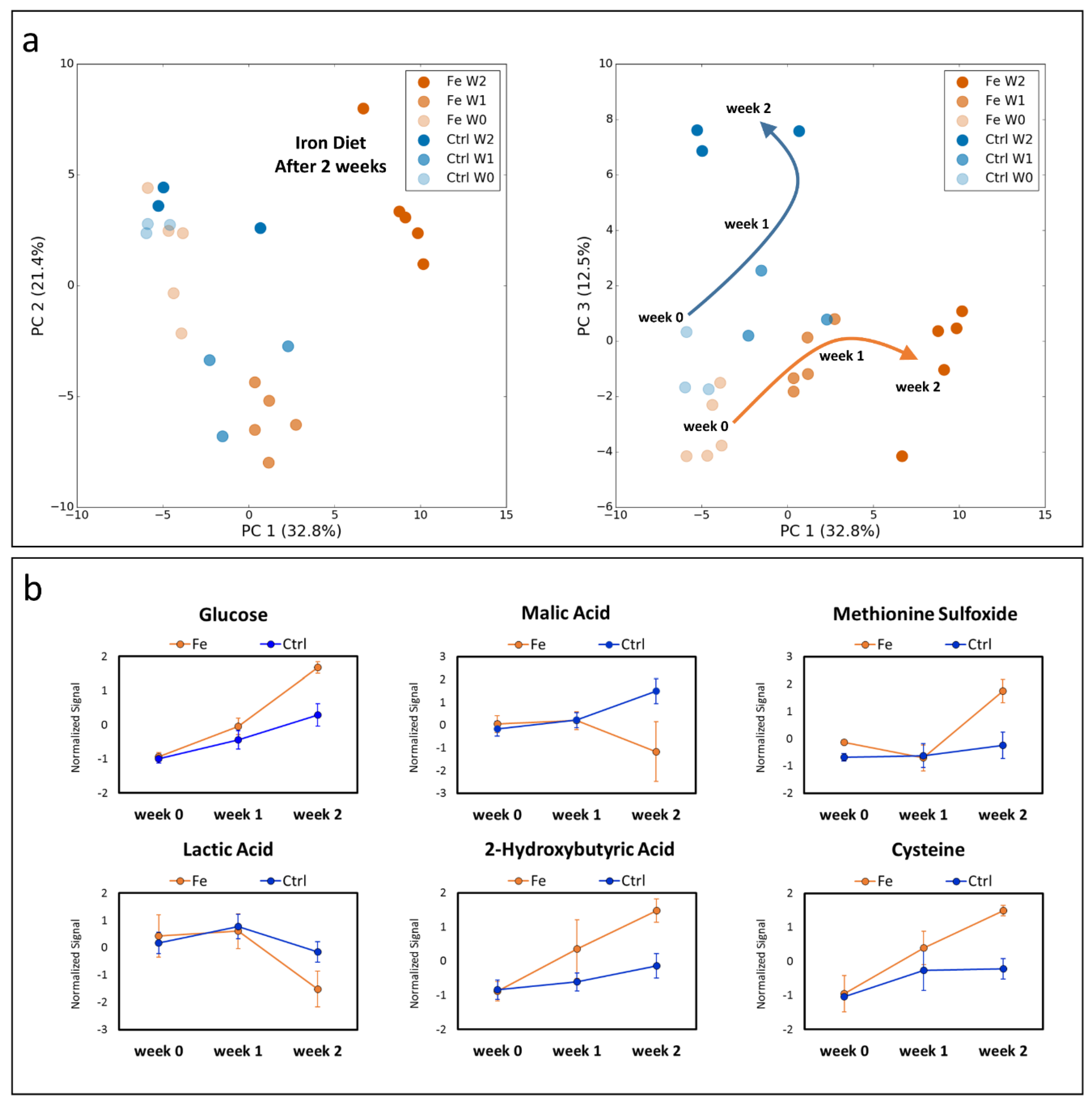
3.1. Dietary Iron Overload Changes the Metabolic Signature over the Time
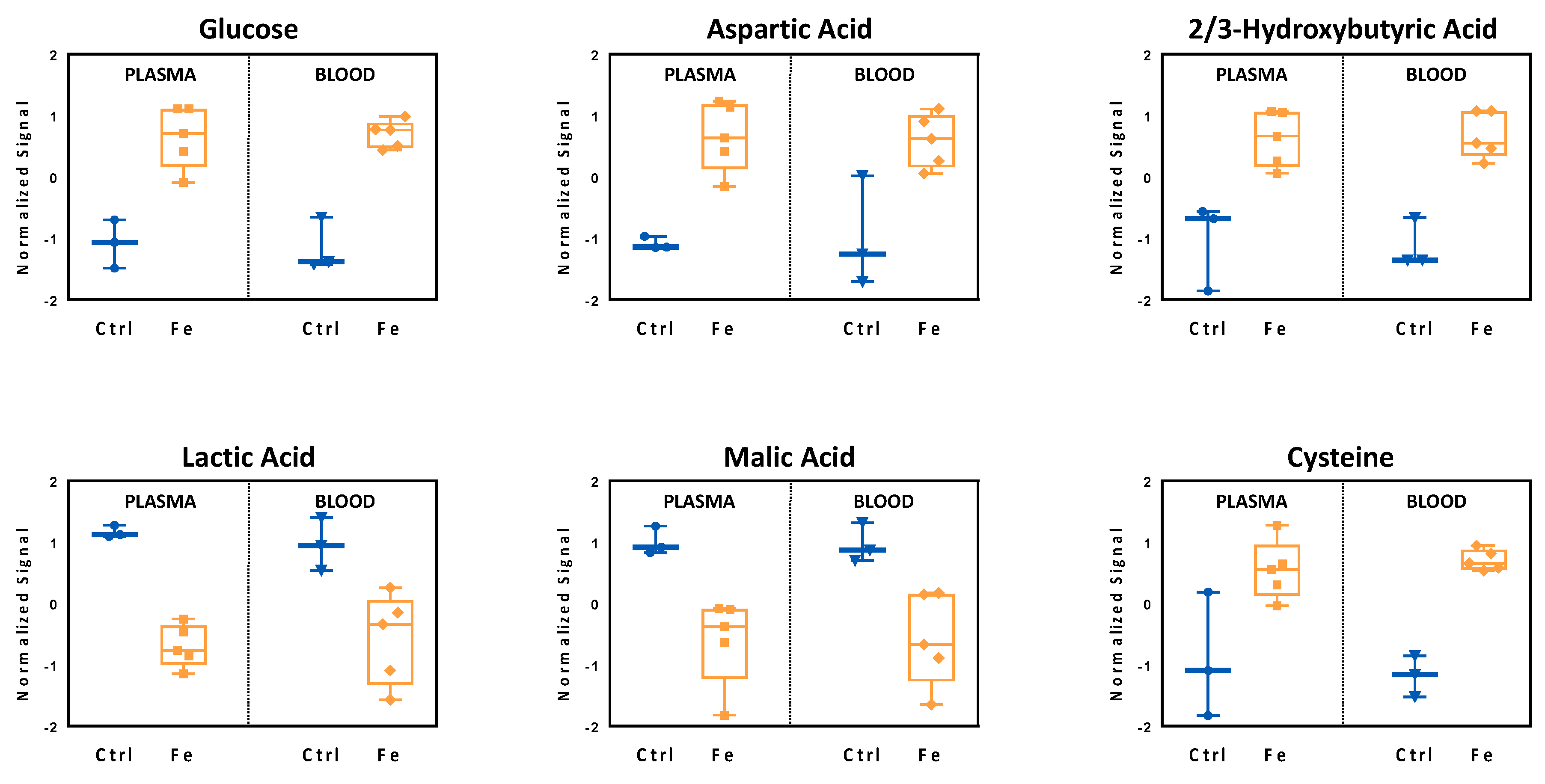
3.2. Validation of the Systemic Metabolic Signature
3.3. Analysis of Liver Metabolome Reflects Iron-Induced Changes Observed in the Circulation
4. Discussion
5. Conclusions
6. Limitations of the Study
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hentze, M.W.; Muckenthaler, M.U.; Galy, B.; Camaschella, C. Two to tango: regulation of mammalian iron metabolism. Cell 2010, 142, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Systemic iron homeostasis. Physiol. Rev. 2013, 93, 1721–1741. [Google Scholar] [CrossRef] [PubMed]
- Rouault, T.A. Mitochondrial iron overload: Causes and consequences. Curr. Opin. Genet. Dev. 2016, 38, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Oexle, H.; Gnaiger, E.; Weiss, G. Iron-dependent changes in cellular energy metabolism: Influence on citric acid cycle and oxidative phosphorylation. Biochim. Biophys. Acta 1999, 1413, 99–107. [Google Scholar] [CrossRef]
- Sheftel, A.D.; Lill, R. The power plant of the cell is also a smithy: The emerging role of mitochondria in cellular iron homeostasis. Ann. Med. 2009, 41, 82–99. [Google Scholar] [CrossRef] [PubMed]
- Lakhal-Littleton, S.; Wolna, M.; Carr, C.A.; Miller, J.J.J.; Christian, H.C.; Ball, V.; Santos, A.; Diaz, R.; Biggs, D.; Stillion, R.; et al. Cardiac ferroportin regulates cellular iron homeostasis and is important for cardiac function. Proc. Natl. Acad. Sci. USA 2015, 112, 3164–3169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koskenkorva-Frank, T.S.; Weiss, G.; Koppenol, W.H.; Burckhardt, S. The complex interplay of iron metabolism, reactive oxygen species, and reactive nitrogen species: Insights into the potential of various iron therapies to induce oxidative and nitrosative stress. Free Radical Biol. Med. 2013, 65, 1174–1194. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G. Genetic mechanisms and modifying factors in hereditary hemochromatosis. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 50–58. [Google Scholar] [CrossRef]
- Volani, C.; Doerrier, C.; Demetz, E.; Haschka, D.; Paglia, G.; Lavdas, A.A.; Gnaiger, E.; Weiss, G. Dietary iron loading negatively affects liver mitochondrial function. Metallomics 2017, 9. [Google Scholar] [CrossRef]
- Allen, K.J.; Gurrin, L.C.; Constantine, C.C.; Osborne, N.J.; Delatycki, M.B.; Nicoll, A.J.; McLaren, C.E.; Bahlo, M.; Nisselle, A.E.; Vulpe, C.D.; et al. Iron-overload–related disease in HFE hereditary hemochromatosis. N. Engl. J. Med. 2008, 358, 221–230. [Google Scholar] [CrossRef]
- Camaschella, C. New insights into iron deficiency and iron deficiency anemia. Blood Rev. 2017, 31, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G. Iron metabolism in the anemia of chronic disease. Biochim. Biophys. Acta 2009, 1790, 682–693. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Evans, K.J.; Sexton, A.E.; Schofield, L.; Creek, D.J. Metabolomics-based elucidation of active metabolic pathways in erythrocytes and HSC-derived reticulocytes. J. Proteome Res. 2017, 16, 1492–1505. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Dunn, W.; Schmidt, M.A.; Gross, S.S.; Kirwan, J.A.; Cascante, M.; Brennan, L.; Wishart, D.S.; Oresic, M.; Hankemeier, T.; et al. Metabolomics Society Initiative Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics 2016, 12. [Google Scholar] [CrossRef]
- Paglia, G.; Stocchero, M.; Cacciatore, S.; Lai, S.; Angel, P.; Alam, M.T.; Keller, M.; Ralser, M.; Astarita, G. Unbiased metabolomic investigation of alzheimer’s disease brain points to dysregulation of mitochondrial aspartate metabolism. J. Proteome Res. 2016, 15, 608–618. [Google Scholar] [CrossRef]
- Volani, C.; Caprioli, G.; Calderisi, G.; Sigurdsson, B.B.; Rainer, J.; Gentilini, I.; Hicks, A.A.; Pramstaller, P.P.; Weiss, G.; Smarason, S.V.; et al. Pre-analytic evaluation of volumetric absorptive microsampling and integration in a mass spectrometry-based metabolomics workflow. Anal. Bioanal.Chem. 2017, 409, 6263–6276. [Google Scholar] [CrossRef] [Green Version]
- Paglia, G.; Del Greco, F.M.; Sigurdsson, B.B.; Rainer, J.; Volani, C.; Hicks, A.A.; Pramstaller, P.P.; Smarason, S.V. Influence of collection tubes during quantitative targeted metabolomics studies in human blood samples. Clin. Chim. Acta 2018, 486, 320–328. [Google Scholar] [CrossRef]
- Guthrie, R.; Susi, A. A simple phenylalanine method for detecting phenylketonuria in large populations of infants. Pediatrics 1963, 32, 338–343. [Google Scholar] [CrossRef]
- Mcdade, T.W.; Williams, S.; Snodgrass, J.J. What a drop can do: Dried blood spots as a minimally invasive method for integrating biomarkers into population-based research. Demography 2007, 44, 899–925. [Google Scholar] [CrossRef]
- Corso, G.; D’Apolito, O.; Gelzo, M.; Paglia, G.; Dello Russo, A. A powerful couple in the future of clinical biochemistry: In situ analysis of dried blood spots by ambient mass spectrometry. Bioanalysis 2010, 2, 1883–1891. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; D’Apolito, O.; Gelzo, M.; Dello Russo, A.; Corso, G. Direct analysis of sterols from dried plasma/blood spots by an atmospheric pressure thermal desorption chemical ionization mass spectrometry (APTDCI-MS) method for a rapid screening of Smith-Lemli-Opitz syndrome. The Analyst 2010, 135, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Corso, G.; Paglia, G.; Garofalo, D.; D’Apolito, O. Neutral loss analysis of amino acids by desorption electrospray ionization using an unmodified tandem quadrupole mass spectrometer. Rapid Commun. Mass Spectrom. 2007, 21, 3777–3784. [Google Scholar] [CrossRef] [PubMed]
- De Kesel, P.M.M.; Lambert, W.E.; Stove, C.P. Does volumetric absorptive microsampling eliminate the hematocrit bias for caffeine and paraxanthine in dried blood samples? A comparative study. Anal. Chim. Acta 2015, 881, 65–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denniff, P.; Spooner, N. Volumetric absorbtive micro Sampling (VAMS): A novel dried sample collection technique for quantitative bioanalysis. Anal. Chem. 2014, 86, 8489–8495. [Google Scholar] [CrossRef] [PubMed]
- Tannenbaum, J.; Bennett, B.T. Russell and burch’s 3Rs then and now: The need for clarity in definition and purpose. J. Am. Assoc. Lab. Anim. Sci. 2015, 54, 120–132. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The human metabolome database in 2013. Nucleic Acids Res. 2012, 41, D801–D807. [Google Scholar] [CrossRef]
- Zhu, Z.-J.; Schultz, A.W.; Wang, J.; Johnson, C.H.; Yannone, S.M.; Patti, G.J.; Siuzdak, G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 2013, 8, 451–460. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Wishart, D.S. Using Metaboanalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinf. 2016, 55. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Sigurjõnsson, Ó.E.; Rolfsson, Ó.; Valgeirsdottir, S.; Hansen, M.B.; Brynjõlfsson, S.; Gudmundsson, S.; Palsson, B.O. Comprehensive metabolomic study of platelets reveals the expression of discrete metabolic phenotypes during storage. Transfusion 2014, 54, 2911–2923. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; D’Alessandro, A.; Rolfsson, Ó.; Sigurjónsson, Ó.E.; Bordbar, A.; Palsson, S.; Nemkov, T.; Hansen, K.C.; Gudmundsson, S.; Palsson, B.O. Biomarkers defining the metabolic age of red blood cells during cold storage. Blood 2016, 128, e43–e50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paglia, G.; Sigurjónsson, Ó.E.; Rolfsson, Ó.; Hansen, M.B.; Brynjólfsson, S.; Gudmundsson, S.; Palsson, B.O. Metabolomic analysis of platelets during storage: A comparison between apheresis- and buffy coat-derived platelet concentrates. Transfusion 2015, 55, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Gall, W.E.; Beebe, K.; Lawton, K.A.; Adam, K.-P.; Mitchell, M.W.; Nakhle, P.J.; Ryals, J.A.; Milburn, M.V.; Nannipieri, M.; Camastra, S.; et al. Alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PloS ONE 2010, 5, e10883. [Google Scholar] [CrossRef] [PubMed]
- Stechemesser, L.; Eder, S.K.; Wagner, A.; Patsch, W.; Feldman, A.; Strasser, M.; Auer, S.; Niederseer, D.; Huber-Schönauer, U.; Paulweber, B.; et al. Metabolomic profiling identifies potential pathways involved in the interaction of iron homeostasis with glucose metabolism. Mol. Metab. 2016, 6, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Alexeev, E.E.; He, X.; Slupsky, C.M.; Lönnerdal, B. Effects of iron supplementation on growth, gut microbiota, metabolomics and cognitive development of rat pups. PloS ONE 2017, 12, e0179713. [Google Scholar] [CrossRef]
- Huang, J.; Jones, D.; Luo, B.; Sanderson, M.; Soto, J.; Abel, E.D.; Cooksey, R.C.; Mcclain, D.A. Iron overload and diabetes risk : A shift rrom glucose. Diabetes 2011, 60. [Google Scholar] [CrossRef]
- Nielsen, T.T.; Stottrup, N.B.; Lofgren, B.; Botker, H.E. Metabolic fingerprint of ischaemic cardioprotection: Importance of the malate-aspartate shuttle. Cardiovasc. Res. 2011, 91, 382–391. [Google Scholar] [CrossRef]
- Bene, J.; Hadzsiev, K.; Melegh, B. Role of carnitine and its derivatives in the development and management of type 2 diabetes. Nutr. Diabetes 2018, 1–10. [Google Scholar] [CrossRef]
- Brissot, P.; Pietrangelo, A.; Adams, P.C.; De Graaff, B.; McLaren, C.E.; Loreál, O. Haemochromatosis. Nat. Rev. Dis. Primers 2018, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sohn, Y.S.; Mitterstiller, A.M.; Breuer, W.; Weiss, G.; Cabantchik, Z.I. Rescuing iron-overloaded macrophages by conservative relocation of the accumulated metal. Br. J. Pharmacol. 2011, 164, 406–418. [Google Scholar] [CrossRef] [PubMed]
- Dongiovanni, P.; Ruscica, M.; Rametta, R.; Recalcati, S.; Steffani, L.; Gatti, S.; Girelli, D.; Cairo, G.; Magni, P.; Fargion, S.; et al. Dietary iron overload induces visceral adipose tissue insulin resistance. Am. J. Pathol. 2013, 182, 2254–2263. [Google Scholar] [CrossRef] [PubMed]
- Aigner, E.; Theurl, I.; Theurl, M.; Lederer, D.; Haufe, H.; Dietze, O.; Strasser, M.; Datz, C.; Weiss, G. Pathways underlying iron accumulation in human nonalcoholic fatty liver disease. Am. J. Clin. Nutr. 2008, 87, 1374–1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lord, R.S.; Bralley, J.A. Clinical applications of urinary organic acids. Part 1 : Detoxification markers. Altern. Med. Rev. 2008, 13, 205–215. [Google Scholar]
- Bonfils, L.; Ellervik, C.; Friedrich, N.; Linneberg, A.; Sandholt, C.H.; Jørgensen, M.E.; Jørgensen, T.; Hansen, T.; Pedersen, O.; Allin, K.H. Fasting serum levels of ferritin are associated with impaired pancreatic beta cell function and decreased insulin sensitivity: A population-based study. Diabetologia 2014, 58, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Raffield, L.M.; Louie, T.; Sofer, T.; Jain, D.; Ipp, E.; Taylor, K.D.; Papanicolaou, G.J.; Avilés-Santa, L.; Lange, L.A.; Laurie, C.C.; et al. Genome-wide association study of iron traits and relation to diabetes in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL): Potential genomic intersection of iron and glucose regulation? Hum. Mol. Genet. 2017, 26, 1966–1978. [Google Scholar] [CrossRef] [PubMed]
- Gelaye, B.; Clish, C.B.; Denis, M.; Larrabure, G.; Tadesse, M.G.; Deik, A.; Pierce, K.; Bullock, K.; Dennis, C.; Enquobahrie, D.A.; et al. Metabolomics signatures associated with an oral glucose challenge in pregnant women. Diabetes Metab. 2018. [Google Scholar] [CrossRef]
- Flanagan, J.L.; Simmons, P.A.; Vehige, J.; Willcox, M.D.; Garrett, Q. Role of carnitine in disease. Nutr. Metab. 2010, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- Ringseis, R.; Keller, J.; Eder, K. Role of carnitine in the regulation of glucose homeostasis and insulin sensitivity: Evidence from in vivo and in vitro studies with carnitine supplementation and carnitine deficiency. Eur. J. Nutr. 2012, 51, 1–18. [Google Scholar] [CrossRef]


| Metabolite | HMDB | Fold Change | p Value | Trend in Fe |
|---|---|---|---|---|
| AMP | HMDB00045 | 3.9 | 7.9 × 10−6 | ↑ |
| ADP | HMDB01341 | 2.9 | 8.1 × 10−4 | ↑ |
| 2/3-Hydroxybutyric acid | HMDB0000008/HMDB00357 | 2.5 | 7.0 × 10−4 | ↑ |
| Glucose | HMDB00122 | 2.5 | 1.8 × 10−4 | ↑ |
| N1-Methyl-2-pyridone-5-carboxamide | HMDB0004193 | 2.4 | 1.3 × 10−3 | ↑ |
| Gluconic acid | HMDB00625 | 2.3 | 3.0 × 10−3 | ↑ |
| l-Aspartyl-4-phosphate | HMDB0012250 | 2.3 | 1.2 × 10−2 | ↑ |
| Hydroxyproline | HMDB00725 | 2.3 | 6.7 × 10−3 | ↑ |
| Methionine sulfoxide | HMDB0002005 | 2.1 | 8.6 × 10−4 | ↑ |
| l-Aspartic acid | HMDB00191 | 2.1 | 1.4 × 10−2 | ↑ |
| γ-Glutamylcysteine | HMDB0001049 | 2.0 | 3.4 × 10−3 | ↑ |
| l-Cysteine | HMDB00574 | 1.9 | 3.6 × 10−5 | ↑ |
| Hydroxyisocaproic acid | HMDB0000746 | 1,8 | 8.0 × 10−3 | ↑ |
| Pyroglutamic acid | HMDB0000267 | 1.7 | 2.6 × 10−3 | ↑ |
| ADP-Glucose | HMDB06557 | 1.7 | 1.7 × 10−2 | ↑ |
| Adenosine | HMDB00050 | 1.7 | 2.4 × 10−2 | ↑ |
| 2-Phosphoglyceric acid | HMDB00362 | 1.6 | 1.9 × 10−5 | ↑ |
| Phosphorylethanolamine | HMDB00224 | 1.5 | 3.8 × 10−4 | ↑ |
| GMP | HMDB01397 | 1.5 | 2.5 × 10−2 | ↑ |
| UDP-glucose | HMDB00286 | 1.5 | 5.3 × 10−5 | ↑ |
| UDP-N-acetylglucosamine | HMDB0000290 | 1.5 | 1.1 × 10−4 | ↑ |
| Glucose 6-phosphate | HMDB01401 | 1.5 | 2.3 × 10−4 | ↑ |
| Acetylmethionine | HMDB11745 | 1.5 | 2.3 × 10−3 | ↑ |
| l-Threonine | HMDB00167 | 1.5 | 1.7 × 10−2 | ↑ |
| CDP-ethanolamine | HMDB01564 | 1.5 | 3.7 × 10−3 | ↑ |
| N-Acetyl-beta-alanine | HMDB0061880 | 1.4 | 2.4 × 10−3 | ↑ |
| Glyceric acid | HMDB00139 | 1.4 | 4.6 × 10−2 | ↑ |
| Phosphoenolpyruvic acid | HMDB00263 | 1.4 | 2.9 × 10−3 | ↑ |
| 2-Octenedioic acid | HMDB0000341 | 1.4 | 5.0 × 10−2 | ↑ |
| Pyruvic acid | HMDB00243 | 1.4 | 1.2 × 10−2 | ↑ |
| l-Lysine | HMDB00182 | 1.4 | 8.5 × 10−5 | ↑ |
| Pipecolic acid | HMDB00070 | 1.4 | 8.1 × 10−5 | ↑ |
| PABA | HMDB01392 | 1.4 | 2.8 × 10−2 | ↑ |
| l-Proline | HMDB00162 | 1.3 | 4.3 × 10−2 | ↑ |
| Glycolic acid | HMDB00115 | 1.3 | 3.2 × 10−2 | ↑ |
| Glutaric acid | HMDB00661 | 1.3 | 3.5 × 10−3 | ↑ |
| l-Glutamic acid | HMDB00148 | 1.3 | 1.2 × 10−2 | ↑ |
| l-Asparagine | HMDB00168 | 1.2 | 2.9 × 10−2 | ↑ |
| Creatinine | HMDB00562 | 1.2 | 1.7 × 10−2 | ↑ |
| Urea | HMDB0000294 | 1.2 | 2.3 × 10−3 | ↑ |
| ADMA/SDMA | HMDB0001539/HMDB0003334 | 1.2 | 3.3 × 10−2 | ↑ |
| Choline | HMDB00097 | 1.1 | 1.3 × 10−3 | ↑ |
| N6,N6,N6-Trimethyl-L-lysine | HMDB0001325 | 1.1 | 2.7 × 10−2 | ↑ |
| IMP | HMDB00175 | 0.3 | 1.1 × 10−2 | ↓ |
| N4-Acetylcytidine | HMDB0005923 | 0.4 | 4.5 × 10−2 | ↓ |
| l-Acetylcarnitine | HMDB00201 | 0.4 | 5.0 × 10−6 | ↓ |
| Stearoylcarnitine | HMDB0000848 | 0.5 | 1.8 × 10−2 | ↓ |
| α-ketoisovaleric acid | HMDB00019 | 0.5 | 9.9 × 10−3 | ↓ |
| Propionylcarnitine | HMDB0000824 | 0.7 | 3.5 × 10−2 | ↓ |
| Malic acid | HMDB00156 | 0.7 | 1.8 × 10−2 | ↓ |
| Ornithine | HMDB00214 | 0.7 | 4.4 × 10−2 | ↓ |
| (Iso)leucine | HMDB00172 | 0.7 | 2.6 × 10−2 | ↓ |
| Decanoylcarnitine | HMDB0000651 | 0.7 | 2.4 × 10−2 | ↓ |
| l-Arginine | HMDB00517 | 0.8 | 4.3 × 10−3 | ↓ |
| l-Tryptophan | HMDB00929 | 0,8 | 9.5 × 10−3 | ↓ |
| Lactic acid | HMDB00190 | 0.8 | 1.8 × 10−2 | ↓ |
| l-Palmitoylcarnitine | HMDB0000222 | 0.8 | 2.2 × 10−2 | ↓ |
| Pyrrolidonecarboxylic acid | HMDB0000805 | 0.9 | 4.4 × 10−2 | ↓ |
| Metabolite | HMDB | Fold Change | p Value | Trend in Fe |
|---|---|---|---|---|
| l-Aspartic acid | HMDB00191 | 3.4 | 2.2 × 10−3 | ↑ |
| l-Cysteine | HMDB00574 | 2.2 | 3.0 × 10−2 | ↑ |
| N1-Methyl-2-pyridone-5-carboxamide | HMDB0004193 | 2.1 | 1.2 × 10−2 | ↑ |
| Hydroxyproline | HMDB00725 | 2.0 | 6.1 × 10−3 | ↑ |
| 2-Octenedioic acid | HMDB0000341 | 2.0 | 3.3 × 10−2 | ↑ |
| Glucose | HMDB00122 | 1.9 | 2.3 × 10−3 | ↑ |
| Hydroxyisocaproic acid | HMDB0000746 | 1.8 | 1.3 × 10−3 | ↑ |
| Glutathione | HMDB0000125 | 1.7 | 2.7 × 10−2 | ↑ |
| 2/3-Hydroxybutyric acid | HMDB0000008/HMDB00357 | 1.7 | 6.5 × 10−3 | ↑ |
| Pipecolic acid | HMDB00070 | 1.4 | 1.3 × 10−2 | ↑ |
| l-Lysine | HMDB00182 | 1.4 | 8.2 × 10−3 | ↑ |
| γ-Glutamylcysteine | HMDB0001049 | 1.4 | 4.0 × 10−4 | ↑ |
| Choline | HMDB00097 | 1.4 | 1.0 × 10−2 | ↑ |
| 1-Methylhistidine | HMDB00001 | 1.3 | 3.2 × 10−2 | ↑ |
| Adenosine | HMDB00050 | 1.3 | 8.1 × 10−3 | ↑ |
| Acetylcarnosine | HMDB0012881 | 1.3 | 4.8 × 10−2 | ↑ |
| l-Serine | HMDB00187 | 1.2 | 4.0 × 10−2 | ↑ |
| l-Histidine | HMDB00177 | 1.2 | 1.1 × 10−2 | ↑ |
| l-Acetylcarnitine | HMDB00201 | 0.3 | 8.0 × 10−5 | ↓ |
| Propionylcarnitine | HMDB0000824 | 0.3 | 1.8 × 10−3 | ↓ |
| Malic acid | HMDB00156 | 0.3 | 1.1 × 10−2 | ↓ |
| IMP | HMDB00175 | 0.4 | 5.7 × 10−3 | ↓ |
| Gamma-linolenyl carnitine | HMDB0006318 | 0.5 | 1.6 × 10−2 | ↓ |
| Lactic acid | HMDB00190 | 0.7 | 1.2 × 10−4 | ↓ |
| Metabolite | HMDB | Fold Change | p Value | Trend in Fe |
|---|---|---|---|---|
| Ascorbic Acid | HMDB00044 | 2.0 | 0.0208 | ↑ |
| ADP-Glucose | HMDB06557 | 1.7 | 0.0169 | ↑ |
| Aconitic acid | HMDB0000072 | 1.7 | 0.0206 | ↑ |
| γ-Glutamylcysteine | HMDB0001049 | 1.6 | 0.0010 | ↑ |
| Ureidopropionic acid | HMDB00026 | 1.6 | 0.0159 | ↑ |
| 2/3-Hydroxybutyric acid | HMDB0000008/HMDB00357 | 1.6 | 0.0050 | ↑ |
| Folic acid | HMDB00121 | 1.3 | 0.0402 | ↑ |
| Glucosamine | HMDB01514 | 1.3 | 0.0164 | ↑ |
| Hydroxyproline | HMDB00725 | 1.2 | 0.0138 | ↑ |
| Glutathione | HMDB0000125 | 1.2 | 0.0000 | ↑ |
| Taurine | HMDB00251 | 1.1 | 0.0140 | ↑ |
| CDP-ethanolamine | HMDB01564 | 0.5 | 0.0350 | ↓ |
| Propionylcarnitine | HMDB0000824 | 0.5 | 0.0015 | ↓ |
| l-Aspartyl-4-phosphate | HMDB0012250 | 0.5 | 0.0086 | ↓ |
| l-Serine | HMDB00187 | 0.5 | 0.0005 | ↓ |
| Acetylhistidine | HMDB32055 | 0.5 | 0.0030 | ↓ |
| Xanthosine | HMDB00299 | 0.5 | 0.0030 | ↓ |
| l-Arginine | HMDB00517 | 0.5 | 0.0000 | ↓ |
| N6-Methyllysine | HMDB0002038 | 0.5 | 0.0000 | ↓ |
| l-Valine | HMDB00883 | 0.6 | 0.0050 | ↓ |
| Hypoxanthine | HMDB00157 | 0.6 | 0.0492 | ↓ |
| l-Proline | HMDB00162 | 0.6 | 0.0252 | ↓ |
| Carnosine | HMDB00033 | 0.6 | 0.0003 | ↓ |
| Creatinine | HMDB00562 | 0.6 | 0.0400 | ↓ |
| IMP | HMDB00175 | 0.6 | 0.0024 | ↓ |
| Allantoin | HMDB00462 | 0.6 | 0.0221 | ↓ |
| Glycerol-3-Phosphate | HMDB00126 | 0.6 | 0.0043 | ↓ |
| UDP-glucose | HMDB00286 | 0.6 | 0.0309 | ↓ |
| Guanine | HMDB00132 | 0.6 | 0.0425 | ↓ |
| Citrulline | HMDB00904 | 0.6 | 0.0007 | ↓ |
| l-Carnitine | HMDB00062 | 0.6 | 0.0005 | ↓ |
| Choline | HMDB00097 | 0.6 | 0.0183 | ↓ |
| SAH | HMDB00939 | 0.6 | 0.0001 | ↓ |
| Inosine | HMDB00195 | 0.6 | 0.0434 | ↓ |
| Uric acid | HMDB00289 | 0.7 | 0.0470 | ↓ |
| Creatine | HMDB00064 | 0.7 | 0.0462 | ↓ |
| l-Acetylcarnitine | HMDB00201 | 0.7 | 0.0292 | ↓ |
| l-Asparagine | HMDB00168 | 0.7 | 0.0104 | ↓ |
| Glycerylphosphorylethanolamine | HMDB0000114 | 0.7 | 0.0037 | ↓ |
| (Iso)leucine | HMDB00172 | 0.7 | 0.0018 | ↓ |
| Ornithine | HMDB00214 | 0.7 | 0.0007 | ↓ |
| Pentose-Phosphate | HMDB0000098/HMDB0001489/HMDB0001548 | 0.7 | 0.0297 | ↓ |
| Glycine | HMDB00123 | 0.7 | 0.0018 | ↓ |
| l-Phenylalanine | HMDB00159 | 0.8 | 0.0017 | ↓ |
| l-Threonine | HMDB00167 | 0.8 | 0.0023 | ↓ |
| l-Glutamine | HMDB00641 | 0.9 | 0.0398 | ↓ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volani, C.; Paglia, G.; Smarason, S.V.; Pramstaller, P.P.; Demetz, E.; Pfeifhofer-Obermair, C.; Weiss, G. Metabolic Signature of Dietary Iron Overload in a Mouse Model. Cells 2018, 7, 264. https://doi.org/10.3390/cells7120264
Volani C, Paglia G, Smarason SV, Pramstaller PP, Demetz E, Pfeifhofer-Obermair C, Weiss G. Metabolic Signature of Dietary Iron Overload in a Mouse Model. Cells. 2018; 7(12):264. https://doi.org/10.3390/cells7120264
Chicago/Turabian StyleVolani, Chiara, Giuseppe Paglia, Sigurdur V. Smarason, Peter P. Pramstaller, Egon Demetz, Christa Pfeifhofer-Obermair, and Guenter Weiss. 2018. "Metabolic Signature of Dietary Iron Overload in a Mouse Model" Cells 7, no. 12: 264. https://doi.org/10.3390/cells7120264






