Identification of Novel Hemangioblast Genes in the Early Chick Embryo
Abstract
:1. Introduction
2. Materials and Methods
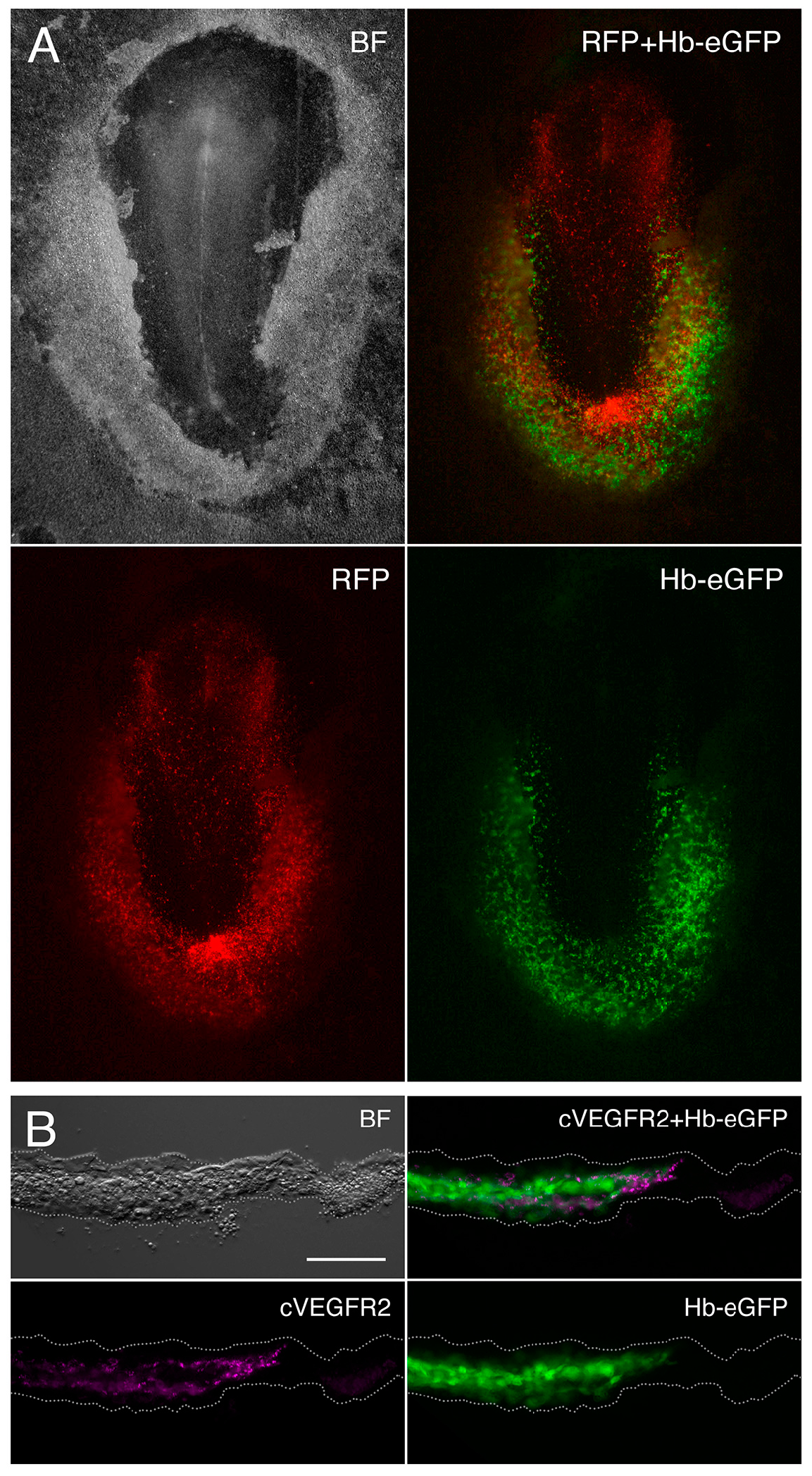
2.1. Embryo Ex Ovo Electroporation
2.2. Immunohistochemistry
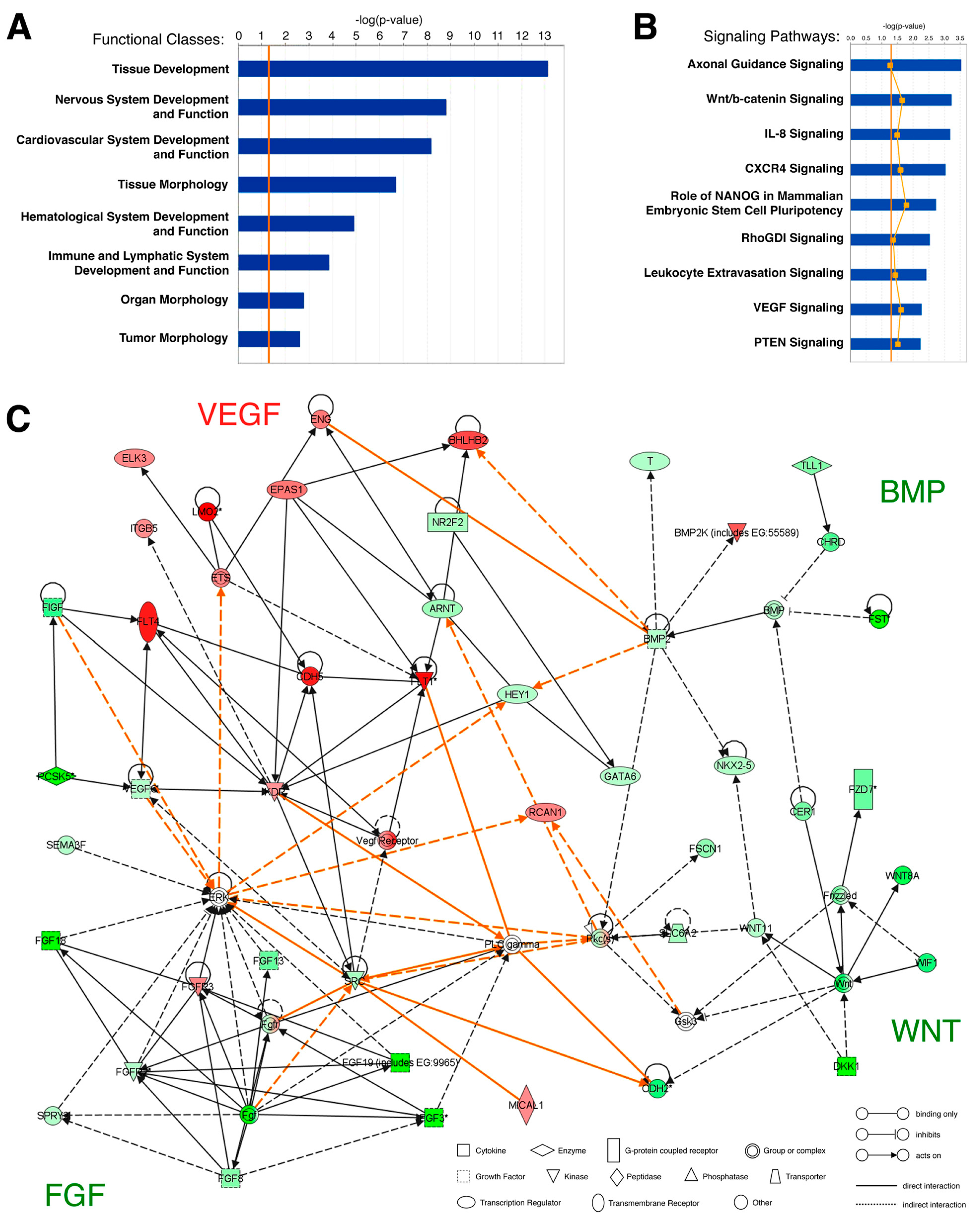
2.3. Microarray Data Analysis
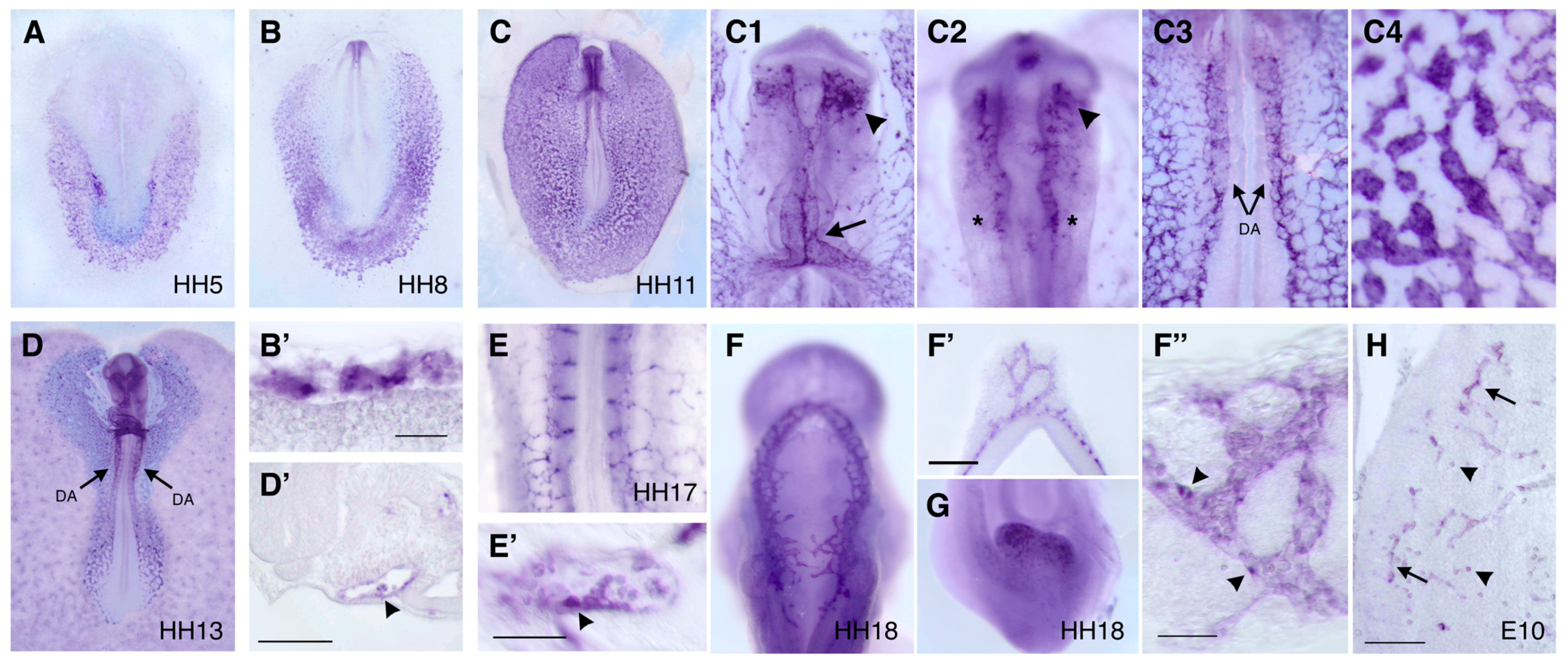
2.4. In Situ Hybridization
3. Results and Discussion
3.1. Gene Expression Analysis of the Hemangioblast Transcriptome
3.2. Identification of Novel Hemangioblast Genes
3.3. Expression Pattern of DIA1R in the Chick Embryo
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sabin, F.R. Studies on the origin of blood vessels and of red corpuscles as seen in the living blastoderm of the chick during the second day of incubation. Contrib. Embryol. 1920, 9, 213–262. [Google Scholar]
- Murray, P.D.F. The development in vitro of the blood of the early chick embryo. Proc. R. Soc. Lond. B 1932, 111, 497–521. [Google Scholar] [CrossRef]
- Gritz, E.; Hirschi, K.K. Specification and function of hemogenic endothelium during embryogenesis. Cell. Mol. Life Sci. 2016, 73, 1547–1567. [Google Scholar] [CrossRef] [PubMed]
- Lacaud, G.; Kouskoff, V. Hemangioblast, hemogenic endothelium, and primitive versus definitive hematopoiesis. Exp. Hematol. 2017, 49, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Palis, J.; Robertson, S.; Kennedy, M.; Wall, C.; Keller, G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 1999, 126, 5073–5084. [Google Scholar] [PubMed]
- Tober, J.; Koniski, A.; McGrath, K.E.; Vemishetti, R.; Emerson, R.; de Mesy-Bentley, K.K.; Waugh, R.; Palis, J. The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood 2007, 109, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Guilliams, M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 2016, 44, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Tay, T.L.; Hagemeyer, N.; Prinz, M. The force awakens: Insights into the origin and formation of microglia. Curr. Opin. Neurobiol. 2016, 39, 30–37. [Google Scholar] [CrossRef] [PubMed]
- De Bruijn, M.F.; Speck, N.A.; Peeters, M.C.; Dzierzak, E. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 2000, 19, 2465–2474. [Google Scholar] [CrossRef] [PubMed]
- Samokhvalov, I.M.; Samokhvalova, N.I.; Nishikawa, S. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature 2007, 446, 1056–1061. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sanchez, V.; Takata, N.; Yokomizo, T.; Yamanaka, Y.; Kataoka, H.; Hoppe, P.S.; Schroeder, T.; Nishikawa, S. Circulation-independent differentiation pathway from extraembryonic mesoderm toward hematopoietic stem cells via hemogenic angioblasts. Cell Rep. 2014, 8, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Patterson, L.J.; Gering, M.; Eckfeldt, C.E.; Green, A.R.; Verfaillie, C.M.; Ekker, S.C.; Patient, R. The transcription factors Scl and Lmo2 act together during development of the hemangioblast in zebrafish. Blood 2007, 109, 2389–2398. [Google Scholar] [CrossRef] [PubMed]
- Stanulovic, V.S.; Cauchy, P.; Assi, S.A.; Hoogenkamp, M. LMO2 is required for TAL1 DNA binding activity and initiation of definitive haematopoiesis at the haemangioblast stage. Nucleic Acids Res. 2017, 45, 9874–9888. [Google Scholar] [CrossRef] [PubMed]
- Gering, M.; Rodaway, A.R.; Göttgens, B.; Patient, R.K.; Green, A.R. The SCL gene specifies haemangioblast development from early mesoderm. EMBO J. 1998, 17, 4029–4045. [Google Scholar] [CrossRef] [PubMed]
- Lacaud, G.; Gore, L.; Kennedy, M.; Kouskoff, V.; Kingsley, P.; Hogan, C.; Carlsson, L.; Speck, N.; Palis, J.; Keller, G. Runx1 is essential for hematopoietic commitment at the hemangioblast stage of development in vitro. Blood 2002, 100, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Lilly, A.J.; Costa, G.; Largeot, A.; Fadlullah, M.Z.; Lie-A-Ling, M.; Lacaud, G.; Kouskoff, V. Interplay between SOX7 and RUNX1 regulates hemogenic endothelial fate in the yolk sac. Development 2016, 143, 4341–4351. [Google Scholar] [CrossRef] [PubMed]
- Cannon, J.E.; Place, E.S.; Eve, A.M.; Bradshaw, C.R.; Sesay, A.; Morrell, N.W.; Smith, J.C. Global analysis of the haematopoietic and endothelial transcriptome during zebrafish development. Mech. Dev. 2013, 130, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Nasrallah, R.; Fast, E.M.; Solaimani, P.; Knezevic, K.; Eliades, A.; Patel, R.; Thambyrajah, R.; Unnikrishnan, A.; Thoms, J.; Beck, D.; et al. Identification of novel regulators of developmental hematopoiesis using Endoglin regulatory elements as molecular probes. Blood 2016, 128, 1928–1939. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, V.; Arede, N.; Gardner, R.; Rodríguez-León, J.; Tavares, A.T. Targeting the hemangioblast with a novel cell type-specific enhancer. BMC Dev. Biol. 2011, 11, 76. [Google Scholar] [CrossRef] [PubMed]
- Gordon-Keylock, S.; Medvinsky, A. Endothelio-hematopoietic relationship: Getting closer to the beginnings. BMC Biol. 2011, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Hamburger, V.; Hamilton, H.L. A series of normal stages in the development of the chick embryo. J. Morphol. 1951, 88, 49–92. [Google Scholar] [CrossRef] [PubMed]
- New, D.A.T. A New Technique for the Cultivation of the Chick Embryo in vitro. Development 1955, 3, 326–331. [Google Scholar]
- Eichmann, A.; Corbel, C.; Nataf, V.; Vaigot, P.; Bréant, C.; Le Douarin, N.M. Ligand-dependent development of the endothelial and hemopoietic lineages from embryonic mesodermal cells expressing vascular endothelial growth factor receptor 2. Proc. Natl. Acad. Sci. USA 1997, 94, 5141–5146. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.G. Whole mount in situ hybridisation of vertebrate embryos. In In Situ Hybridization; Wilkinson, D.G., Ed.; Oxford University Press: Oxford, UK, 1992; pp. 75–83. [Google Scholar]
- Braissant, O.; Wahli, W. A simplified in situ hybridization protocol using non-radioactively labelled probes to detect abundant and rare mRNAs on tissue sections. Biochemica 1998, 1, 10–16. [Google Scholar]
- Tavares, A.T.; Andrade, S.; Silva, A.C.; Belo, J.A. Cerberus is a feedback inhibitor of Nodal asymmetric signaling in the chick embryo. Development 2007, 134, 2051–2060. [Google Scholar] [CrossRef] [PubMed]
- Minko, K.; Bollerot, K.; Drevon, C.; Hallais, M.F.; Jaffredo, T. From mesoderm to blood islands: Patterns of key molecules during yolk sac erythropoiesis. Gene Expr. Patterns 2003, 3, 261–272. [Google Scholar] [CrossRef]
- Olsson, A.K.; Dimberg, A.; Kreuger, J.; Claesson-Welsh, L. VEGF receptor signaling—In control of vascular function. Nat. Rev. Mol. Cell Biol. 2006, 7, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Afrin, F.; Satija, N.; Tripathi, R.P.; Gangenahalli, G.U. Stromal-derived factor-1/CXCR4 signaling: Indispensable role in homing and engraftment of hematopoietic stem cells in bone marrow. Stem Cells Dev. 2011, 20, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Tsuji-Tamura, K.; Ogawa, M. CXCR4 Signaling Negatively Modulates the Bipotential State of Hemogenic Endothelial Cells Derived from Embryonic Stem Cells by Attenuating the Endothelial Potential. Stem Cells 2016, 34, 2814–2824. [Google Scholar] [CrossRef] [PubMed]
- Carmeliet, P.; Tessier-Lavigne, M. Common mechanisms of nerve and blood vessel wiring. Nature 2005, 436, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, O.; Sánchez-Madrid, F. Molecular Basis of Leukocyte–Endothelium Interactions during the Inflammatory Response. Rev. Española Cardiol. 2009, 62, 552–562. [Google Scholar] [CrossRef]
- Costa, G.; Mazan, A.; Gandillet, A.; Pearson, S.; Lacaud, G.; Kouskoff, V. SOX7 regulates the expression of VE-cadherin in the haemogenic endothelium at the onset of haematopoietic development. Development 2012, 139, 1587–1598. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Warren, A.J.; Dobson, C.; Forster, A.; Pannell, R.; Rabbitts, T.H. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc. Natl. Acad. Sci. USA 1998, 95, 3890–3895. [Google Scholar] [CrossRef] [PubMed]
- Connolly, D.J.; Patel, K.; Seleiro, E.A.; Wilkinson, D.G.; Cooke, J. Cloning, sequencing, and expressional analysis of the chick homologue of follistatin. Dev. Genet. 1995, 17, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Kurose, H.; Bito, T.; Adachi, T.; Shimizu, M.; Noji, S.; Ohuchi, H. Expression of Fibroblast growth factor 19 (Fgf19) during chicken embryogenesis and eye development, compared with Fgf15 expression in the mouse. Gene Expr. Patterns 2004, 4, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Chapman, S.C.; Schubert, F.R.; Schoenwolf, G.C.; Lumsden, A. Analysis of spatial and temporal gene expression patterns in blastula and gastrula stage chick embryos. Dev. Biol. 2002, 245, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.E.; Clémençon, B.; Hediger, M.A. Proton-coupled oligopeptide transporter family SLC15: Physiological, pharmacological and pathological implications. Mol. Asp. Med. 2013, 34, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Schiöth, H.B.; Roshanbin, S.; Hägglund, M.G.; Fredriksson, R. Evolutionary origin of amino acid transporter families SLC32, SLC36 and SLC38 and physiological, pathological and therapeutic aspects. Mol. Asp. Med. 2013, 34, 571–585. [Google Scholar] [CrossRef] [PubMed]
- Prakash, S.K.; Paylor, R.; Jenna, S.; Lamarche-Vane, N.; Armstrong, D.L.; Xu, B.; Mancini, M.A.; Zoghbi, H.Y. Functional analysis of ARHGAP6, a novel GTPase-activating protein for RhoA. Hum. Mol. Genet. 2000, 9, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R.D.; Connolly, N.L.; Burnett, D.; Campbell, E.J.; Senior, R.M.; Ley, T.J. Developmental regulation of the human cathepsin G gene in myelomonocytic cells. J. Biol. Chem. 1990, 265, 1524–1530. [Google Scholar] [PubMed]
- Heutinck, K.M.; ten Berge, I.J.M.; Hack, C.E.; Hamann, J.; Rowshani, A.T. Serine proteases of the human immune system in health and disease. Mol. Immunol. 2010, 47, 1943–1955. [Google Scholar] [CrossRef] [PubMed]
- Hermant, B.; Bibert, S.; Concord, E.; Dublet, B.; Weidenhaupt, M.; Vernet, T.; Gulino-Debrac, D. Identification of Proteases Involved in the Proteolysis of Vascular Endothelium Cadherin during Neutrophil Transmigration. J. Biol. Chem. 2003, 278, 14002–14012. [Google Scholar] [CrossRef] [PubMed]
- East, L.; Isacke, C.M. The mannose receptor family. Biochim. Biophys. Acta 2002, 1572, 364–386. [Google Scholar] [CrossRef]
- Wong, K.S.; Proulx, K.; Rost, M.S.; Sumanas, S. Identification of vasculature-specific genes by microarray analysis of etsrp/etv2 overexpressing zebrafish embryos. Dev. Dyn. 2009, 238, 1836–1850. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Donovan, M.J.; Rogers, R.A.; Ezekowitz, R.A. Distribution of murine mannose receptor expression from early embryogenesis through to adulthood. Cell Tissue Res. 1998, 292, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Marin, O.; Hermesz, E.; Powell, A.; Flames, N.; Palkovits, M.; Rubenstein, J.L.R.; Westphal, H. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc. Natl. Acad. Sci. USA 2003, 100, 9005–9010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miake, J.; Notsu, T.; Higaki, K.; Hidaka, K.; Morisaki, T.; Yamamoto, K.; Hisatome, I. Cited4 is related to cardiogenic induction and maintenance of proliferation capacity of embryonic stem cell-derived cardiomyocytes during in vitro cardiogenesis. PLoS ONE 2017, 12, e0183225. [Google Scholar] [CrossRef] [PubMed]
- Szeto, D.P.; Rodriguez-Esteban, C.; Ryan, A.K.; O’Connell, S.M.; Liu, F.; Kioussi, C.; Gleiberman, A.S.; Izpisúa-Belmonte, J.C.; Rosenfeld, M.G. Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 1999, 13, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Pinto do O, P.; Richter, K.; Carlsson, L. Hematopoietic progenitor/stem cells immortalized by Lhx2 generate functional hematopoietic cells in vivo. Blood 2002, 99, 3939–3946. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Yang, Y.-C. Cited2 in hematopoietic stem cell function. Curr. Opin. Hematol. 2013, 20, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Kieusseian, A.; Chagraoui, J.; Kerdudo, C.; Mangeot, P.E.; Gage, P.J.; Navarro, N.; Izac, B.; Uzan, G.; Forget, B.G.; Dubart-Kupperschmitt, A. Expression of Pitx2 in stromal cells is required for normal hematopoiesis. Blood 2006, 107, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Aziz, A.; Harrop, S.P.; Bishop, N.E. DIA1R is an X-linked gene related to Deleted in Autism-1. PLoS ONE 2011, 6, e14534. [Google Scholar] [CrossRef] [PubMed]
- Aziz, A.; Harrop, S.P.; Bishop, N.E. Characterization of the deleted in autism 1 protein family: Implications for studying cognitive disorders. PLoS ONE 2011, 6, e14547. [Google Scholar] [CrossRef] [PubMed]
- Dudkiewicz, M.; Lenart, A.; Pawłowski, K. A novel predicted calcium-regulated kinase family implicated in neurological disorders. PLoS ONE 2013, 8, e66427. [Google Scholar] [CrossRef] [PubMed]
- Swiers, G.; Rode, C.; Azzoni, E.; de Bruijn, M.F. A short history of hemogenic endothelium. Blood Cells Mol. Dis. 2013, 51, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Ignacio, A.R.; Muller, Y.M.; Carvalho, M.S.; Nazari, E.M. Distribution of microglial cells in the cerebral hemispheres of embryonic and neonatal chicks. Braz. J. Med. Biol. Res. 2005, 38, 1615–1621. [Google Scholar] [CrossRef] [PubMed]
- Gay, L.; Miller, M.R.; Ventura, P.B.; Devasthali, V.; Vue, Z.; Thompson, H.L.; Temple, S.; Zong, H.; Cleary, M.D.; Stankunas, K.; et al. Mouse TU tagging: A chemical/genetic intersectional method for purifying cell type-specific nascent RNA. Genes Dev. 2013, 27, 98–115. [Google Scholar] [CrossRef] [PubMed]
- Bhasin, M.; Yuan, L.; Keskin, D.B.; Otu, H.H.; Libermann, T.A.; Oettgen, P. Bioinformatic identification and characterization of human endothelial cell-restricted genes. BMC Genom. 2010, 11, 342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prinz, M.; Priller, J. Microglia and brain macrophages in the molecular age: From origin to neuropsychiatric disease. Nat. Rev. Neurosci. 2014, 15, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Edmonson, C.A.; Ziats, M.N.; Rennert, O.M. A Non-inflammatory Role for Microglia in Autism Spectrum Disorders. Front. Neurol. 2016, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Mosser, C.A.; Baptista, S.; Arnoux, I.; Audinat, E. Microglia in CNS development: Shaping the brain for the future. Prog. Neurobiol. 2017, 149–150, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Emanuele, E.; Orsi, P.; Barale, F.; di Nemi, S.U.; Bertona, M.; Politi, P. Serum levels of vascular endothelial growth factor and its receptors in patients with severe autism. Clin. Biochem. 2010, 43, 317–319. [Google Scholar] [CrossRef] [PubMed]
| FC b | Gene Symbol | Gene Name | Gene ID | Molecular Function | Biological Function | Expression in Early Embryos |
|---|---|---|---|---|---|---|
| +5.1 | SLC15A1 | Solute carrier family 15, member 1 | 378789 | Membrane transporter | Oligopeptide transport | - |
| +4.2 | C1HXorf36 (DIA1R) | Chromosome 1 open reading frame, human CXorf36 (Deleted in Autism 1 Related) | 418555 | - | - | (this study) |
| +4.19 | SLC32A1 | Solute carrier family 32, member 1 | 419167 | Vesicular transporter | GABA vesicular transporter | - |
| +4.15 | SOX7 | SRY (sex determining region Y)-box 7 | 771337 | Transcription factor | Vasculogenesis and hematopoiesis | Angioblasts |
| +3.73 | LMO2 | LIM domain only 2 | 374129 | Transcription factor | Hematopoiesis | Hematopoietic progenitors |
| +3.7 | CTSG | Cathepsin G | 426049 | Serine protease | Tissue remodeling | Myeloid progenitors |
| +3.62 | TAL1 (SCL) | T-cell acute lymphocytic leukemia 1 (stem cell leukemia) | 396298 | Transcription factor | Hematopoiesis | Hematopoietic progenitors |
| +3.51 | RUNX1 | Runt-related transcription factor 1 | 396152 | Transcription factor | Hematopoiesis | Blood islands |
| +3.44 | EGR1 | Early growth response 1 | 373931 | Transcription factor | HSPC proliferation | Vasculogenic mesoderm |
| +3.2 | MRC1 | Mannose receptor C-type 1 | 420516 | Membrane receptor | Endocytosis | Blood islands |
| +3.17 | KLHL6 | Kelch-like 6 | 424762 | Transcription factor | Lymphocyte differentiation | - |
| +3.3 | SPI1 (PU.1) | Spleen focus forming virus (SFFV) proviral integration oncogene spi1 | 395879 | Transcription factor | Hematopoiesis | Hematopoietic progenitors |
| +3.07 | RhoGap6 | Similar to Rho-GTPase-activating protein 6 (LOC422284 locus) | 422284 | Cytoskeleton regulator | Actin remodeling | - |
| +2.77 | LHX8 | LIM homeobox 8 | 424721 | Transcription factor | Neurogenesis | Blood islands |
| +2.71 | FLT1 (VEGFR1) | Fms-related tyrosine kinase 1 (vascular endothelial growth factor receptor 1) | 374100 | Receptor tyrosine kinase | Vasculogenesis/Angiogenesis | Hemangioblasts and endothelial cells |
| +2.63 | SOX18 | SRY (sex determining region Y)-box 18 | 374200 | Transcription factor | Vasculogenesis | Blood islands |
| +2.49 | CDH5 | Cadherin 5, type 2, VE-cadherin (vascular epithelium) | 374068 | Cell adhesion molecule | Vasculogenesis/Angiogenesis | Endothelial cells |
| +2.39 | CD34 | Hematopoietic progenitor cell antigen CD34 | 419856 | Cell surface antigen | - | Hematopoietic progenitors |
| +2.29 | FLT4 (VEGFR3) | Fms-related tyrosine kinase 4 (vascular endothelial growth factor receptor 3) | 395742 | Receptor tyrosine kinase | Angiogenesis | Blood islands and endothelial cells |
| +2.23 | CITED4 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 | 395465 | Transcription regulator | in vitro cardiogenesis | Blood islands |
| +2.2 | PITX1 | Paired-like homeodomain 1 | 374201 | Transcription factor | Pituitary and hindlimb development | Posterior extraembryonic mesoderm |
| +2.09 | Fli1 | Friend leukemia virus integration 1 gene | 419723 | Transcription factor | Vasculogenesis and hematopoiesis | Endothelial and erythroid progenitors |
| +1.96 | HHEX | Hematopoietically expressed homeobox | 396182 | Transcription factor | Vasculogenesis and hematopoiesis | Blood islands |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serrado Marques, J.; Teixeira, V.; Jacinto, A.; Tavares, A.T. Identification of Novel Hemangioblast Genes in the Early Chick Embryo. Cells 2018, 7, 9. https://doi.org/10.3390/cells7020009
Serrado Marques J, Teixeira V, Jacinto A, Tavares AT. Identification of Novel Hemangioblast Genes in the Early Chick Embryo. Cells. 2018; 7(2):9. https://doi.org/10.3390/cells7020009
Chicago/Turabian StyleSerrado Marques, José, Vera Teixeira, António Jacinto, and Ana Teresa Tavares. 2018. "Identification of Novel Hemangioblast Genes in the Early Chick Embryo" Cells 7, no. 2: 9. https://doi.org/10.3390/cells7020009
APA StyleSerrado Marques, J., Teixeira, V., Jacinto, A., & Tavares, A. T. (2018). Identification of Novel Hemangioblast Genes in the Early Chick Embryo. Cells, 7(2), 9. https://doi.org/10.3390/cells7020009





