Apelin Effects Migration and Invasion Abilities of Colon Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cell Culture
2.3. Proliferation Analysis
2.4. Migration and Invasion Assay
2.5. Cytosolic Fraction Isolation
2.6. Actin Polymerization State Determination
2.7. Cell Extracts Isolation
2.8. Western Blotting Assay
2.9. Fluorescent Staining
2.10. Fluorescent-Substrate Degradation Assay
2.11. Statistical Analysis
3. Results
3.1. Apelin Increases Proliferation, Migration and Invasion of Colon Cancer Cells
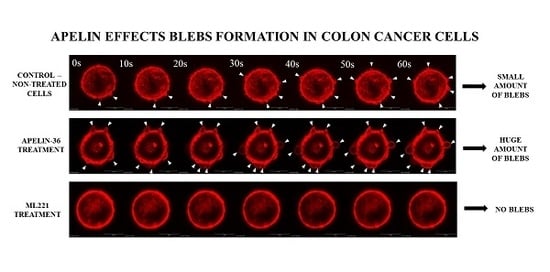
3.2. Apelin Stimulates Blebs Formation in Colon Cancer Cells
3.3. Apelin Influences Actin Polymerisation State and the Level of Cofilin
3.4. Apelin Effects the Proteolytic Abilities of Colon Cancer Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflict of Interest
References
- Siegel, R.L.; Miller, K.D.; Fedewa, S.A.; Ahnen, D.J.; Meester, R.G.S.; Barzi, A.; Jemal, A. Colorectal cancer statistics. 2017. CA Cancer J. Clin. 2017, 67, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Kubiak, A.; Kycler, W.; Trojanowski, M. Epidemiologia i profilaktyka raka jelita grubego w Polsce. Probl. Hig. Epidemiol. 2014, 95, 636–642. [Google Scholar]
- Jochem, C.; Leitzmann, M. Obesity and colorectal cancer. In Recent Results in Cancer Research; Springer: New York, NY, USA, 2016; Volume 208, pp. 17–41. [Google Scholar]
- Tatemoto, K.; Hosoya, M.; Habata, Y.; Fujii, R.; Kakegawa, T.; Zou, M.-X.; Kawamata, Y.; Fukusumi, S.; Hinuma, S.; Kitada, C.; et al. Isolation and characterization of a novel endogenous peptide ligand for the human APJ receptor. Biochem. Biophys. Res. Commun. 1998, 251, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Masri, B.; Knibiehler, B.; Audigier, Y. Apelin signalling: A promising pathway from cloning to pharmacology. Cell. Signal. 2005, 17, 415–426. [Google Scholar] [CrossRef] [PubMed]
- O’Carroll, A.M.; Lolait, S.J.; Harris, L.E.; Pope, G.R. The apelin receptor APJ: Journey from an orphan to a multifaceted regulator of homeostasis. J. Endocrinol. 2013, 219. [Google Scholar] [CrossRef] [PubMed]
- Wysocka, M.B.; Pietraszek-gremplewicz, K.; Nowak, D. The role of apelin in cardiovascular diseases, obesity and cancer. Front. Physiol. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sorli, S.C.; Le Gonidec, S.; Knibiehler, B.; Audigier, Y. Apelin is a potent activator of tumour neoangiogenesis. Oncogene 2007, 26, 7692–7699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorli, S.C.; van den Berghe, L.; Masri, B.; Knibiehler, B.; Audigier, Y. Therapeutic potential of interfering with apelin signalling. Drug Discov. Today 2006, 11, 1100–1106. [Google Scholar] [CrossRef] [PubMed]
- Berta, J.; Kenessey, I.; Dobos, J.; Tovari, J.; Klepetko, W.; Jan Ankersmit, H.; Hegedus, B.; Renyi-Vamos, F.; Varga, J.; Lorincz, Z.; et al. Apelin expression in human non-small cell lung cancer: Role in angiogenesis and prognosis. J. Thorac. Oncol. 2010, 5, 1120–1129. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.Y.; Chuang, W.L. Hepatocellular carcinoma cells cause different responses in expressions of cancer-promoting genes in different cancer-associated fibroblasts. Kaohsiung J. Med. Sci. 2013, 29, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Picault, F.X.; Chaves-Almagro, C.; Projetti, F.; Prats, H.; Masri, B.; Audigier, Y. Tumour co-expression of apelin and its receptor is the basis of an autocrine loop involved in the growth of colon adenocarcinomas. Eur. J. Cancer 2014, 50, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Liu, N.; Xu, G.; Liu, T.; Liu, Y.; Zhou, Y. Apelin13/APJ promotes proliferation of colon carcinoma by activating Notch3 signaling pathway. Oncotarget. 2017, 8, 101697–101706. [Google Scholar] [CrossRef] [PubMed]
- Blanchoin, L.; Boujemaa-Paterski, R.; Sykes, C.; Plastino, J. Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 2014, 94, 235–263. [Google Scholar] [CrossRef] [PubMed]
- Khaitlina, S.Y. Mechanisms of spatial segregation of actin isoforms. Tsitologiya 2007, 49, 345–354. [Google Scholar] [CrossRef]
- Simiczyjew, A.; Pietraszek-Gremplewicz, K.; Mazur, A.J.; Nowak, D. Are non-muscle actin isoforms functionally equivalent? Histol. Histopathol. 2017, 32, 1125–1139. [Google Scholar] [PubMed]
- Gross, S.R. Actin binding proteins: Their ups and downs in metastatic life. Cell Adhes. Migr. 2013, 7, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Cordero, J.J.; Magalhaes, M.A.O.; Eddy, R.J.; Hodgson, L.; Condeelis, J. Functions of cofilin in cell locomotion and invasion. Nat. Rev. Mol. Cell Biol. 2013, 14, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Li, L.; Lu, Q.; Li, Y.; Xie, F.; Li, H.; Cao, J.; Liu, M.; Wu, D.; He, L.; et al. PAK1-cofilin phosphorylation mediates human lung adenocarcinoma cells migration induced by apelin-13. Clin. Exp. Pharmacol. Physiol. 2016, 43, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Gialeli, C.; Theocharis, A.D.; Karamanos, N.K. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011, 278, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Yao, G.; Yu, H.; Qing, Y.; Wang, K. Tumor apelin, not serum apelin, is associated with the clinical features and prognosis of gastric cancer. BMC Cancer 2016, 16, 794. [Google Scholar] [CrossRef] [PubMed]
- Heo, K.; Kim, Y.H.; Sung, H.J.; Li, H.Y.; Yoo, C.W.; Kim, J.Y.; Park, J.Y.; Lee, U.L.; Nam, B.H.; Kim, E.O.; et al. Hypoxia-induced up-regulation of apelin is associated with a poor prognosis in oral squamous cell carcinoma patients. Oral Oncol. 2012, 48, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Li, F.; Wang, P.; Jia, S.; Sun, L.; Huo, H. Apelin-13 induces MCF-7 cell proliferation and invasion via phosphorylation of ERK1/2. Int. J. Mol. Med. 2015, 36, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Nowak, D.; Krawczenko, A.; Duś, D.; Malicka-Błaszkiewicz, M. Actin in human colon adenocarcinoma cells with different metastatic potential. Acta Biochim. Pol. 2002, 49, 823–828. [Google Scholar] [PubMed]
- Opolski, A.; Wietrzyk, J.; Duś, D.; Kieda, C.; Matejuk, A.; Makowska, A.; Wojdat, E.; Ugorski, M.; Laskowska, A.; KŁopocki, A.; et al. Metastatic potential and saccharide antigens expression of human colon cancer cells xenotransplanted into athymic nude mice. Folia Microbiol. 1998, 43, 507–510. [Google Scholar] [CrossRef]
- Malicka-Blaszkiewicz, M.; Roth, J.S. Some factors affecting the interaction between actin in leukemic L1210 cells and DNASE I. Biochem. Biophys. Res. Commun. 1981, 102, 594–601. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the for the quantitation of microgram quantities of protein utilizing the principle of protein dye-binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horzum, U.; Ozdil, B.; Pesen-Okvur, D. Step-by-step quantitative analysis of focal adhesions. MethodsX 2014, 1, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Artym, V.V.; Zhang, Y.; Seillier-Moiseiwitsch, F.; Yamada, K.M.; Mueller, S.C. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: Defining the stages of invadopodia formation and function. Cancer Res. 2006, 66, 3034–3043. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Zheng, X.; Jiang, Y. Apelin-13 induces proliferation, migration, and collagen I mRNA expression in human RPE cells via PI3K/Akt and MEK/Erk signaling pathways. Mol. Vis. 2013, 19, 2227–2236. [Google Scholar] [PubMed]
- Berta, J.; Hoda, M.A.; Laszlo, V.; Rozsas, A.; Garay, T.; Torok, S.; Grusch, M.; Berger, W.; Paku, S.; Renyi-Vamos, F.; et al. Apelin promotes lymphangiogenesis and lymph node metastasis. Oncotarget 2014, 5, 4426–4437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diz-Muñoz, A.; Romanczuk, P.; Yu, W.; Bergert, M.; Ivanovitch, K.; Salbreux, G.; Heisenberg, C.P.; Paluch, E.K. Steering cell migration by alternating blebs and actin-rich protrusions. BMC Biol. 2016, 14, 74. [Google Scholar] [CrossRef] [PubMed]
- Paluch, E.K.; Raz, E. The role and regulation of blebs in cell migration. Curr. Opin. Cell Biol. 2013, 25, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Nowak, D.; Mazur, A.J.; Popow-Woźniak, A.; Radwańska, A.; Mannherz, H.G.; Malicka-Błaszkiewicz, M. Subcellular distribution and expression of cofilin and ezrin in human colon adenocarcinoma cell lines with different metastatic potential. Eur. J. Histochem. 2010, 54, e14. [Google Scholar] [CrossRef] [PubMed]
- Barros, L.F.; Kanaseki, T.; Sabirov, R.; Morishima, S.; Castro, J.; Bittner, C.X.; Maeno, E.; Ando-Akatsuka, Y.; Okada, Y. Apoptotic and necrotic blebs in epethelial cells display similar neck diameters but different kinase dependency. Cell Death Differ. 2003, 10, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Shuang, L.; Jidong, W.; Hongjuan, P.; Zhenwei, Y. Effects of apelin on proliferation and apoptosis in rat ovarian granulosa cells. Clin. Exp. Obs. Gynecol. 2016, 43, 409–413. [Google Scholar]
- Antushevich, H.; Krawczynska, A.; Kapica, M.; PrzemyslawHerman, A.; Zabielski, R. Effect of apelin on mitosis, apoptosis and DNA repair enzyme OGG 1/2 expression in intestinal cell lines IEC-6 and Caco-2. Folia Histochem. Cytobiol. 2014, 52, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Winder, S.J.; Ayscough, K.R. Actin-binding proteins. J. Cell Sci. 2005, 118, 651–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duffy, M.J.; Maguire, T.M.; Hill, A.; McDermott, E.; O’Higgins, N. Metalloproteinases: Role in breast carcinogenesis, invasion and metastasis. Breast Cancer Res. 2000, 2, 252–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poincloux, R.; Lizarraga, F.; Chavrier, P. Matrix invasion by tumour cells: A focus on MT1-MMP trafficking to invadopodia. J. Cell Sci. 2009, 122, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Leal, V.; Mafra, D. Adipokines in obesity. Clin. Chim. Acta 2013, 419, 87–94. [Google Scholar] [CrossRef] [PubMed]
- El Husseny, M.W.A.; Mamdouh, M.; Shaban, S.; Ibrahim Abushouk, A.; Zaki, M.M.M.; Ahmed, O.M.; Abdel-Daim, M.M. Adipokines: Potential therapeutic targets for vascular dysfunction in type II diabetes mellitus and obesity. J. Diabetes Res. 2017, 2017, 8095926. [Google Scholar] [CrossRef] [PubMed]
- Ba, H.J.; Chen, H.S.; Su, Z.; Du, M.L.; Chen, Q.L.; Li, Y.H.; Ma, H.M. Associations between serum apelin-12 levels and obesity-related markers in Chinese children. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Al-harithy, R.N.; Al-otaibi, W.A. Apelin-12 levels in obese patients with colon cancer. Cancer Immunol. Immunother. 2015, 1, 1–5. [Google Scholar]
- Narayanan, S.; Harris, D.L.; Maitra, R.; Runyon, S.P. Regulation of the apelinergic system and its potential in cardiovascular disease: Peptides and small molecules as tools for discovery. J. Med. Chem. 2015, 58, 7913–7927. [Google Scholar] [CrossRef] [PubMed]
- Maloney, P.R.; Khan, P.; Hedrick, M.; Gosalia, P.; Milewski, M.; Li, L.; Roth, G.P.; Sergienko, E.; Suyama, E.; Sugarman, E.; et al. Discovery of 4-oxo-6-((pyrimidin-2-ylthio)methyl)-4H-pyran-3-yl 4-nitrobenzoate (ML221) as a functional antagonist of the apelin (APJ) receptor. Bioorg. Med. Chem. Lett. 2012, 22, 6656–6660. [Google Scholar] [CrossRef] [PubMed]
- Petrie, R.J.; Yamada, K.M. At the leading edge of three-dimensional cell migration. J. Cell Sci. 2012, 125, 5917–5926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fackler, O.T.; Grosse, R. Cell motility through plasma membrane blebbing. J. Cell Biol. 2008, 181, 879–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, S.; Yamamoto, H.; Mukaisho, K.I.; Sato, S.; Higo, T.; Hattori, T.; Yamamoto, G.; Sugihara, H. Mechanisms underlying cancer progression caused by ezrin overexpression in tongue squamous cell carcinoma. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Lappalainen, P. Actin-binding proteins: The long road to understanding the dynamic landscape of cellular actin networks. Mol. Biol. Cell 2016, 27, 2519–2522. [Google Scholar] [CrossRef] [PubMed]
- Popow-Wozniak, A.; Mazur, A.J.; Mannherz, H.G.; Malicka-Blaszkiewicz, M.; Nowak, D. Cofilin overexpression affects actin cytoskeleton organization and migration of human colon adenocarcinoma cells. Histochem. Cell Biol. 2012, 138, 725–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madsen, D.H.; Bugge, T.H. The source of matrix-degrading enzymes in human cancer: Problems of research reproducibility and possible solutions. J. Cell Biol. 2015, 209, 195–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, C.; Ehrlich, L.; Venter, J.; O’Brien, A.; White, T.; Zhou, T.; Dang, T.; Meng, F.; Invernizzi, P.; Bernuzzi, F.; et al. Inhibition of the apelin/apelin receptor axis decreases cholangiocarcinoma growth. Cancer Lett. 2017, 386, 179–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jassal, B.; Jupe, S.; Caudy, M.; Birney, E.; Stein, L.; Hermjakob, H.; D’Eustachio, P. The systematic annotation of the three main GPCR families in Reactome. Database 2010, 2010. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wan, Y.; Fang, C.; Chen, J.; Ouyang, W.; Li, J.; Wang, Y. The orphan G protein-coupled receptor 25 (GPR25) is activated by Apelin and Apela in non-mammalian vertebrates. Biochem. Biophys. Res. Commun. 2018, 501, 408–414. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Podgórska, M.; Pietraszek-Gremplewicz, K.; Nowak, D. Apelin Effects Migration and Invasion Abilities of Colon Cancer Cells. Cells 2018, 7, 113. https://doi.org/10.3390/cells7080113
Podgórska M, Pietraszek-Gremplewicz K, Nowak D. Apelin Effects Migration and Invasion Abilities of Colon Cancer Cells. Cells. 2018; 7(8):113. https://doi.org/10.3390/cells7080113
Chicago/Turabian StylePodgórska, Marta, Katarzyna Pietraszek-Gremplewicz, and Dorota Nowak. 2018. "Apelin Effects Migration and Invasion Abilities of Colon Cancer Cells" Cells 7, no. 8: 113. https://doi.org/10.3390/cells7080113
APA StylePodgórska, M., Pietraszek-Gremplewicz, K., & Nowak, D. (2018). Apelin Effects Migration and Invasion Abilities of Colon Cancer Cells. Cells, 7(8), 113. https://doi.org/10.3390/cells7080113